- Sep 2016
-
scienceintheclassroom.org scienceintheclassroom.org
-
Morris water maze
A maze in which mice learn the location of a platform. The platform is visible at first and then hidden under the water surface once the mice learn its location.
This type of maze requires the hippocampus, one of the first parts of the brain to become damaged in Alzheimer’s disease.
See a video about the Morris Water maze here:
-
familial AD mutations
Specific genetic mutations that cause Alzheimer’s disease.
These mutations are very rare and only account for about 2% of all cases of Alzheimer’s disease. If an individual has one of these rare mutations s/he is destined to get Alzheimer’s disease (100% chance of developing Alzheimer's).
-
haplotypes
A set of genes inherited together.
In this case, the authors discuss the idea that several genes related to the tau protein seem to affect the probability that a person will develop Alzheimer’s disease later in life.
-
posttranslationally
This means that molecular changes are made to tau proteins after they are being made.
Some types of abnormal tau appear in cases of Alzheimer’s disease.
-
Alzheimer's disease
Alzheimer's disease is the most common type of dementia. It's a disease of aging in which the proteins amyloid-β and tau build up or aggregate in the brain, causing neurons to die.
Amyloid-β proteins build up into clumps called plaques, and abnormal tau proteins fall off of their microtubules, which kills the neurons. These tau proteins later aggregate into clumps called tangles.
Symptoms of Alzheimer’s disease include memory and cognitive problems, and patients eventually die from the disorder.
-
amyloid-β peptide
This peptide consists of a strand of up to about 40 amino acids that are the main component of the amyloid plaques found in the brains of Alzheimer's patients.
-
excitotoxicity
A process in which neurons are damaged or die as a result of too much stimulation (or excitation, hence "excito-toxicity").
For example, excitotoxicity can occur from a seizure.
-
transgenic mice expressing human amyloid precursor protein
Mice that have been genetically engineered to produce the amyloid precursor protein, which is thought to give rise to amyloid-β.
-
endogenous
Originating from within the body. In this case, "endogenous" refers to the tau proteins that occur naturally in the body of the mice.
-
microtubule-associated protein
A protein that is bound to microtubules.
Microtubules are part of a neuron’s inner cytoskeleton that maintains the structure and stability of the cell.
-
tau
Tau proteins are primarily found in neurons of the central nervous system.
They stabilize the neuron's microtubules, which are components of the cell's cytoskeleton, the inner stabilizing network of filaments and tubules.
-
peptides
A small protein.
-
-
scienceintheclassroom.org scienceintheclassroom.org
-
pithed
pithing is a method used while preparing specimen for experimental procedures and dissection. It is done by inserting a metal rod into the brain or spinal chord of a living specimen as to kill or immobilise in the least painful way. This method is useful in that it allows for the testing and examination of the physiology of the organs of the prey because they still work, but the animal feels no pain and has no control over their muscles.
-
tetanus
Tetanus could refer to the disease caused by a bacterial infection or to rapid/continuous muscle contraction. In this context it is referring to the latter definition;muscle contraction. Each contraction produces an involuntary twitch. Because the electric shocks produced by the electric eel are very frequent the fish ends up constantly twitching with no breaks and no control over its movement .
for more information about muscle contraction; Mann MD (2011). "Chapter 14: Muscle Contraction: Twitch and tetanic contractions". The Nervous System In Action. Michael D. Mann
-
motor neurons
Cells that carry messages and transmit signals. It is those cells that allow us to perform basic motor functions such as walking, eating, holding things etc.
-
Plexiglas
Is a sturdy alternative to glass. made out of acrylic it is shatter proof as well as sound proof. this is ideal in this experiment because it is clear enough for the eel to se through but does not allow noise or movement of prey to go through, or electric shocks of eel.
-
agar
jelly like gelatine but made from algae
-
neuromuscular junction
The connection point between the motor neuron (carrying signal from the spinal chord) and a muscle fibre. This is the communication channel between the two where messages are transmitted using calcium ions. This is necessary to control muscles.
-
latency
Latency refers to the time thats elapses between an action and a reaction in a system. In this context it refers to the time it takes for the activation of the motor neurone efferents of the prey after the release of the eel electric discharge.
-
latency
length of a time step of delay
-
suction-feeding strike
sucking in food (a fish) to catch and eat it, like you suck through a straw
-
volleys
in this context volleys refers to multiple back-to-back electric discharges
-
Electrophorus electricus
While they are referred to as Eels, Electric Eels actually belong to the nocturnal family of knifefish which consists of electric fish that have organs that generate electric current. Eels are the largest Gymnotiformes and use electric current to defend and hunt.
-
efferent
when a neutron is described as being efferent it means that it carries orders away from the central nervous system, which acts like a control centre, towards organs, mainly muscles and glands
-
providing a combined discharge of up to 600 V
This is almost five times more voltage than what you would get out of an Electrical outlet in a US household (120V) and more than twice the voltage of an outlet in Europe (220-240 V)
-
electrolocation
Location by electricity. In using this tools the eels can explore extreme environment such as muddy water.
-
mechanosensory
mechano referes to mechanical stimulus, which is a physical change such as direct contact, change in pressure or vibration. This change can be sensed by special sensory cells that are found all over the body. These cells allow us to sense vibrations from and contact with other objects.
