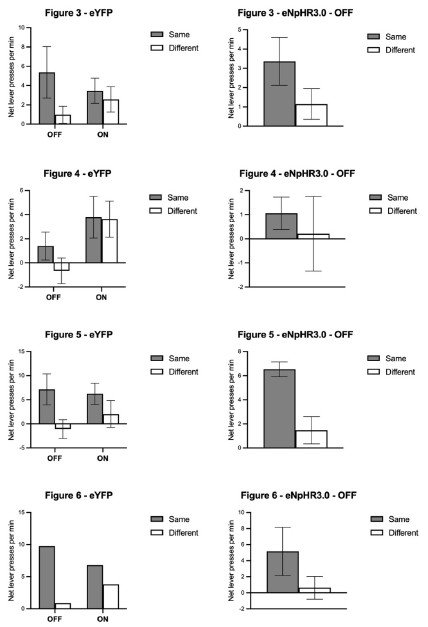
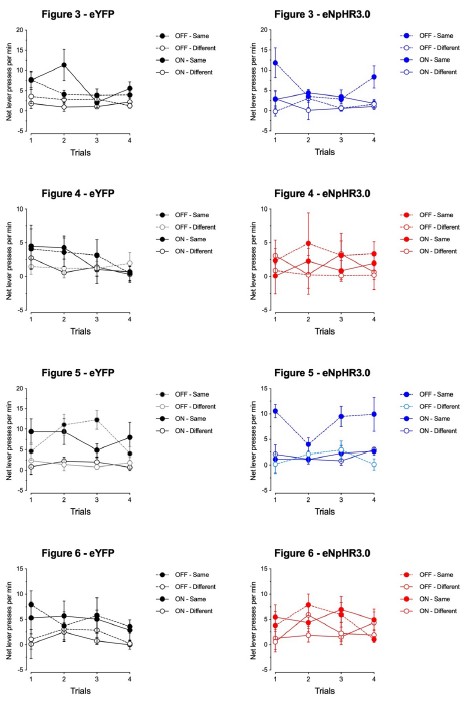
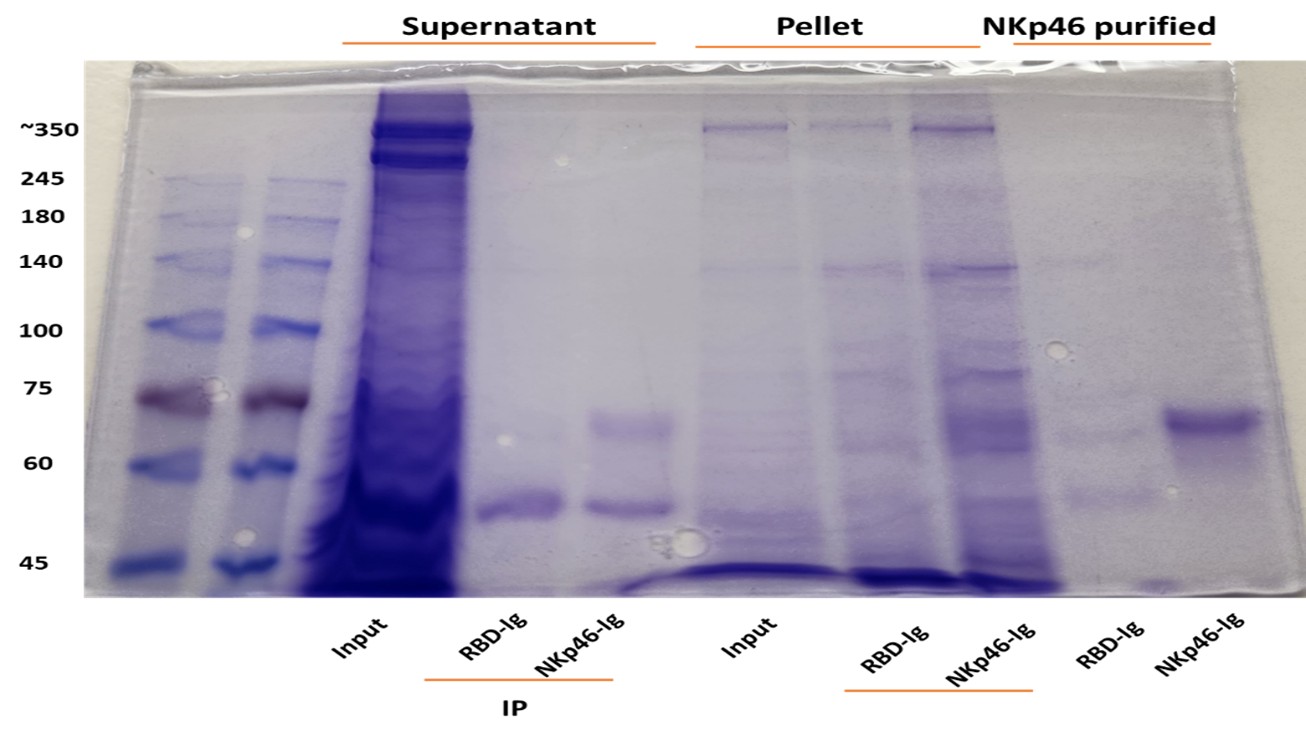
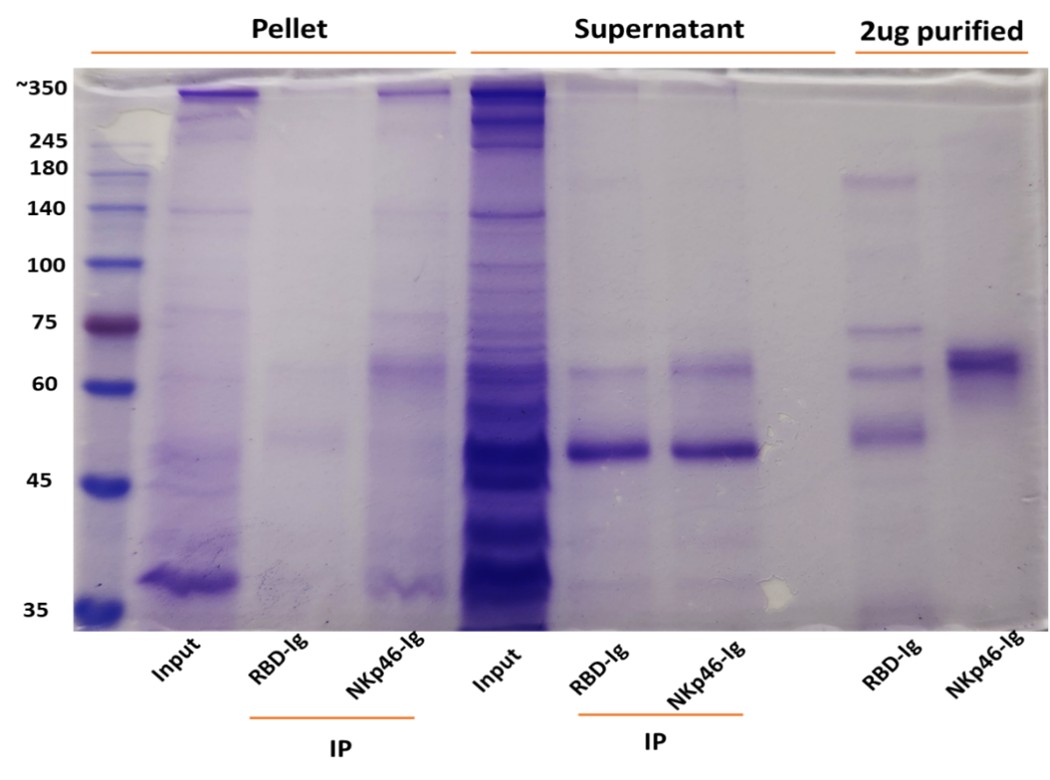
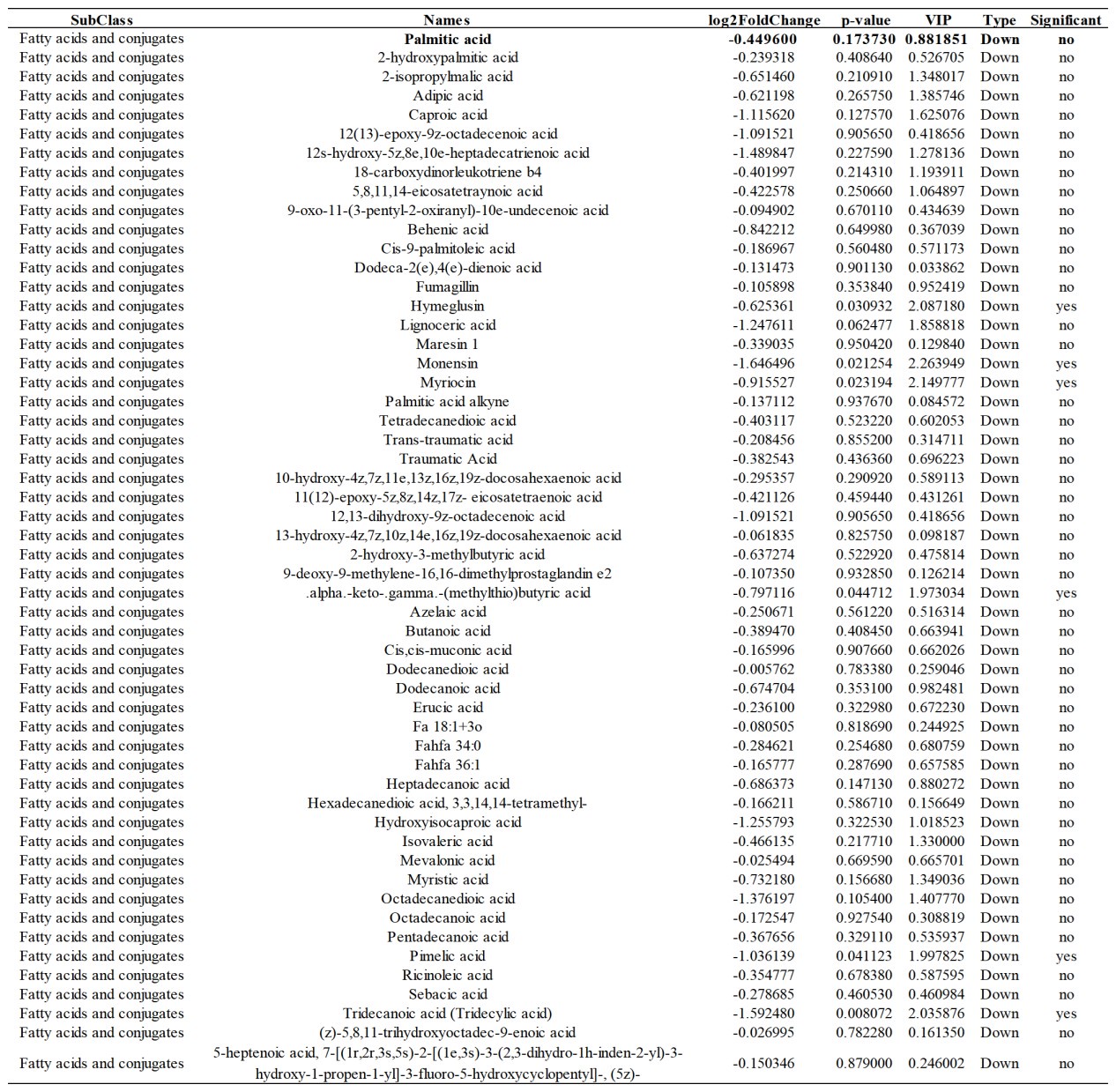
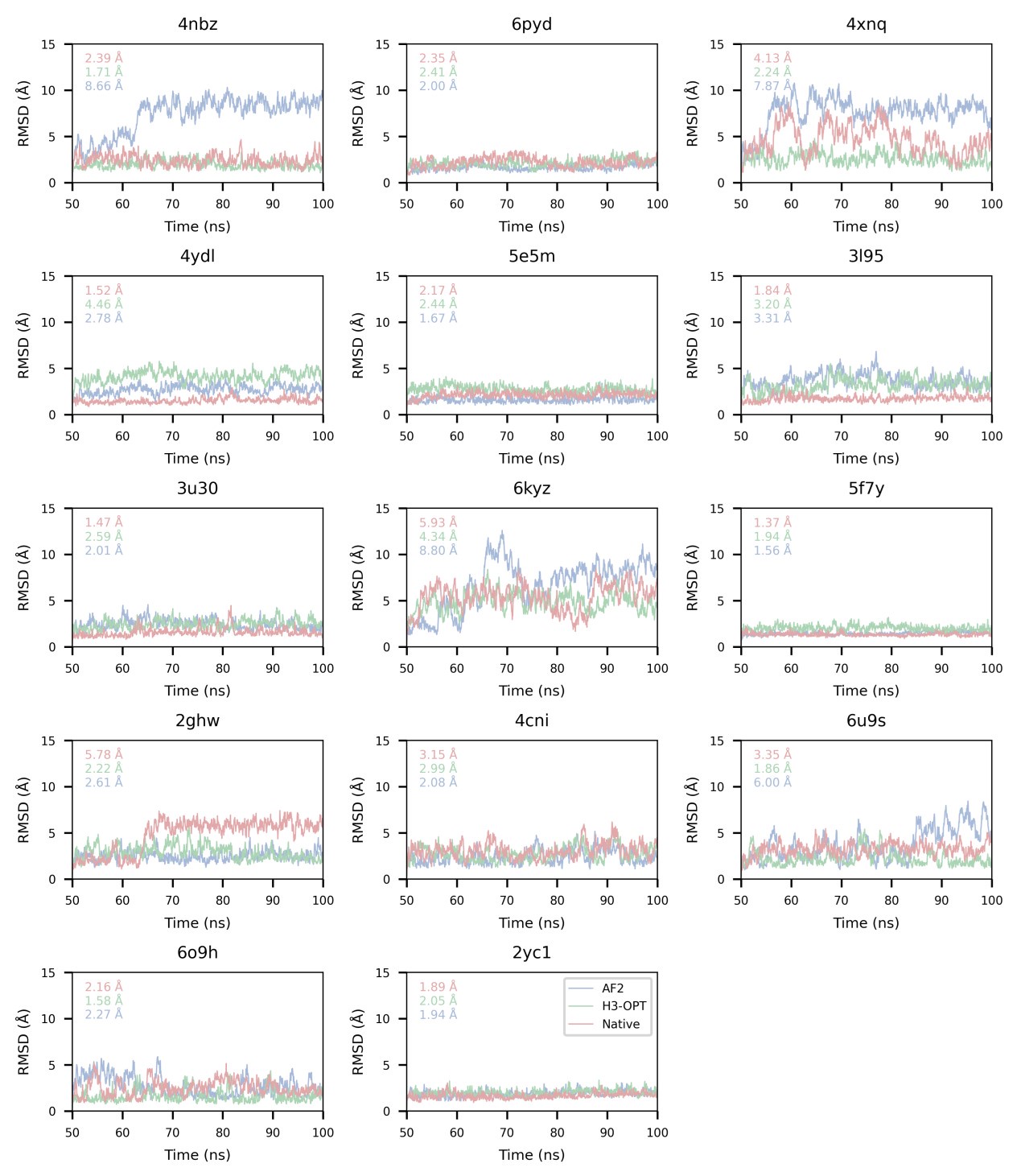
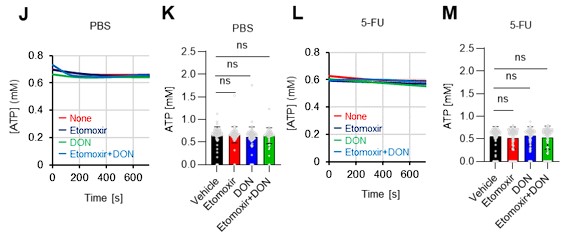
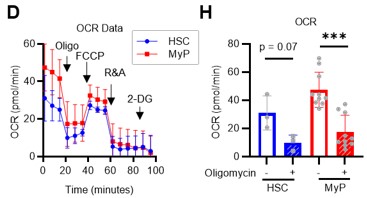
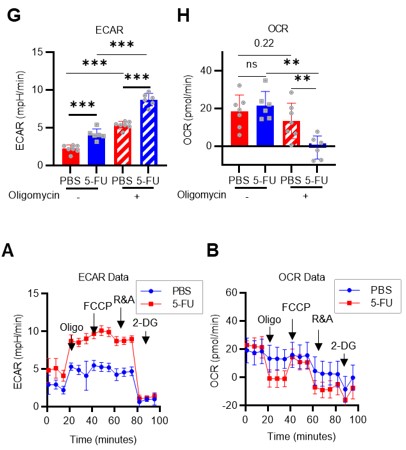
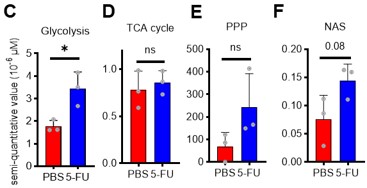
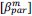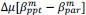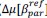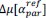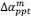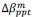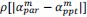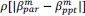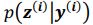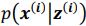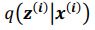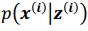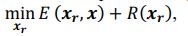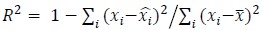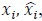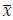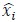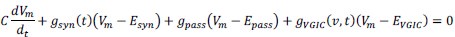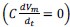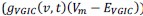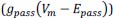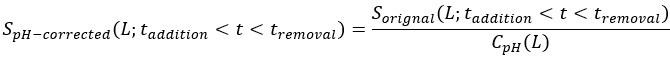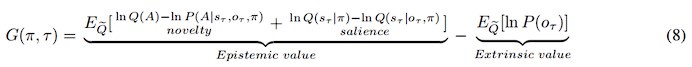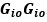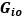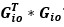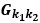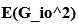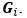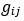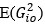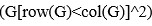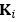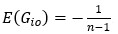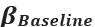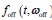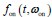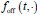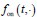