-
curare (an acetylcholine antagonist)
poison that stops a chemical messenger
-
electrocytes
electrical cells
-
acetylcholine
Neurotransmitter (molecule, chemical messenger) that takes part in muscular contraction
-
cyanoacrylate
very strong glue
-
electrogenic
able to produce a change in the electrical potential of a cell
-
-
scienceintheclassroom.org scienceintheclassroom.org
-
glutamatergic transmission
mechanisms that involve the release of glutamate (a common neurotransmitter)
-
homozygous
Humans have chromosomes by pairs. An individual receives one chromosome from each parent. Therefore, he receives two copies of each gene. One says that an individual is homozygous for a certain gene when both of its chromosomes carry the same version of the gene.
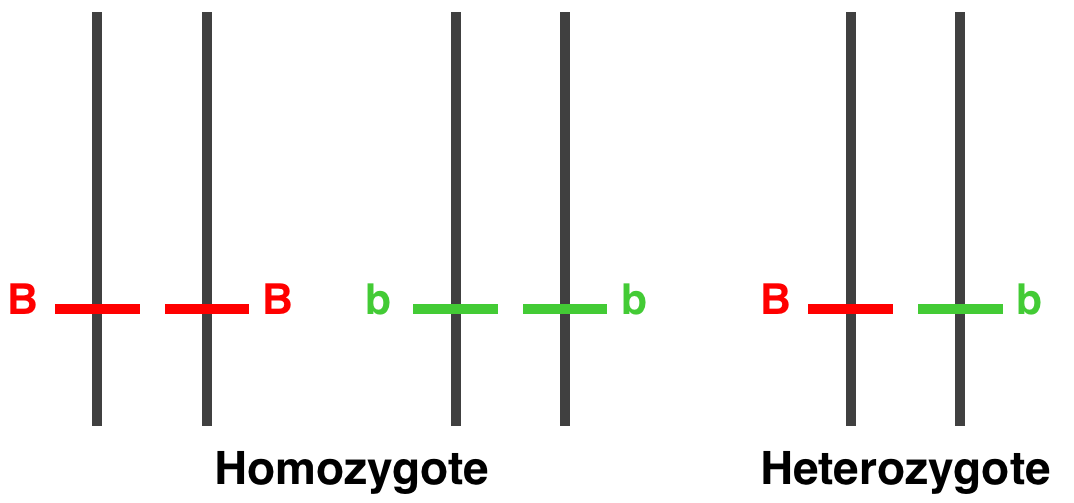
-
PCDH10
Generally PCDH10 belongs to a proto cadherin protein family and it acts as a potential tumor suppressor protein, as the dysregulation of PCDH10 gene frequently existed in multiple human tumors.
PCDH10 is a transcriptional target of p53 and exhibits inhibitory roles in cancer cell motility and cell migration.
-
MEF2 transcription factor
The myocyte enhancer factor 2 (MEF2) transcription factor holds together the transcriptional circuits and controls cell differentiation and organogenesis. In adult tissues, Mef2 proteins are also reported to regulate the stress-response during cardiac hypertrophy and tissue remodeling in cardiac and skeletal muscle.
-
transcription
At the beginning of the transcription process, the genetic information of a strand of DNA is copied onto a new molecule, called messenger RNA (mRNA). The mRNA can then exit the nucleus of the cell to be translated into a protein.
-
hemizygous
This deletion was only present on one chromosome from each parent.
-
simplex and
In simplex families, only one person is affected by the disease. Multiplex families have more that one of their members affected by the disease.
-
single-nucleotide polymorphism
A variation of a single nucleotide in a DNA sequence. When less than 1% of a population carries the nucleotide at this position, this variation is classified as a SNP. Some SNPs are associated with certain diseases.
-
HMCA
Reminder : Homozygosity Mapping Collaborative for Autism
-
Identifying Autism
DNA is a macromolecule forming a long double helical shape and containing the “blueprint” of an organism. It gives the instructions for the production of all the proteins of the organism. Each long portion of DNA is called a chromosome.
-
Endosomal trafficking and protein turnover
Other genes the authors identified are involved in how cells move proteins around. Cells usually do this by packing proteins into endosomes (small spheres made out of membrane) to send proteins to specific places.
-
stop codon
A stop codon (or termination codon) is a nucleotide triplet, which stops the synthesis of a protein molecule. It's placed within messenger RNA and signals a termination of translation into proteins.
This termination is taking place due to binding release factors, which cause the ribosomal subunits to disassociate, releasing the amino acid chain.

-
transcriptome
The set of all messenger RNA transcripts in the considered cell.
-
centromere to telomere
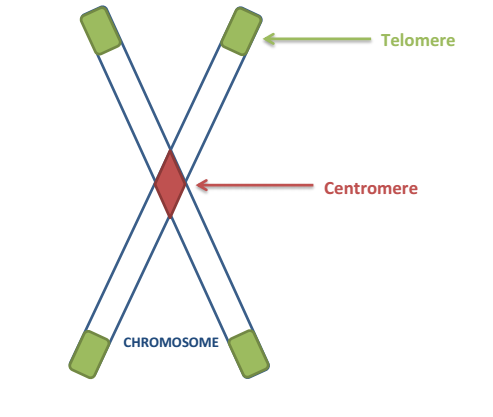
-
neuronal cell adhesion molecules
Some mutations can affect a neuron's ability to adhere to other neurons. This difficulty can result in autism symptoms.
-
-
scienceintheclassroom.org scienceintheclassroom.org
-
caudal
-
spinal
pertaining to spine.

https://upload.wikimedia.org/wikipedia/commons/a/a4/Spinal_column_curvature_2011.png
-
cervical
-
ventral
the underside or the abdominal part of the body
https://upload.wikimedia.org/wikipedia/commons/6/64/Horse_Axes.JPG
-
humeral
an anatomical term which refers to a bone called humerus

https://upload.wikimedia.org/wikipedia/commons/e/e4/Humerus_-_anterior_view.png
-
femoral
an anatomical term which refers to a bone called femur![]https://upload.wikimedia.org/wikipedia/commons/8/8f/Femur_-_anterior_view2.png
https://upload.wikimedia.org/wikipedia/commons/8/8f/Femur_-_anterior_view2.png
-
lateral
-
sebocytes
any kind of cell that secretes sebum- an oily substance which contains fat-
-
nested subpopulation
a population that fits into a larger one!
https://upload.wikimedia.org/wikipedia/commons/4/41/Floral_matryoshka_set_2_smallest_doll_nested.JPG
-
integument
the outer layer that mainly serves as protection
-
keratinized
to change to a form that contains keratin- a fibrous protein which mostly found in hair, nail, hoof, etc.-
-
superposed
superposing means to put or lay something over something else
-
histological
pertaining to histology-the science of studying tissues-
-
in-frame deletion
As you know from molecular biology, protein-coding region of DNA transcribe into mRNAs and then, mRNAs, in their turn, translate into proteins on ribosomes. Basically three nucleotides are required to code for any aminoacid, so the part of mRNA that translates into proteins has a "frame" which is divisible by three; if a deletion cause removal of three or a multiplicate of three then the frame will stop at the previous stop codon and other aminoiacs remain untouched, otherwise the deletion may turn the protein to anything at all.

-
homologous
homologous properties in biology have the same evolutionary origins but not necessarily a same function. like human and bat's arm.

https://upload.wikimedia.org/wikipedia/commons/5/5b/Evolution_pl.png
-
crocodiles
-
reptiles
-
scutate
covered with scutate-A horny or bony external plate.!

https://upload.wikimedia.org/wikipedia/commons/1/17/Alligator_foot_detail.jpg
-
lineage
A sequence of species that have evolved from a same ancestor.
 a sequence of species that have evolved from a same ancestor.
a sequence of species that have evolved from a same ancestor.https://upload.wikimedia.org/wikipedia/en/6/68/Diagram_of_Eusocial_Species.png
-
cryosections
sections that are made in a cryostat- a device for keeping the temperature low-.
-
PCNA
A protein of DNA polymerase delta. it is involved in the control of eukaryotic DNA replication. PCNA is important for replication and participates in cell division. so it's present in cell shows that the cell is proliferating.
-
parasagittal
an imaginary plane that divides the body into right and left halves.
-
BMP
A group of chemicals that are growth factors.
-
dermoepidermal elevations
An area of tissue that joins the epidermal (outer layer) and the dermal layers (middle layer) of the skin.
Elevations refer to bumps found in this layer, leading to areas that are slightly raised.

https://upload.wikimedia.org/wikipedia/commons/e/ec/Anatomy_of_the_skin.jpg
-
columnar cells
Cell's shape that are similar to the columns and it's height is at least four times their width.

https://upload.wikimedia.org/wikipedia/commons/c/c5/Simple_columnar_epithelial_cells.png
-
common ancestry
-
parasagittal
An imaginary plane that divides the body into right and left halves.
-
nonconcurrently
Not happening at the same time
-
signaling pathways
Refers to a group of molecules in a cell that work together to control one or more cell functions.
-
(TNF)
The tumor necrosis factor (TNF) superfamily : a group of cytokines that take place in apoptosis.(cell death)
-
codominant
Two alleles of a gene in a heterozygote that are both fully expressed.
-
pleurodont
Tooth fused to the inner edge of the jaw.
This means it is loosely attached and can regenerate if it is lost.
-
autopod
futherest part of the limb, such as the hand or foot.
-
co-option
A structure or system with an original function evolves so that the it adds or changes to a new function.
-
follicular organs
A small spherical group of cells containing a cavity in which materials are contained and can grow.
Example: hair, teeth, feathers
-
homology
In biology, similarity of the structure, physiology, or development of different species of organisms based upon their descent from a common evolutionary ancestor.
-
molecular markers
Basically, in biology each cellular function achieves by corporation of protein complexes; for example in mammals, DNA replication happens by DNA polymerase and a dozen of other molecules( for example PCNA); so if we recognise any of these molecules that make DNA replication happens; it shows that the cell is proliferating;
-
wild-type
A type of the typical form of a species in which seen in nature.
-
avian
Relating to, or characteristic of birds.
-
placode
A platelike structure, especially a thickening of the ectoderm marking the site of future development in the early embryo
-
squamates
The order Squamata, or the scaled reptiles, are the largest recent order of reptiles, comprising all lizards and snakes.
-
morphogenesis
A biological process in which embryo develops to adult in some organisms.
-
fossil intermediate
A transitional fossile that gives us information about a transition from one species to another. Actually is one that falls between "before transition forms" and "after transition forms"
-
Extant
Life form that is currently in existence.
Antonym: extinct
-
cDNA
DNA (which stores genetic information) makes RNA (which transports genetic information) which is then read to make a protein which is functional
cDNA is a copy of the RNA which is tells us what protein will be created.
-
ectodermal dysplasia syndrome
A genetic disease which affects the growth of hair, teeth, nails and sweat glands.
-
-
scienceintheclassroom.org scienceintheclassroom.org
-
Student’s t test,
A statistical test that is used to determine if two sets of data are significantly different from each other.