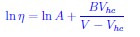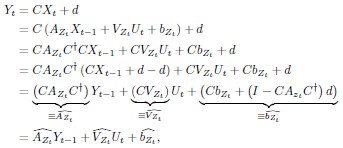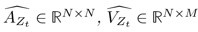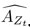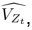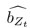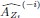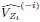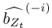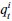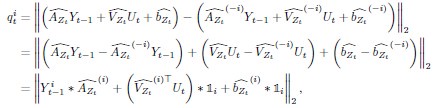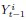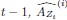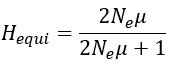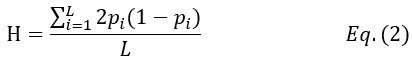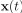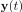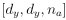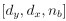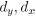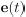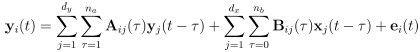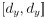Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public review):
This paper describes a number of patterns of epistasis in a large fitness landscape dataset recently published by Papkou et al. The paper is motivated by an important goal in the field of evolutionary biology to understand the statistical structure of epistasis in protein fitness landscapes, and it capitalizes on the unique opportunities presented by this new dataset to address this problem.
The paper reports some interesting previously unobserved patterns that may have implications for our understanding of fitness landscapes and protein evolution. In particular, Figure 5 is very intriguing. However, I have two major concerns detailed below. First, I found the paper rather descriptive (it makes little attempt to gain deeper insights into the origins of the observed patterns) and unfocused (it reports what appears to be a disjointed collection of various statistics without a clear narrative. Second, I have concerns with the statistical rigor of the work.