-
-
scienceintheclassroom.org scienceintheclassroom.org
-
probabilistic algorithm
The probabilistic algorithm is algorithm that, providing circulation on the certain stages of its work to the random number generator in order to obtain savings in work time.
-
correlation
A correlation describes how two variables are linked one to the other.
-
National Science Foundation
The National Science Foundation (NSF) is an independent agency of the U.S. government that is responsible for the development of science and technology. The Foundation carries out it's mission by providing, in general, temporary grants.
-
-
www.scienceintheclassroom.org www.scienceintheclassroom.org
-
bat-detector
A piece of equipment used to record and identify high frequency calls beyond the range of human hearing.
These detectors have a frequency dial. Once a frequency is chosen, the detector only detects sounds at that frequency. Because the dial was set at 23 kHz, sounds far above or below that frequency would not be detected.
-
Ultrasonic
Used to describe sounds at a frequency above the human hearing range.
-
empathic concern
Empathic concern (identifying with the emotions of another person) in humans is thought to be hard-wired into our brains. It becomes evident as early as age 2 and is dependent on our social interactions.
There are several contributors to empathy including:
(A) Shared neural representations (activity in the brain in response to an experience in your environment). For example, the neural networks that are active when you feel sad would also be active when you see someone else expressing sadness and feel empathy for them.
(B) Self-awareness (conscious knowledge of one's own character, feelings, motives, and desires).
(C) Mental flexibility, or the ability to shift a course of thought or action according to the changing demands of a situation. It allows an individual to abandon a previous response set or pattern in order to generate an alternative that is better suited to the requirements of the situation at hand.
(D) Emotion regulation, or the ability to respond to the ongoing demands of an emotional experience in a manner that is socially tolerable and sufficiently flexible to permit spontaneous reactions.
-
homolog
Behavior in different species that arose from a common ancestral gene.
-
most parsimonious
Simplest.
-
palatable
Acceptable or agreeable to the taste.
-
latency
The amount of time between the free rat being placed in the arena and the door opening.
-
liberate
To free.
-
congruent
In agreement.
-
other-oriented
The opposite of selfish. Thinking or acting in the interest of someone else. Putting the well-being of another above your own.
-
-
www.scienceintheclassroom.org www.scienceintheclassroom.org
-
single-nucleotide polymorphism
A single-nucleotide polymorphism is a variation in a single nucleotide at a particular location in the genome.
The authors are referring to a position in the human BDNF gene. Variation in the DNA sequence at this position is associated with a reduced ability to recall extinction.
-
genetic knockdown
Genetic knockdown is a technique that researchers use to reduce the expression of specific genes in order to study their function.
-
putative
Based on previous research (see references 14–16), the authors chose to test the amygdala and hippocampus because they were the most likely contributors of BDNF to the IL mPFC.
-
in vitro
When something is said to occur "in vitro," it means that it has been tested or observed in parts of an organism that have been removed from their original biological surroundings.
Examples include most studies done in test tubes or petri dishes, where some tissues or cells are isolated so they can be studied more carefully.
-
in vivo
When something is said to occur "in vivo," it means that it has been tested or observed in whole, living organisms.
-
antagonist
A receptor antagonist is a drug or compound that can bind a receptor and inhibit its activation.
-
habituation
A type of learning where repeated presentation of a stimulus leads to a decrease in response. If you put an animal into a new environment for behavioral testing, the animal may become agitated or fearful.
However, if you expose the animal to that environment a few times before the experiment begins, the animal will not be as nervous because it has been “habituated” to the testing environment.
-
latent inhibition
Pre-exposure to a stimulus can make it difficult to form new associations with that stimulus. In other words, it takes longer for a familiar stimulus to acquire meaning than a new stimulus.
In this case, the authors wanted to see whether BDNF was changing freezing behavior by enhancing the effects of habituation trials.
-
potentiation
BDNF does not appear to enhance the process of latent inhibition.
-
locomotion, anxiety, or motivation to seek food reward
The authors performed additional behavioral tests in order to rule out nonspecific behavioral effects of the BDNF injection.
-
Conditioned freezing
Freezing is a behavioral response that many animals exhibit when scared. This is a measurable response that researchers use to study the fear learning process in rodents. If an animal is conditioned to fear a high-pitched sound, for instance, it will freeze when the sound is played.
During fear extinction, as the animal learns to no longer fear the sound, less freezing will be observed.
-
bilateral IL mPFC infusion
The authors are interested in the function of the IL mPFC, and how it is affected by the injection of BDNF.
The targeted injection is achieved by using a stereotaxic setup, which is a highly precise, 3D guiding system that allows researchers to place a canula into specific regions of the brain. The canula is just a hollow tube that allows for the delivery of drugs or other fluids to that brain region.
Once the canula is in place, BDNF can be directly injected into the region of interest at any time.
-
memory consolidation
Consolidation is a series of events following the initial acquisition of a memory trace that leads to its retention.
-
Epigenetic regulation
Epigenetic regulation is the process of altering gene expression levels without mutating the DNA itself.
For instance, if a cell needs to increase the expression of a specific gene, it can change the microstructure of the DNA in the area of that gene, making it more accessible for transcription into RNA.
-
N-methyl-D-aspartate receptors
NMDA receptors are found in nerve cells and are activated by the excitatory neurotransmitter glutamate. These receptors are involved in memory function in the brain.