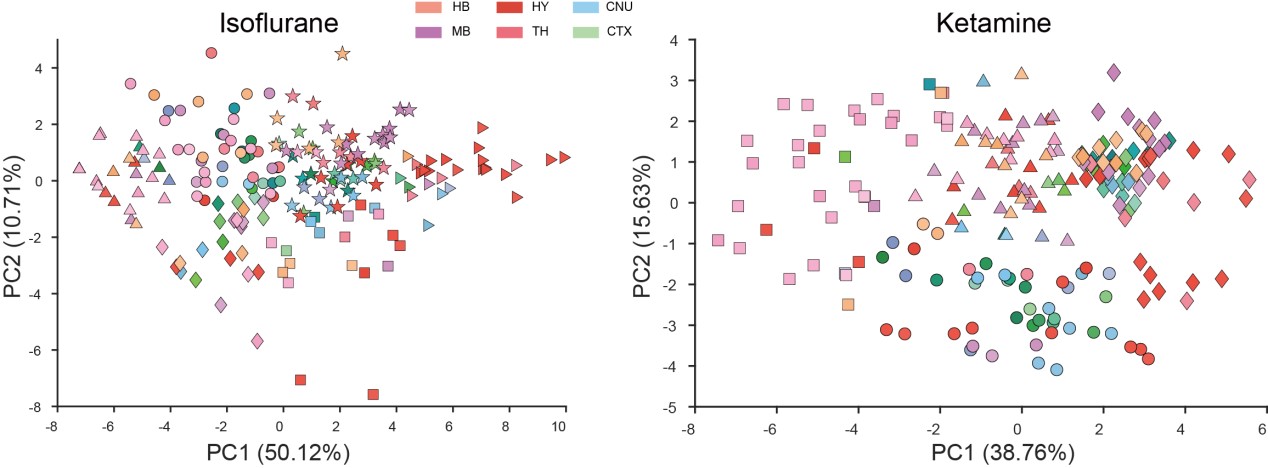
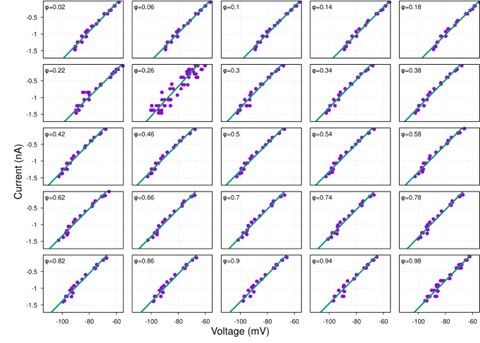
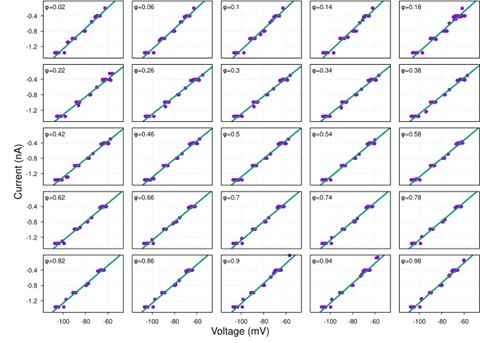
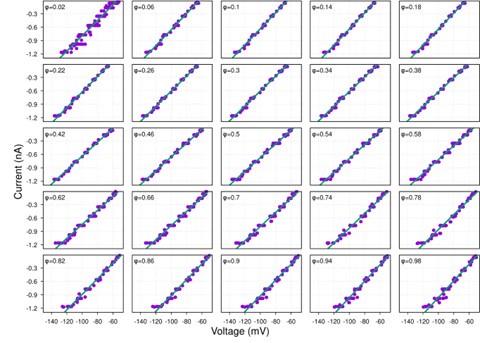
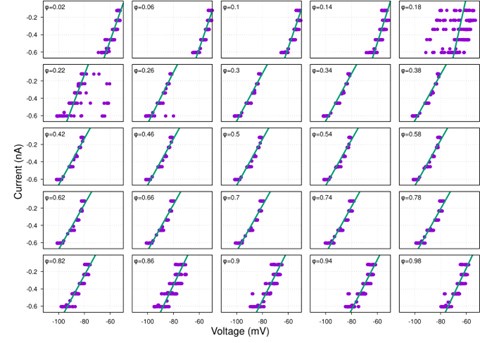
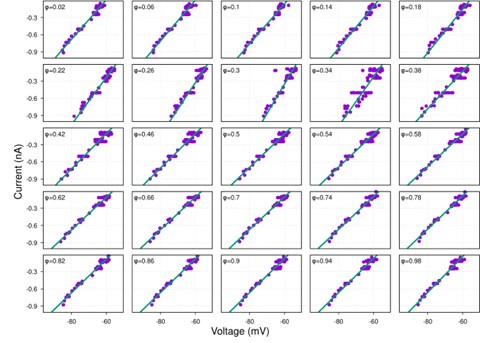
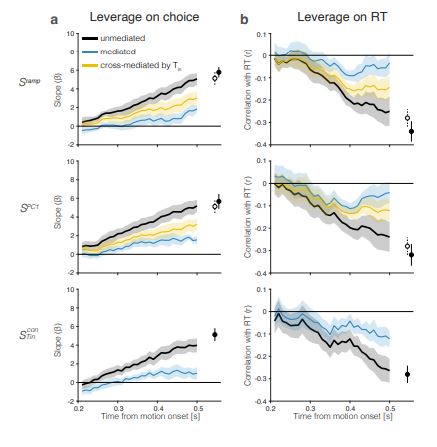
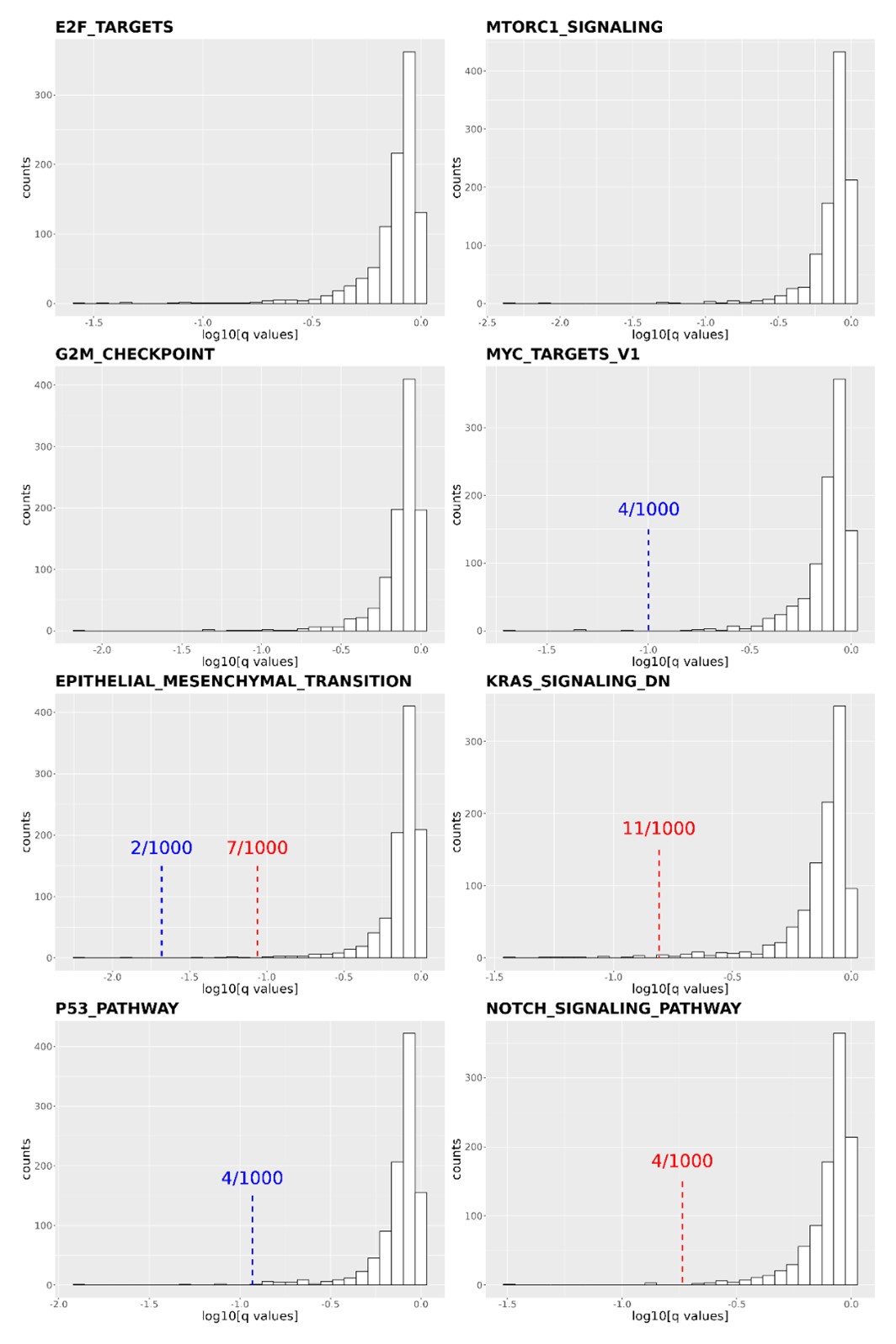
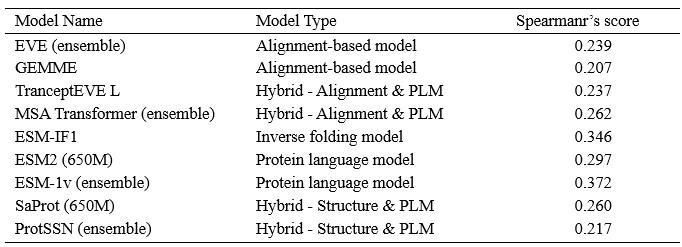
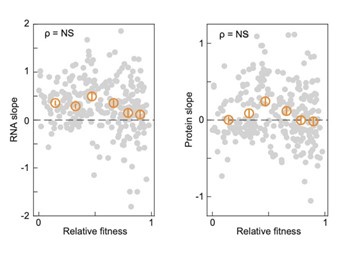
(1) I think Figures 5 and 7 are the main, most interesting, and novel results of the paper. However, I don't think that the statement "Only a small fraction of mutations exhibit global epistasis" accurately describes what we see in Figure 5. To me, the most striking feature of this figure is that the effects of most mutations at all sites appear to be a mixture of three patterns. The most interesting pattern noted by the authors is of course the "strong" global epistasis, i.e., when the effect of a mutation is highly negatively correlated with the fitness of the background genotype. The second pattern is a "weak" global epistasis, where the correlation with background fitness is much weaker or non-existent. The third pattern is the vertically spread-out cluster at low-fitness backgrounds, i.e., a mutation has a wide range of mostly positive effects that are clearly not correlated with fitness. What is very interesting to me is that all background genotypes fall into these three groups with respect to almost every mutation, but the proportions of the three groups are different for different mutations. In contrast to the authors' statement, it seems to me that almost all mutations display strong global epistasis in at least a subset of backgrounds. A clear example is C>A mutation at site 3.