-
extinction training
After the animal has formed an association between the tone and the shock, it will freeze when it hears the tone. However, if the tone is presented enough times in the absence of the shock, the mouse will eventually stop showing a fear response to the tone through a process known as fear extinction.
-
auditory fear conditioning
A rodent model in which a neutral auditory cue (high-pitched beep) is paired with an aversive stimulus (footshock), resulting in the formation of an associative fear memory.
After repeated presentations of the paired stimuli, the animal will experience fear in response to the tone alone.
-
synaptic plasticity
A term used to describe the ability of neuronal synapses to change their strength, becoming stronger or weaker in response to changes in their activity levels.
-
Brain-derived neurotrophic factor (BDNF)
BDNF is a protein in the brain (and peripheral nervous system) that supports the survival, growth, and differentiation of neurons involved in memory.
-
infralimbic medial prefrontal cortex
The medial prefrontal cortex is a region of the brain that is associated with cognitive and executive processes such as working memory and decision-making.
The infralimbic subregion of the medial prefrontal cortex is necessary for the inhibition of conditioned fear after extinction.
-
- Aug 2016
-
www.scienceintheclassroom.org www.scienceintheclassroom.org
-
cohort
A group used in a study that have something in common.
-
salient
Meaningful, noteworthy, important.
-
perforated
Pierced with holes.
-
learned helplessness
Conditioned behavior in which an individual gives up trying to escape a painful situation after repeatedly failing to escape. http://dictionary.reference.com/browse/learned+helplessness
Learned helplessness occurs when an animal is repeatedly subjected to an aversive stimulus that it cannot escape. Eventually, the animal will stop trying to avoid the stimulus and behave as if it is utterly helpless to change the situation.
Even when opportunities to escape are presented, this learned helplessness will prevent any action http://psychology.about.com/od/lindex/f/earned-helplessness.htm
Learned helplessness is also a model for depression in rodents.
-
down-regulated
Reduced or suppressed.
-
pro-socially
Voluntarily helping others.
-
empathically motivated pro-social behavior
Actions that help others caused by a feeling of empathy.
-
motivational state
Condition of the mind that drives a behavior
-
-
scienceintheclassroom.org scienceintheclassroom.org
-
Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES)
The Convention on International Trade in Endangered Species (CITES) of Wild Fauna and Flora is an international United Nations agreement between governments.
The goal is to ensure that international trade with specimens of wild animals and plants does not threaten their survival. CITES implements controls in the international trade with specimens of selected species.
-
Voronoi prior
Voronoi diagrams or polygons divide a plane into regions based on distance to certain points of interest. They have distance information that can help answer questions like “which object is closest to point P?” and “where is the nearest hospital from point Q?”
-
Markov chain Monte Carlo algorithm
A Markov chain refers to a sequence of random events or variables, where each event is dependent on the previous event.
In biology, Markov chain algorithms are often used to model population processes or to describe gene frequencies in populations.
Here, the researchers used a statistical method to model complicated allele distributions and assign geographic locations to seized ivory.
-
microsatellite DNA loci
A stretch of repetitive DNA in which sequences of base pairs are repeated, typically two to seven times.
Example: GATC GATC GATC GATC GATC
-
allele frequency
Allele frequency is the relative frequency of a particular allele (or gene variant) in a population. Allele frequencies are shown as fractions or percentages.
An allele is one of two or more alternative forms of a gene that are located at the same position (genetic locus) on a chromosome.
-
genotypes
The genotype is the genetic makeup of an organism. It can also refer to the two inherited alleles of a particular gene.
-
keystone species
A species that is crucial to how an ecosystem functions. If the keystone species were removed, the ecosystem would dramatically change or cease to exist.
-
ivory
Ivory is a hard, creamy white material that is derived from tusks and incisor teeth of animals, including elephants.
-
African elephant
There are two subspecies of African elephants, the African forest elephant and the African savanna elephant.
-
transnational organized crime
Transnational organized crime involves associations or groups of individuals that profit from the sale of illegal goods across national borders, creating international illegal markets.
Activities of transnational organized crime include drug trafficking, human trafficking, money laundering, sale of counterfeit goods, and illegal wildlife trade.
-
hotspots
A place of significant activity or danger.
-
Poaching
Illegal hunting, killing, or capturing of wild animals.
-
-
scienceintheclassroom.org scienceintheclassroom.org
-
conspecific alarms
What are conspecific alarms? Conspecific means that the individuals belong to the same species.
Thus these alarms are signals that one individual calls out to warn other individuals in the same species. Think of meerkats.
-
La la la la la la la la la la la la la la la la la la la la la la la la la
Here the editor summarizes the key findings ("la la la") and outlines the general impact of the work ("la").
Melissa can edit as SitC
Shelby can also edit as SitC
-
- Jul 2016
-
scienceintheclassroom.org scienceintheclassroom.org
-
Acknowledgments
It is common for a paper to contain an "Acknowledgments" section to thank the people and organizations without whom the study could not have occurred, but who were not sufficiently involved in the study to be included as authors on the paper.
Commonly included in this section are funding agencies that provided the money for the study, core facilities that (in exchange for a fee) provided services or instruments to perform assays included in the study, and collaborators who provided reagents or materials for the study.
-
liquid chromatography–mass spectrometry
A technique that first separates individual chemicals present in a sample (via liquid chromatography) and then identifies what those chemicals are by measuring the masses of the particles in those separated chemicals (via mass spectrometry).
Liquid chromatography separates out the different chemicals in the sample by forcing a liquid containing the sample at a very high pressure through a column. This column contains a solid material that the particles in the sample will cling to.