(1a) I think the authors ought to try to dissect these patterns and investigate them separately rather than lumping them all together and declaring that global epistasis is rare. For example, I would like to know whether those backgrounds in which mutations exhibit strong global epistasis are the same for all mutations or whether they are mutation- or perhaps positionspecific. Both answers could be potentially very interesting, either pointing to some specific site-site interactions or, alternatively, suggesting that the statistical patterns are conserved despite variation in the underlying interactions.
(1b) Another rather remarkable feature of this plot is that the slopes of the strong global epistasis patterns seem to be very similar across mutations. Is this the case? Is there anything special about this slope? For example, does this slope simply reflect the fact that a given mutation becomes essentially lethal (i.e., produces the same minimal fitness) in a certain set of background genotypes?
(1c) Finally, how consistent are these patterns with some null expectations? Specifically, would one expect the same distribution of global epistasis slopes on an uncorrelated landscape? Are the pivot points unusually clustered relative to an expectation on an uncorrelated landscape?
(1d) The shapes of the DFE shown in Figure 7 are also quite interesting, particularly the bimodal nature of the DFE in high-fitness (HF) backgrounds. I think this bimodality must be a reflection of the clustering of mutation-background combinations mentioned above. I think the authors ought to draw this connection explicitly. Do all HF backgrounds have a bimodal DFE? What mutations occupy the "moving" peak?
(1e) In several figures, the authors compare the patterns for HF and low-fitness (LF) genotypes. In some cases, there are some stark differences between these two groups, most notably in the shape of the DFE (Figure 7B, C). But there is no discussion about what could underlie these differences. Why are the statistics of epistasis different for HF and LF genotypes? Can the authors at least speculate about possible reasons? Why do HF and LF genotypes have qualitatively different DFEs? I actually don't quite understand why the transition between bimodal DFE in Figure 7B and unimodal DFE in Figure 7C is so abrupt. Is there something biologically special about the threshold that separates LF and HF genotypes? My understanding was that this was just a statistical cutoff. Perhaps the authors can plot the DFEs for all backgrounds on the same plot and just draw a line that separates HF and LF backgrounds so that the reader can better see whether the DFE shape changes gradually or abruptly.
(1f) The analysis of the synonymous mutations is also interesting. However I think a few additional analyses are necessary to clarify what is happening here. I would like to know the extent to which synonymous mutations are more often neutral compared to non-synonymous ones. Then, synonymous pairs interact in the same way as non-synonymous pair (i.e., plot Figure 1 for synonymous pairs)? Do synonymous or non-synonymous mutations that are neutral exhibit less epistasis than non-neutral ones? Finally, do non-synonymous mutations alter epistasis among other mutations more often than synonymous mutations do? What about synonymous-neutral versus synonymous-non-neutral. Basically, I'd like to understand the extent to which a mutation that is neutral in a given background is more or less likely to alter epistasis between other mutations than a non-neutral mutation in the same background.
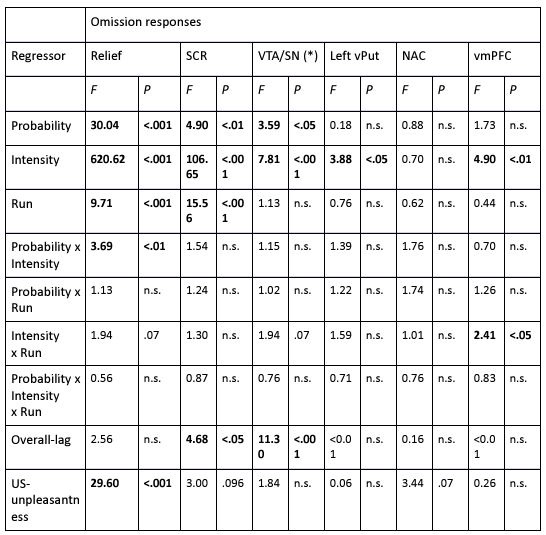
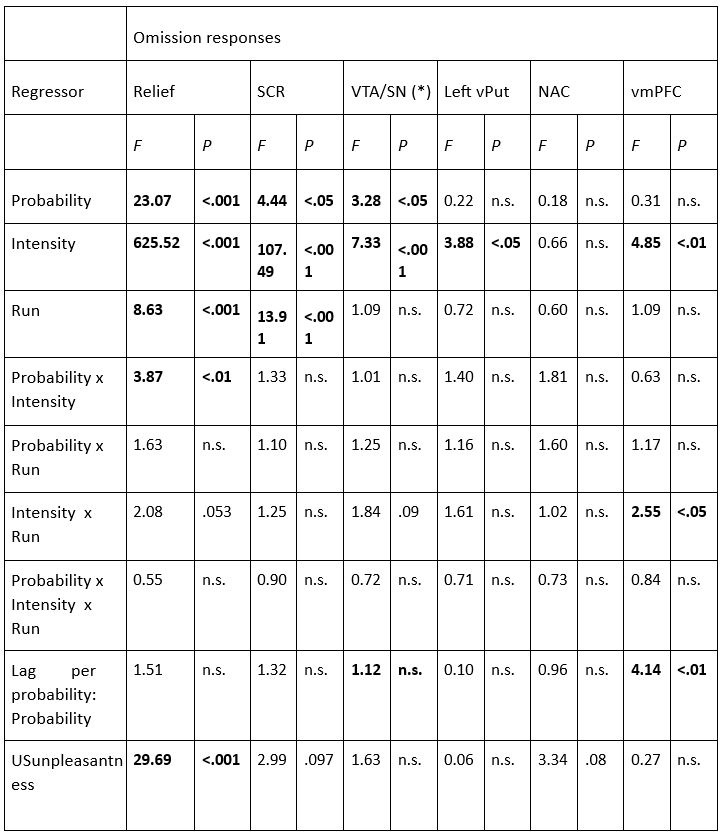
(2) I have two related methodological concerns. First, in several analyses, the authors employ thresholds that appear to be arbitrary. And second, I did not see any account of measurement errors. For example, the authors chose the 0.05 threshold to distinguish between epistasis and no epistasis, but why this particular threshold was chosen is not justified. Another example: is whether the product s12 × (s1 + s2) is greater or smaller than zero for any given mutation is uncertain due to measurement errors. Presumably, how to classify each pair of mutations should depend on the precision with which the fitness of mutants is measured. These thresholds could well be different across mutants. We know, for example, that low-fitness mutants typically have noisier fitness estimates than high-fitness mutants. I think the authors should use a statistically rigorous procedure to categorize mutations and their epistatic interactions. I think it is very important to address this issue. I got very concerned about it when I saw on LL 383-388 that synonymous stop codon mutations appear to modulate epistasis among other mutations. This seems very strange to me and makes me quite worried that this is a result of noise in LF genotypes.
Thank you for your review of the manuscript. In the revised version, we have addressed both major criticisms, as detailed below.
When carefully examining the plots in Figure 5 independently, we indeed observe that the fitness effect of a mutation on different genetic backgrounds can be classified into three characteristic patterns. Our reasoning for these patterns is as follows:
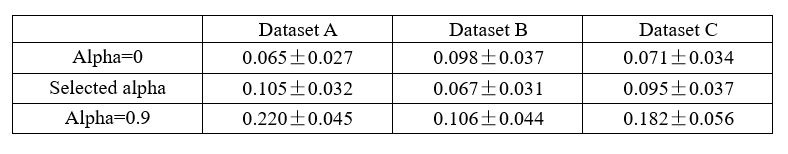
Strong correlation: Typically observed when the mutation is lethal across backgrounds. Linear regression of mutations exhibiting strong global epistasis shows slopes close to −1 and pivot points near −0.7 (Table S4). Since the reported fitness threshold is −0.508, these mutations push otherwise functional backgrounds into the non-functional range, consistent with lethal effects.
Weak correlation: Observed when a mutation has no significant effect on fitness across backgrounds, consistent with neutrality.
No correlation: Out of the 261,333 reported variants, 243,303 (93%) lie below the fitness threshold of −0.508, indicating that the low-fitness region is densely populated by nonfunctional variants. The “strong correlation” and “weak correlation” lines intersect in this zone. Most mutations in this region have little effect (neutral), but occasional abrupt fitness increases correspond to “resurrecting” mutations, the converse of lethal changes. For example, mutations such as X→G at locus 4 or X→A at locus 5 restore function, while the reverse changes (e.g. C→A at locus 3) are lethal.
Thus, the “no-correlation” pattern is largely explained by mutations that reverse the effect of lethal changes, effectively resurrecting non-functional variants. In the revised manuscript, we highlight these nuances within the broader classification of fitness effect versus background fitness (pp. 10–13).