Different chemicals in the sample will interact slightly differently with that solid material, and so they will flow through the column at different rates. This causes them to separate out so the investigators can look at each of these chemicals individually in the next step and try to figure out what they are.
Mass spectrometry allows investigators to identify what the now-separated chemicals are by measuring the mass-charge ratio of the particles that make up the chemicals.
In order to do this, the chemicals are bombarded with electrons, causing some of their molecules to break apart into charged fragments. Those charged fragments are then run through an electromagnetic field, and naturally separate according to their mass-charge ratio.
The pattern of separation that results is called a "mass spectrum." By looking at the masses of the individual particles that make up a chemical, we can guess what that chemical is.
For example, if the mass spectrum told us that we had 2 particles the mass of a hydrogen atom and 1 particle the mass of an oxygen atom, we would deduce that we had a water molecule on our hands.
-
pollinator
An organism that moves pollen from the male reproductive organs of a flower to the female reproductive organs of flower, resulting in fertilization.
Without pollinators, many types of flowers would not be able to reproduce. Types of pollinators include honey bees, butterflies, and hummingbirds.
See more about pollinators in this video from National Geographic: People, Plants, and Pollinators
-
-
www.scienceintheclassroom.org www.scienceintheclassroom.org
-
myosin
Myosin is the motor protein for the cytoskeletal protein actin. These two proteins make up much of the cytoskeleton in muscle cells and thus myosin is good marker for skeletal muscle.
-
representative
The authors can not possibly show a picture of every single animal they performed this experiment on so they show two "representative" animals.
That is, two animals that best show what they saw in the rest of the animals.
-
abrogated
Prevented.
-
anterior gradient protein family
The grouping of all anterior gradient proteins (see below).
-
proximodistal
The axis extending from the shoulder (proximal) to the tips of the fingers (distal).
In other words, "proximal or distal identity." Proximal meaning near the point of attachment versus distal meaning away from the point of attachment.
-
Prod 1
A cell surface protein studied for its roles in salamander limb regeneration. To date, the Prod 1 gene is only present in salamander species.
-
-
www.science.org www.science.org
-
Cre
CRE Integrases/recombinases (causes recombination) are a family of enzymes which catalyze recombination, i.e., exchange of genetic material either between multiple chromosomes or between different regions of the same chromosome.
They delete segments of DNA flanked by LoxP sites (floxed) in experimental animals, and can generate animals with mutations limited to certain cell types (tissue-specific knockouts) or animals with chemically inducible mutations (time-specific inducible knockouts).
Tags
Annotators
URL
-
-
scienceintheclassroom.org scienceintheclassroom.org
-
herd immunity
Herd immunity is a key principle in infectious disease vaccination. Epidemiologists, scientists who study patterns in disease, have discovered that if you vaccinate the majority of people in a population even those who are not vaccinated don't catch the infectious disease. This effect is because the likelihood of coming into contact with anyone else who could also get sick is so small (because everyone else is vaccinated).
Further thoughts: Does herd immunity apply to NON-infectious disease vaccines, like a hypothetical vaccine that could prevent heart disease?
Hint: does your risk for heart disease depend on if your close contacts have heart disease?
Still confused or want to learn more? VIsit http://www.vaccines.gov/basics/protection (vaccines.gov) for a more detailed explanation and pictures.
-
high-income countries
High-Income countries are those countries whose residents make good living wages. Examples include the US, England, Japan, and many others. This is in contrast to low-income countries where there is more poverty and thus more disease and higher mortality rates (more death) for many diseases.
It is important to know that this study is considering high-income countries because the outcomes of many of the nonmeasles infectious diseases are worse, and more children die in low-income countries. This makes the ability to study how measles effects these outcomes more difficult.
This topic will be discussed more later in the paper in a Previous Work annotation.
-
nonmeasles infectious
Infectious diseases are illnesses that can be passed from one person to another, like the flu. These are the diseases whose outcomes are examined in this paper.
Non-infectious diseases are like heart disease or diabetes which you cannot catch from someone else. Non-infectious diseases are NOT important for this paper and have not been shown to be effected by measles.
Importantly, these nonmeasles infectious diseases include opportunistic infections mentioned previously.
-
Immunosuppression
Immunosuppression is when the body's defense mechanism against disease, called the immune system no longer works as well as it should
-
In practice, we weighted the quarters using a gamma function. Dividing S by the total population of interest thus provides the prevalence of immunomodulation
Gamma Transformation
The number of immunomodulated individuals (after measles infection) was diveded by the total population of interest. This means that the data is presented as a percent of the whole rather than absolute numbers.
It is important to look at data such as this as a percent of the whole and not just total numbers so that the numbers can be compared across time and across populations. For example, if a total of 50 people out of 100 people get sick and die from some illness, that is a very high number or rate of death (50%). However, if 50 people die out of 1 million who got sick, then even though it is the same number of people who died, it is a much much lower rate of death per person (0.005%). When the number of people who are ill is divided by the total population at risk for the illness, the answer to the division problem is essentially rate or the prevalence of the illness in the population.
Once the 'baseline prevalence' of measles immune-amnesia was calculated, the authors used a tool from statistics, called a gamma function, to make theoretical calculations of how many people from this 'baseline prevalence' would truly still have immune amnesia, at a certain point in time since the measles infection.
The purpose of using the gamma function was to create a distribution of immune-amnesia with time following measles. For example, if the immune amnesia would last for 3 years, it wouldn't make sense, biologically, to assume that 100% of children have immune-amnesia at the end of three years since getting measles and then 0% have immune-amnesia 1 day later, at the beginning of the fourth year since getting measles. The gamma function allowed a smooth gradient where 100% of children had immune amnesia directly following measles infection and this gradually dropped over time to 0% of children with immune amnesia by, for example, four years following measles. In the study, how long it took to reach 0% was what was defined by the gamma function.
-
measles incidence
Incidence is how much of something first happens within a given amount of time. There is another term called prevalence that is similar, but has an important difference. In diseases, for example, incidence is 'how much new disease there is over a certain time-period,' while prevalence is how much disease there is at a given point in time. This distinction is especially important for diseases that may last a long time. For example, if 100 people each year get HIV, the incidence is 100 new cases of HIV per year. However, because HIV does not go away, the prevalence, or number of people with HIV in a population at a certain time, will be much higher because it will include not just the 100 people who just got HIV, but also all of the people who still have HIV due to infections in previous years.

For example, you can see a graph of measles incidence in the US here. You will notice that it is plotted by when on the X-axis, and how many on the Y-axis.
The incidence of measles significantly drops after vaccination because this provides host resistance to the disease
-
host resistance
Host resistance describes how well an individual (the host for the disease) can resist getting a disease (resistance).
For example, if you get the chickenpox vaccine, it is extremely unlikely you will ever get the chickenpox disease. This means that you have a lot of host resistance against chickenpox (because you got the vaccine). It would also be true if you got the chickenpox disease once, you would be resistant to getting it again. Both are examples of host resistance.
In this context, they are describing that the opposite is true. If you get the measles disease there is a prolonged effect decreasing your host resistance to other diseases for 2-3 years.
Keep reading to learn about how this could be the case, and why it is so important around the world.
-
opportunistic infections
Opportunistic infections describe pathogens (things that make you sick like bacteria, viruses and fungi) that take advantage of when people's immune systems are weak. Under normal conditions these opportunistic pathogens DO NOT make you sick, but if you have some problem that reduces your immune system's ability to protect you, like a recent sickness or malnutrition, then opportunistic pathogens can make you sick.
Opportunistic pathogens/infections are important in medicine because they can make treating sick people very complicated. For example, AIDS patients don't actually die because the HIV virus made them sick. Victims of AIDS most often die because their immune system is so damaged they cannot fight off opportunistic pathogens. Learn more about why these opportunistic infections are so devastating at AIDS.gov.
-
-
scienceintheclassroom.org scienceintheclassroom.org
-
social-emotional learning (SEL)
Social-emotional learning is a particular approach to teaching people how to manage and process their emotions.
This study is randomly assigning half of the treatment group to social-emotional learning to see if this kind of curriculum has any effect on rates of violent activity.
The link below provides more detail on what social-emotional learning entails and can help you consider why this would be an important part of this kind of study:
http://greatergood.berkeley.edu/article/item/how_social_emotional_learning_transforms_classrooms
-
As in any social experiment, external validity is not guaranteed
External validity refers to the ability to say that we could expect these same results in other contexts.
When experiments are conducted in social settings, external validity is not something we can expect to acquire because different social, economic and cultural contexts would have different effects on the outcomes.
-
proximal
Proximal refers to something that is near to, or in close proximity to, a point of reference. Proximal is a term commonly used in research because research is always striving to explain cause and effect relationships.
Proximal causes are those causes that are thought to be part of the relationship, but are perhaps not the ultimate source of the relationship.
In this case, hostile attribution bias, or the problem of assuming other people's intent is hostile, even when it isn't, is thought to be the proximal cause.
-
randomized controlled trial
A randomized controlled trial is one of many ways to conduct a scientific experiment.
Many people are selected for the experiment, but only some people are randomly selected to receive the intervention that is being tested for.
These kinds of studies are often referred to as the gold standard. When pharmaceuticals are still in the development stage, randomized controlled trials are used to test the drug's effectiveness.
-
This “public health crisis
Violence can be considered a public health crisis not only because it can lead to mortality, but because those involved often end up needing health care services.
The Centers for Disease Control has identified youth violence as a public health crisis. They report that in 2008 more than 656,000 young adults between the ages of 10 and 24 were treated in emergency rooms for injuries related to violence.
In terms of mortality, homicide kills more young black males than the 9 leading causes of death combined.
-
-
scienceintheclassroom.org scienceintheclassroom.org
-
Polymerase chain reaction (PCR)
A lab technique that uses short DNA sequences to amplify a specific region of the genome. These short DNA sequences are called primers. They anneal to genomic DNA and serve as a starting point for DNA polymerase to replicate the DNA at that location.
PCR applies multiple cycles of heating and cooling temperatures, creating the ideal conditions for many rounds of replication. This technique is often used to determine the genotype of an animal at a specific gene.
This YouTube video goes over the technique in more detail.
-
CRISPR/Cas9
CRISPR stands for "Clustered Regularly Interspaced Short Palindromic Repeats" and Cas9 refers to the protein that is associated with CRISPR. CRISPRs are DNA sequences that are transcribed into RNA.
Cas9 protein is guided to a specific DNA sequence by the CRISPR RNA. Because Cas9 is a nuclease, meaning that is cuts DNA, it cleaves the DNA at the sequence specified by the CRISPR RNA.
-
- Jun 2016
-
scienceintheclassroom.org scienceintheclassroom.org
-
mean
A statistical term. Summation of all the elements divided by their number.