Additional analyses included in the revision:
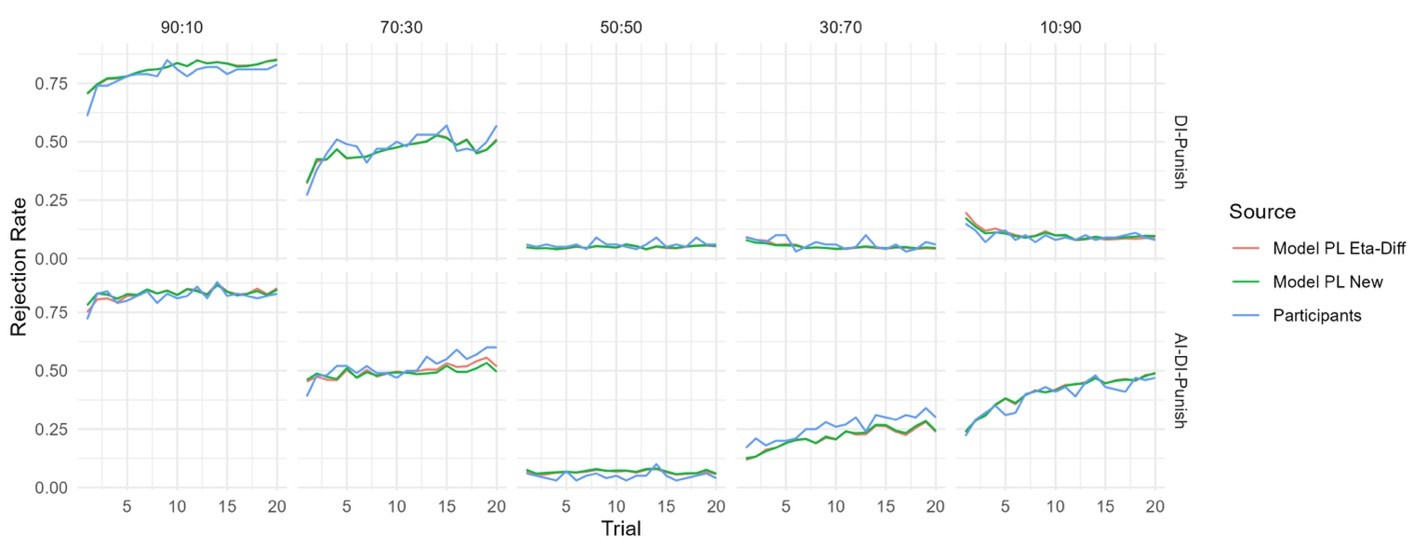
Synonymous vs. non-synonymous pairs: We repeated the Figure 1 analysis for synonymous–synonymous pairs. As expected, synonymous pairs exhibit lower overall frequencies of epistasis, consistent with their greater neutrality. However, the qualitative spectrum remains similar: positive and negative epistasis dominate, while sign epistasis is rare (Supplementary Figs. S6–S7, S9–S10).
Fitness effect vs. epistasis change: We tested whether the mean fitness effect of a mutation correlates with the percent of cases in which it changes the nature of epistasis. No correlation was found (R² ≈ 0.11), and this analysis is now included in the revised manuscript.
Epistasis-modulating ability: Non-synonymous mutations more frequently alter the interactions between other mutations than synonymous substitutions. Within synonymous substitutions, the subset with measurable fitness effects disproportionately contributes to epistasis modulation. Thus, the ability of synonymous substitutions to modulate epistasis arises primarily from the non-neutral subset.
These analyses clarify the role of synonymous mutations in reshaping epistasis on the folA landscape.
Revision of statistical treatment of epistasis:
In our original submission, we used an arbitrary threshold of 0.05 to classify the presence or absence of epistasis, following Papkou et al., who based conclusions on a single experimental replicate. However, as the reviewer correctly noted, this does not adequately account for measurement variability across different genotypes.
In the revised manuscript, we adopt a statistically rigorous framework that incorporates replicate-based error directly. Specifically, we now use the mean fitness across six independent replicates, together with the corresponding standard deviation, to classify fitness peaks and epistasis. This eliminates arbitrary thresholds and ensures that epistatic classifications reflect the precision of measurements for each genotype.
This revision led to both quantitative and qualitative changes:
For high-fitness genotypes, the core patterns of higher-order (“fluid”) epistasis remain robust (Figures 2–3).
For low-fitness genotypes, incorporating replicate-based error removed spurious fluidity effects, yielding a more accurate characterization of epistasis (Figures 2–3; Supplementary Figs. S6–S7, S9–S10).
We describe these methodological changes in detail in the revised Methods section and provide updated code.
Together, these revisions directly address the reviewer’s concerns. They improve the statistical rigor of our analysis, strengthen the robustness of our conclusions, and underscore the importance of accounting for measurement error in large-scale fitness landscape studies—a point we now emphasize in the manuscript.
Reviewer #2 (Public review):
Significance:
This paper reanalyzes an experimental fitness landscape generated by Papkou et al., who assayed the fitness of all possible combinations of 4 nucleotide states at 9 sites in the E. coli DHFR gene, which confers antibiotic resistance. The 9 nucleotide sites make up 3 amino acid sites in the protein, of which one was shown to be the primary determinant of fitness by Papkou et al. This paper sought to assess whether pairwise epistatic interactions differ among genetic backgrounds at other sites and whether there are major patterns in any such differences. They use a "double mutant cycle" approach to quantify pairwise epistasis, where the epistatic interaction between two mutations is the difference between the measured fitness of the double-mutant and its predicted fitness in the absence of epistasis (which equals the sum of individual effects of each mutation observed in the single mutants relative to the reference genotype). The paper claims that epistasis is "fluid," because pairwise epistatic effects often differs depending on the genetic state at the other site. It also claims that this fluidity is "binary," because pairwise effects depend strongly on the state at nucleotide positions 5 and 6 but weakly on those at other sites. Finally, they compare the distribution of fitness effects (DFE) of single mutations for starting genotypes with similar fitness and find that despite the apparent "fluidity" of interactions this distribution is well-predicted by the fitness of the starting genotype.
The paper addresses an important question for genetics and evolution: how complex and unpredictable are the effects and interactions among mutations in a protein? Epistasis can make the phenotype hard to predict from the genotype and also affect the evolutionary navigability of a genotype landscape. Whether pairwise epistatic interactions depend on genetic background - that is, whether there are important high-order interactions -- is important because interactions of order greater than pairwise would make phenotypes especially idiosyncratic and difficult to predict from the genotype (or by extrapolating from experimentally measured phenotypes of genotypes randomly sampled from the huge space of possible genotypes). Another interesting question is the sparsity of such high-order interactions: if they exist but mostly depend on a small number of identifiable sequence sites in the background, then this would drastically reduce the complexity and idiosyncrasy relative to a landscape on which "fluidity" involves interactions among groups of all sites in the protein. A number of papers in the recent literature have addressed the topics of high-order epistasis and sparsity and have come to conflicting conclusions. This paper contributes to that body of literature with a case study of one published experimental dataset of high quality. The findings are therefore potentially significant if convincingly supported.
Validity:
In my judgment, the major conclusions of this paper are not well supported by the data. There are three major problems with the analysis.
(1) Lack of statistical tests. The authors conclude that pairwise interactions differ among backgrounds, but no statistical analysis is provided to establish that the observed differences are statistically significant, rather than being attributable to error and noise in the assay measurements. It has been established previously that the methods the authors use to estimate high-order interactions can result in inflated inferences of epistasis because of the propagation of measurement noise (see PMID 31527666 and 39261454). Error propagation can be extreme because first-order mutation effects are calculated as the difference between the measured phenotype of a single-mutant variant and the reference genotype; pairwise effects are then calculated as the difference between the measured phenotype of a double mutant and the sum of the differences described above for the single mutants. This paper claims fluidity when this latter difference itself differs when assessed in two different backgrounds. At each step of these calculations, measurement noise propagates. Because no statistical analysis is provided to evaluate whether these observed differences are greater than expected because of propagated error, the paper has not convincingly established or quantified "fluidity" in epistatic effects.
(2) Arbitrary cutoffs. Many of the analyses involve assigning pairwise interactions into discrete categories, based on the magnitude and direction of the difference between the predicted and observed phenotypes for a pairwise mutant. For example, the authors categorize as a positive pairwise interaction if the apparent deviation of phenotype from prediction is >0.05, negative if the deviation is <-0.05, and no interaction if the deviation is between these cutoffs. Fluidity is diagnosed when the category for a pairwise interaction differs among backgrounds. These cutoffs are essentially arbitrary, and the effects are assigned to categories without assessing statistical significance. For example, an interaction of 0.06 in one background and 0.04 in another would be classified as fluid, but it is very plausible that such a difference would arise due to error alone. The frequency of epistatic interactions in each category as claimed in the paper, as well as the extent of fluidity across backgrounds, could therefore be systematically overestimated or underestimated, affecting the major conclusions of the study.
(3) Global nonlinearities. The analyses do not consider the fact that apparent fluidity could be attributable to the fact that fitness measurements are bounded by a minimum (the fitness of cells carrying proteins in which DHFR is essentially nonfunctional) and a maximum (the fitness of cells in which some biological factor other than DHFR function is limiting for fitness). The data are clearly bounded; the original Papkou et al. paper states that 93% of genotypes are at the low-fitness limit at which deleterious effects no longer influence fitness. Because of this bounding, mutations that are strongly deleterious to DHFR function will therefore have an apparently smaller effect when introduced in combination with other deleterious mutations, leading to apparent epistatic interactions; moreover, these apparent interactions will have different magnitudes if they are introduced into backgrounds that themselves differ in DHFR function/fitness, leading to apparent "fluidity" of these interactions. This is a well-established issue in the literature (see PMIDs 30037990, 28100592, 39261454). It is therefore important to adjust for these global nonlinearities before assessing interactions, but the authors have not done this.