-
disinfection by-products
Normally, the last step during drinking water purification is a disinfection step.
Chlorine is the most commonly used chemical.
Although effective in killing harmful pathogens, chlorine can react with natural organic matter and form harmful byproducts (e.g., trihalomethanes [THM], bromate, chlorite, etc.).
-
natural organic matter
Comes from the natural processes in the environment such as decomposition of organic matter.
Examples include proteins, amino acids, polysaccharides, and humic and fulvic acids, etc. (Reference)
-
-
www.scienceintheclassroom.org www.scienceintheclassroom.org
-
conserved
That is, evolutionarily conserved or kept the same throughout the evolution of salamanders and humans.
-
Cys-rich
A protein that contains a high number of cysteine amino acids.
-
three-dimensional structures
The 3D tertiary structure of these proteins has not been solved.
Doing this would allow for one to compare how related their structures are at the tertiary level. This is as opposed to looking just at the amino acid sequence (primary structure), which may mislead one into thinking that they are very different.
However, it could very well be the case that the primary structures are very different, but the tertiary structures very similar.
Of note, the 3D structure of Prod1 has now been solved. See, Garza-Garcia et al. (2009) PLOS ONE e7123, and the interaction between mammalian AG2 and C4.4 appears important in models of pancreatic cancer (Arumugam et al., Molecular Cancer Therapeutics 14, 941-51 [2015]).
-
supernumerary
More than expected or normally found.
A sixth digit on a human hand would be supernumerary.
-
SD
Standard deviation. A statistical measure that helps demonstrate how far values are from the mean.
In this case, the mean is 8.3 and most values are expected to be within the range of five to 11.6 (+/- 3.3 from the mean).
-
fold
How many times over the baseline
For instance, if Jenny has five flowers and Howard has 50, Howard has 10-fold more flowers than Jenny.
For that example, Jenny sets the baseline and Howard has some number-fold greater flowers than her.
-
bromodeoxyuridine
A molecule that can be incorporated into DNA like thymidine (T). This is a nonnaturally occurring molecule, so it is useful for determining how often new nucleotides are being incorporated into DNA.
For an in-depth discussion of bromodeoxyuridine (BrdU) pulse labeling, see the annotation for figure 6.
A picture of BrdU versus dT (thymidine) can be seen here
-
mean stimulation index for S-phase entry
This can be broken into three parts.
Mean: Average
Stimulation index: The ratio of cells in s-phase to the total number of cells counted.
S-phase: The portion of the cell cycle where DNA strands are duplicated or copied.
In other words, the average ratio of number of cells where DNA is being duplicated compared to the total number of cells.
-
maintenance
The cells were "maintained," or kept in medium containing 1% serum for the rest of their life span.
-
serum
In vertebrate cell culture, most cells are grown in a medium containing "serum." Often this is fetal bovine serum.
The blood from a cow fetus is withdrawn and only the serum is kept. The serum contains proteins, nutrients, and other molecules, but does not contain any other cells.
The components of fetal bovine serum can vary greatly, but in general, its addition to cell culture medium is necessary for keeping cells healthy.
-
cultured
Like Cos7 cells, newt blastema cells are able to be kept outside of the living organism in an artificial medium that provides nutrients, or in other words, kept in "culture."
-
bilaterally
On both sides, i.e., both the left and right limb were amputated.
-
sectioned
Often large structures are too large to perform immunostaining or other visualization techniques on.
To get around this the tissue can be "sectioned": cut into very thin layers. Often this tissue will be sliced into micrometer-thick slices (1/1000 millimeter).
-
extracellular space
The space outside the cell.
-
dedifferentiation
You likely have heard of "differentiation," when a stem cell starts turning into a more specialized cell such as a skin cell or muscle cell.
Dedifferentiation is the exact opposite of that. Now, cells that were originally part of the limb start becoming more like progenitor cells.
These dedifferentiated cells can give rise to the cell type of origin.
-
dermis
The layer of skin below the epidermis (the top layer of skin). This holds many different types of living cells including nerves, vasculature, as well as fibroblasts, which make an important contribution to the blastema after limb transection.
For an example using human skin see here
-
glands
A general term for any part of the body that secretes a substance (i.e., proteins). Remember nAG is likely a secreted protein, so a gland would be a reasonable place to find it.
-
PS cells
"Pentosan sulfate" cells, an immortal cell line derived from mouse brain cells.
-
secreted
Remember, nAG is a secreted protein. The myc-tagged nAG they have transfected should be secreted just like the wild-type protein.
-
standard pull-down assay
Often referred to as "coimmunoprecipitation." Protein complexes are able to be separated by binding an antibody to one of the proteins in that complex.
That antibody is usually bound to something heavy (agarose polysaccharides extracted from seaweed) or magnetic to allow for separation.
-
Epitope-tagged
Epitope-tagged proteins have small stretches of amino acids added to the normal, wild-type protein. Often these additional amino acids create an epitope, an antibody-binding region.
This allows for the use of standard antibodies. Now, an antibody doesn't have to be created for each protein, only to the epitope tag, which usually can be added easily to many different proteins.
Note that in this paper the authors use both antibodies to the sequence of nAG itself, as well as antibodies to an epitope tag added to nAG. Why are they using both approaches?
-
Cos 7 cells
An immortal cell line derived from mouse kidney that acts like fibroblasts (the cells that produce collagen in your skin, among other functions).
-
thioredoxin fold
A common type of tertiary structure found in proteins. It gets it's name from protein thioredoxin, which has this type of fold in it.
-