This global nonlinearity could explain much of the fluidity claimed in this paper. It could explain the observation that epistasis does not seem to depend as much on genetic background for low-fitness backgrounds, and the latter is constant (Figure 2B and 2C): these patterns would arise simply because the effects of deleterious mutations are all epistatically masked in backgrounds that are already near the fitness minimum. It would also explain the observations in Figure 7. For background genotypes with relatively high fitness, there are two distinct peaks of fitness effects, which likely correspond to neutral mutations and deleterious mutations that bring fitness to the lower bound of measurement; as the fitness of the background declines, the deleterious mutations have a smaller effect, so the two peaks draw closer to each other, and in the lowest-fitness backgrounds, they collapse into a single unimodal distribution in which all mutations are approximately neutral (with the distribution reflecting only noise). Global nonlinearity could also explain the apparent "binary" nature of epistasis. Sites 4 and 5 change the second amino acid, and the Papkou paper shows that only 3 amino acid states (C, D, and E) are compatible with function; all others abolish function and yield lower-bound fitness, while mutations at other sites have much weaker effects. The apparent binary nature of epistasis in Figure 5 corresponds to these effects given the nonlinearity of the fitness assay. Most mutations are close to neutral irrespective of the fitness of the background into which they are introduced: these are the "non-epistatic" mutations in the binary scheme. For the mutations at sites 4 and 5 that abolish one of the beneficial mutations, however, these have a strong background-dependence: they are very deleterious when introduced into a high-fitness background but their impact shrinks as they are introduced into backgrounds with progressively lower fitness. The apparent "binary" nature of global epistasis is likely to be a simple artifact of bounding and the bimodal distribution of functional effects: neutral mutations are insensitive to background, while the magnitude of the fitness effect of deleterious mutations declines with background fitness because they are masked by the lower bound. The authors' statement is that "global epistasis often does not hold." This is not established. A more plausible conclusion is that global epistasis imposed by the phenotype limits affects all mutations, but it does so in a nonlinear fashion.
In conclusion, most of the major claims in the paper could be artifactual. Much of the claimed pairwise epistasis could be caused by measurement noise, the use of arbitrary cutoffs, and the lack of adjustment for global nonlinearity. Much of the fluidity or higher-order epistasis could be attributable to the same issues. And the apparently binary nature of global epistasis is also the expected result of this nonlinearity.
We thank the reviewer for raising this important concern. We fully agree that the use of arbitrary thresholds in the earlier version of the manuscript, together with the lack of an explicit treatment of measurement error, could compromise the rigor of our conclusions. To address this, we have undertaken a thorough re-analysis of the folA landscape.
(1) Incorporating measurement error and avoiding noise-driven artifacts
In the original version, we followed Papkou et al. in using a single experimental replicate and applying fixed thresholds to classify epistasis. As the reviewer correctly notes, this approach allows noise to propagate from single-mutant measurements to double-mutant effects, and ultimately to higher-order epistasis.
In the revised analysis, we now:
Use the mean fitness across all six independent replicates for each genotype.
Incorporate the corresponding standard deviation as a measure of experimental error.
Classify epistatic interactions only when differences between a genotype and its neighbors exceed combined error margins, rather than using a fixed cutoff.
This ensures that observed changes in epistasis are statistically distinguishable from noise. Details are provided in the revised Methods section and updated code.
(2) Replacing arbitrary thresholds with error-based criteria
Previously, we used an arbitrary ±0.05 cutoff to define the presence/absence of epistasis. As the reviewer notes, this could misclassify interactions (e.g. labeling an effect as “fluid” when the difference lies within error). In the revised framework, these thresholds have been eliminated. Instead, interactions are classified based on whether their distributions overlap within replicate variance.
This approach scales naturally with measurement precision, which differs between high-fitness and low-fitness genotypes, and removes the need for a universal cutoff.
(3) Consequences of re-analysis
Implementing this revised framework produced several important updates:
High-fitness backgrounds: The qualitative picture of higher-order (“fluid”) epistasis remains robust. The patterns reported originally are preserved.
Low-fitness backgrounds: Accounting for replicate variance revealed that part of the previously inferred “fluidity” arose from noise. These spurious effects are now removed, giving a more conservative but more accurate view of epistasis in non-functional regions.
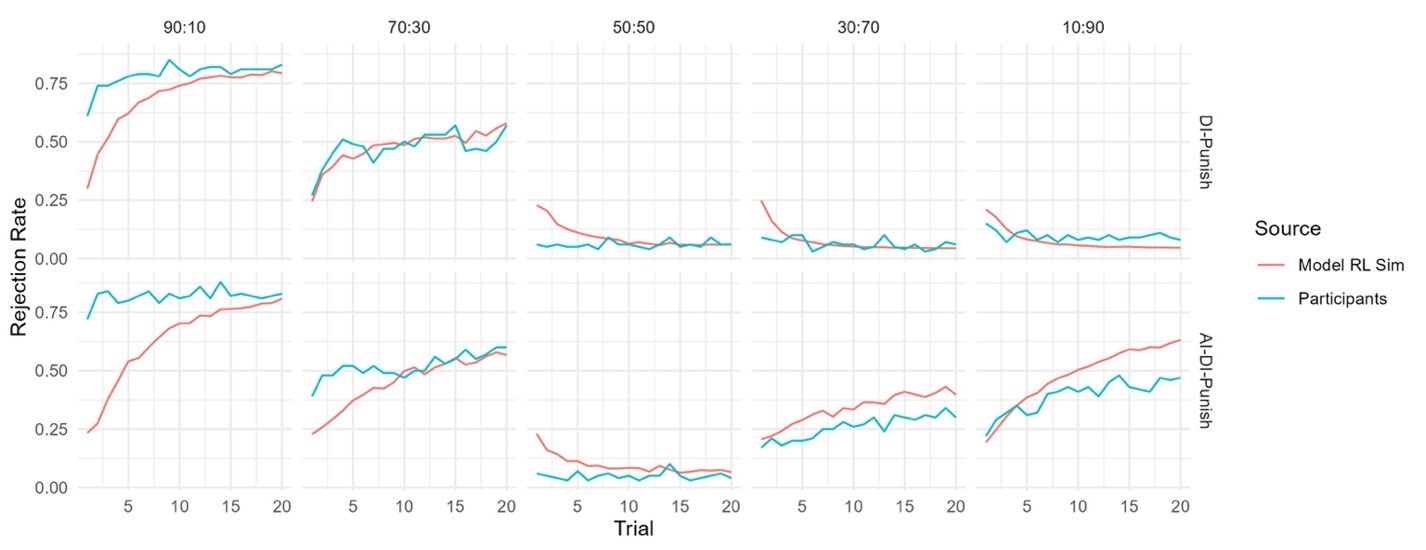
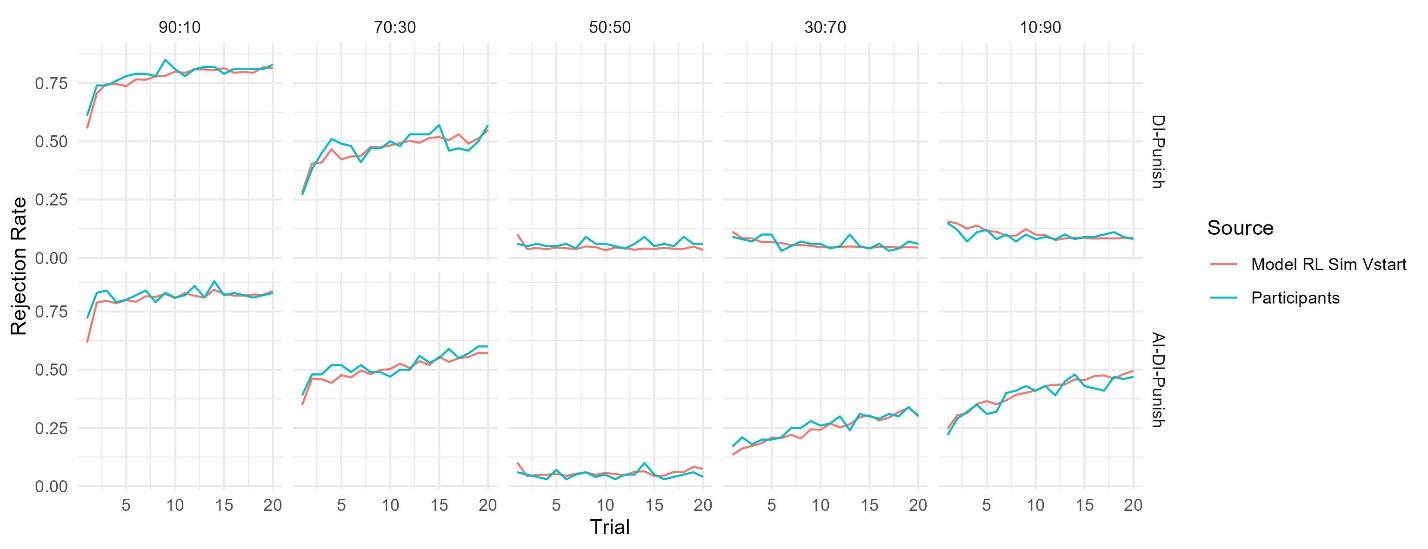
Fitness peaks: Our replicate-aware analysis identifies 127 peaks, compared to 514 in Papkou et al. Importantly, all 127 peaks occur in functional regions of the landscape. This difference highlights the importance of replicate-based error treatment: relying on a single run without demonstrating repeatability can yield artifacts.
(4) Addressing bounding effects and terminology
We also agree with the reviewer that bounding effects, arising from the biological limits of fitness, can create apparent nonlinearities in the genotype–phenotype map. To clarify this, we made the following changes:
Terminology: We now use the term higher-order epistasis instead of fluid epistasis, emphasizing that the observed background-dependence involves more than two mutations and cannot be explained by global nonlinearities alone.
We also clarify the definitions of sign-epistasis used in this work.
By replacing arbitrary cutoffs with replicate-based error estimates and by explicitly considering bounding effects, we have substantially increased the rigor of our analysis. While this reanalysis led to both quantitative and qualitative changes in some regions, the central conclusion remains unchanged: higher-order epistasis is pervasive in the folA landscape, especially in functional backgrounds.
All analysis scripts and codes are provided as Supplementary Material.
Reviewer #3 (Public review):
Summary:
The authors have studied a previously published large dataset on the fitness landscape of a 9 base-pair region of the folA gene. The objective of the paper is to understand various aspects of epistasis in this system, which the authors have achieved through detailed and computationally expensive exploration of the landscape. The authors describe epistasis in this system as "fluid", meaning that it depends sensitively on the genetic background, thereby reducing the predictability of evolution at the genetic level. However, the study also finds two robust patterns. The first is the existence of a "pivot point" for a majority of mutations, which is a fixed growth rate at which the effect of mutations switches from beneficial to deleterious (consistent with a previous study on the topic). The second is the observation that the distribution of fitness effects (DFE) of mutations is predicted quite well by the fitness of the genotype, especially for high-fitness genotypes. While the work does not offer a synthesis of the multitude of reported results, the information provided here raises interesting questions for future studies in this field.
Strengths:
A major strength of the study is its detailed and multifaceted approach, which has helped the authors tease out a number of interesting epistatic properties. The study makes a timely contribution by focusing on topical issues like the prevalence of global epistasis, the existence of pivot points, and the dependence of DFE on the background genotype and its fitness. The methodology is presented in a largely transparent manner, which makes it easy to interpret and evaluate the results.
The authors have classified pairwise epistasis into six types and found that the type of epistasis changes depending on background mutations. Switches happen more frequently for mutations at functionally important sites. Interestingly, the authors find that even synonymous mutations in stop codons can alter the epistatic interaction between mutations in other codons. Consistent with these observations of "fluidity", the study reports limited instances of global epistasis (which predicts a simple linear relationship between the size of a mutational effect and the fitness of the genetic background in which it occurs). Overall, the work presents some evidence for the genetic context-dependent nature of epistasis in this system.
Weaknesses:
Despite the wealth of information provided by the study, there are some shortcomings of the paper which must be mentioned.
(1) In the Significance Statement, the authors say that the "fluid" nature of epistasis is a previously unknown property. This is not accurate. What the authors describe as "fluidity" is essentially the prevalence of certain forms of higher-order epistasis (i.e., epistasis beyond pairwise mutational interactions). The existence of higher-order epistasis is a well-known feature of many landscapes. For example, in an early work, (Szendro et. al., J. Stat. Mech., 2013), the presence of a significant degree of higher-order epistasis was reported for a number of empirical fitness landscapes. Likewise, (Weinreich et. al., Curr. Opin. Genet. Dev., 2013) analysed several fitness landscapes and found that higher-order epistatic terms were on average larger than the pairwise term in nearly all cases. They further showed that ignoring higher-order epistasis leads to a significant overestimate of accessible evolutionary paths. The literature on higher-order epistasis has grown substantially since these early works. Any future versions of the present preprint will benefit from a more thorough contextual discussion of the literature on higher-order epistasis.
(2) In the paper, the term 'sign epistasis' is used in a way that is different from its wellestablished meaning. (Pairwise) sign epistasis, in its standard usage, is said to occur when the effect of a mutation switches from beneficial to deleterious (or vice versa) when a mutation occurs at a different locus. The authors require a stronger condition, namely that the sum of the individual effects of two mutations should have the opposite sign from their joint effect. This is a sufficient condition for sign epistasis, but not a necessary one. The property studied by the authors is important in its own right, but it is not equivalent to sign epistasis.
(3) The authors have looked for global epistasis in all 108 (9x12) mutations, out of which only 16 showed a correlation of R^2 > 0.4. 14 out of these 16 mutations were in the functionally important nucleotide positions. Based on this, the authors conclude that global epistasis is rare in this landscape, and further, that mutations in this landscape can be classified into one of two binary states - those that exhibit global epistasis (a small minority) and those that do not (the majority). I suspect, however, that a biologically significant binary classification based on these data may be premature. Unsurprisingly, mutational effects are stronger at the functional sites as seen in Figure 5 and Figure 2, which means that even if global epistasis is present for all mutations, a statistical signal will be more easily detected for the functionally important sites. Indeed, the authors show that the means of DFEs decrease linearly with background fitness, which hints at the possibility that a weak global epistatic effect may be present (though hard to detect) in the individual mutations. Given the high importance of the phenomenon of global epistasis, it pays to be cautious in interpreting these results.
(4) The study reports that synonymous mutations frequently change the nature of epistasis between mutations in other codons. However, it is unclear whether this should be surprising, because, as the authors have already noted, synonymous mutations can have an impact on cellular functions. The reader may wonder if the synonymous mutations that cause changes in epistatic interactions in a certain background also tend to be non-neutral in that background. Unfortunately, the fitness effect of synonymous mutations has not been reported in the paper.
(5) The authors find that DFEs of high-fitness genotypes tend to depend only on fitness and not on genetic composition. This is an intriguing observation, but unfortunately, the authors do not provide any possible explanation or connect it to theoretical literature. I am reminded of work by (Agarwala and Fisher, Theor. Popul. Biol., 2019) as well as (Reddy and Desai, eLife, 2023) where conditions under which the DFE depends only on the fitness have been derived. Any discussion of possible connections to these works could be a useful addition.
We thank the reviewer for the summary of our work and for highlighting both its strengths and areas for improvement. We have carefully considered the points raised and revised the manuscript accordingly. The revised version:
(1) Clarifies the conceptual framework. We emphasize the distinction between background-dependent, higher-order epistasis and global nonlinearities. To avoid ambiguity, we have replaced the term “fluid” epistasis with higher-order epistasis throughout, in line with prior literature (e.g. Szendro et al., 2013; Weinreich et al., 2013). We now explicitly situate our results in the context of these studies and clarify our definitions of epistasis, correcting the earlier error where “strong sign epistasis” was used in place of “sign epistasis.”
(2) Improves statistical rigor. We now incorporate replicate variance and statistical error criteria in place of arbitrary thresholds. This ensures that classification of epistasis reflects experimental precision rather than fixed, arbitrary cutoffs.
(3) Expands treatment of synonymous mutations. We now explicitly analyze synonymous mutations, separating those that are neutral from those that are non-neutral. Our results show that non-neutral synonymous mutations are disproportionately responsible for altering epistatic interactions, while neutral synonymous mutations rarely do so. We also report the fitness effects of synonymous mutations directly and include new analyses showing that there is no correlation between the mean fitness effect of a synonymous mutation and the frequency with which it alters epistasis (Supplementary Fig. S11).
These revisions strengthen both the rigor and the clarity of the manuscript. We hope they address the reviewer’s concerns and make the significance of our findings, particularly the siteresolved quantification of higher-order epistasis in the folA landscape, including in synonymous mutations, more apparent.
Reviewing Editor Comments:
Key revision suggestions:
(1) Please quantify the impact of measurement noise on your conclusions, and perform statistical analysis to determine whether the observed differences of epistasis due to different backgrounds are statistically significant.
(2) Please investigate how your conclusions depend on the cutoffs, and consider choosing them based on statistical criteria.
(3) Please reconsider the possible role of global epistasis. In particular, the effect of bounds on fitness values. All reviewers are concerned that all claims, including about global epistasis, may be consistent with a simple null model where most low fitness genotypes are non-functional and variation in their fitness is simply driven by measurement noise. Please provide a convincing argument rejecting this model.
More generally, we recommend that you consider all suggestions by reviewers, including those about results, but also those about terminology and citing relevant works.
Thank you for your guidance. We have substantially revised the manuscript to incorporate the reviewers’ suggestions. In addition to addressing the three central issues raised, we have refined terminology, expanded the discussion of prior work, and clarified the presentation of our main results. We believe these changes significantly strengthen both the rigor and the impact of the study. We are grateful to the Reviewing Editor and reviewers for their constructive feedback.
In the revised manuscript, we address the three major points as follows:
(1) Quantifying measurement noise and statistical significance. We now use the average of six independent experimental runs for each genotype, together with the corresponding standard deviations, to explicitly quantify measurement uncertainty. Pairwise and higher-order epistasis are assessed relative to these error estimates, rather than against fixed thresholds. This ensures that differences across genetic backgrounds are statistically distinguishable from noise.
(2) Replacing arbitrary cutoffs with statistical criteria. We have eliminated the use of arbitrary thresholds. Instead, classification of interactions (positive, negative, or neutral epistasis) is based on whether fitness differences exceed replicate variance. This approach scales naturally with measurement precision. While some results change quantitatively for high-fitness backgrounds and qualitatively for low-fitness backgrounds, our central conclusions remain robust.
(3) Analysis of synonymous mutations. We now separately analyze synonymous mutations to test their role in altering epistasis. Our results show that there is no correlation between the average fitness effect of a synonymous mutation and the frequency with which it changes epistatic interactions.
We have revised terminology for clarity (replacing “fluid” with higher-order epistasis) and updated the Discussion to place our work in the broader context of the literature on higher-order epistasis.
Finally, we have rewritten the entire manuscript to improve clarity, refine the narrative flow, and ensure that the presentation more crisply reflects the subject of the study
Reviewer #1 (Recommendations for the authors):
MINOR COMMENTS
(1) Lines 102-107. Papkou's definition of non-functional genotypes makes sense since it is based on the fact that some genotypes are statistically indistinguishable in terms of fitness from mutants with premature stop codons in folA. It doesn't really matter whether to call them low fitness or non-functional, but it would be helpful to explain the basis for this distinction.
Thank you for raising this point. To maintain consistency with the original dataset and analysis, we retain Papkou et al.’s nomenclature and refer to these genotypes as “functional” or “non-functional.”
(2) Lines 111-112. I think the authors need to briefly explain here how they define the absence of epistasis. They do so in the Methods, but this information is essential and needs to be conveyed to the reader in the Results as well.
Thank you for the suggestion. We agree that this definition is essential for readers to follow the Results. In the revised manuscript, we have added a brief explanation at the start of the Results section clarifying how we define the absence of epistasis. Specifically, we now state that two mutations are considered non-epistatic when the observed fitness of the double mutant is statistically indistinguishable (within error of six replicates) from the additive expectation based on the single mutants. This ensures that the Results section is selfcontained, while full details remain in the Methods.
(3) Lines 142 and elsewhere. The authors introduce the qualifier "fluid" to describe the fact that the value or sign of pairwise epistasis changes across genetic backgrounds. I don't see a need for this new terminology, since it is already captured adequately by the term "higher-order epistasis". The epistasis field is already rife with jargon, and I would prefer if new terms were introduced only when absolutely necessary.
Thank you for this helpful suggestion. We agree that introducing new terminology is unnecessary here. In the revised manuscript, we have replaced the term “fluid” epistasis with “higher-order epistasis” throughout, to align with established usage and avoid adding jargon.
(4) Figure 6. I don't think this is the best way of showing that the pivot points are clustered. A histogram would be more appropriate and would take less space. However it would allow the authors to display a null distribution to demonstrate that this clustering is indeed surprising.
(5) Lines 320-321. Mann-Whitney U tests whether one distribution is systematically shifted up or down relative to the other. Please change the language here. It looks like the authors also performed the Kolmogorov-Smirnoff test, which is appropriate, but it doesn't look like the results are reported anywhere. Please report.
(6) Lines 330-334. The fact that HF genotypes seem to have more similar DFEs than LF genotypes is somewhat counterintuitive. Could this be an artifact of the fact that any two random HF genotypes are more similar to each other than any two randomly sampled LF genotypes?
(7) Lines 427. The sentence "The set of these selected variants are assigned their one hamming distance neighbours to construct a new 𝑛-base sequence space" is confusing. I think it is pretty clear how to construct a n-base sequence space, and this sentence adds more confusion than it removes.
Thank you for raising this point. To maintain consistency with the original dataset and analysis, we retain Papkou et al.’s nomenclature and refer to these genotypes as “functional” or “non-functional.”
We now start the results section of the manuscript with a brief description of how each type of epistasis is defined. Specifically, we now state that two mutations are considered non-epistatic when the observed fitness of the double mutant is statistically indistinguishable (within the error of six replicates) from the additive expectation based on the single mutants. This ensures that the Results section is self-contained, while full details remain in the Methods.
We also agree that introducing new terminology is unnecessary. In the revised manuscript, we have replaced the term “fluid” epistasis with “higher-order epistasis” throughout, to align with established usage and avoid adding jargon. Finally, we concur that the identified sentence was unnecessary and potentially confusing; it has been removed from the revised manuscript to improve clarity. In fact, we have rewritten the entire manuscript for better flow and readability.
Reviewer #2 (Recommendations for the authors):
(1) Supplementary Figure S2A and S3 seem to be the same.
(3) The classification scheme for reciprocal sign/single sign/other sign epistasis differs from convention and should be made more explicit or renamed.
(4) Re the claim that high and low fitness backgrounds have different frequencies of the various types of epistasis:
Are the frequency distributions of the different types of epistasis statistically different between high and low fitness backgrounds statistically significant? It seems that they follow similar general patterns, and the sample size is much smaller for high fitness backgrounds so more variance in their distributions is expected.
Do bounding of fitness measurements play a role in generating the differences in types of epistasis seen in high vs. low-fitness backgrounds? If many variants are at the lower bound of the fitness assay, then positive epistasis might simply be less detectable for these backgrounds (which seems to be the biggest difference between high/low fitness backgrounds).
(5) In Figure 4B, points are not independent, because the mutation effects are calculated for all mutations in all backgrounds, rather than with reference to a single background or fluorescence value. The same mutations are therefore counted many times.
(6) It is not clear how the "pivot growth rate" was calculated or what the importance of this metric is.
(7) In the introduction, the justification for reanalyzing the Papkou et al dataset in particular is not clear.
(8) Epistasis at the nucleotide level is expected because of the genetic code: fitness and function are primarily affected by amino acid changes, and nucleotide mutations will affect amino acids depending on the state at other nucleotide sites in the same codon. For the most part, this is not explicitly taken account of in the paper. I recommend separating apparent epistasis due to the genetic code from that attributable to dependence among codons.
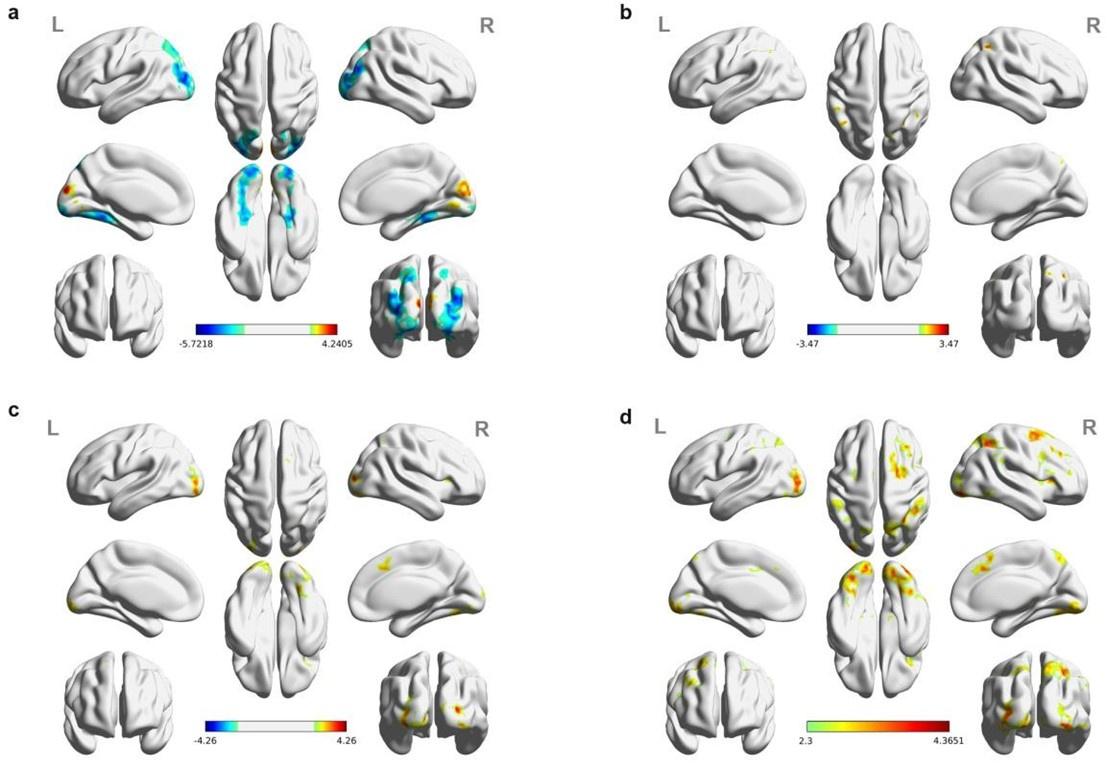
Thank you for noting this. Figure S2A shows results for high-fitness peaks only, whereas Figure S3 shows results for all peaks across the landscape. We have now made this distinction explicit in the figure legends and main text of the revised manuscript.
In the revised analysis, peaks are defined using the average fitness across six experimental replicates along with the corresponding standard deviation. Each genotype is compared with all single-step neighbors, and it is classified as a peak only if its mean fitness is significantly higher than all neighbors (p < 0.05). This procedure explicitly accounts for measurement error and replaces the arbitrary thresholding used previously. Full details are now described in the Methods.
To avoid confusion, we now state our definitions explicitly at the start of the analysis. We have now corrected our definition in the text. We define sign epistasis as a one where at least one mutation switches from being beneficial to deleterious.
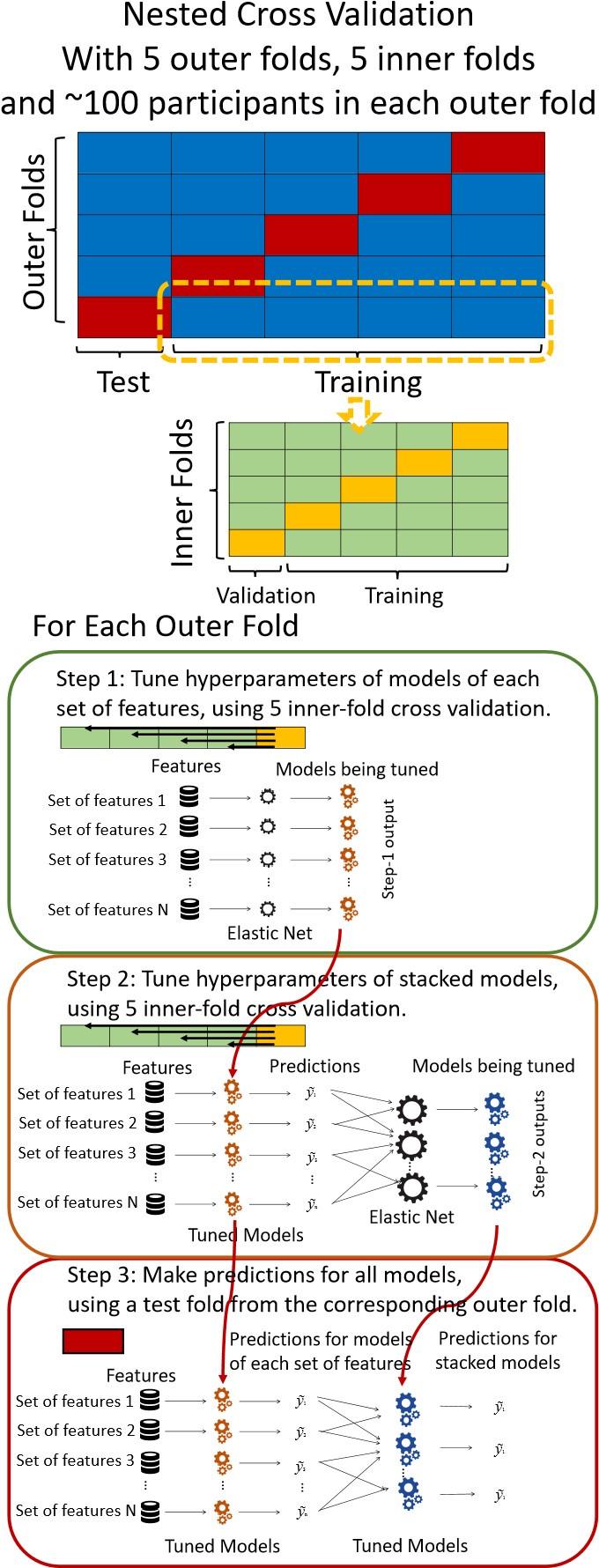
We have clarified our motivation in the Introduction. The Papkou et al. dataset is the most comprehensive experimental map of a complete 9-bp region of folA and provides six independent replicates, making it uniquely suited for testing hypotheses about backgrounddependent epistasis. Importantly, Papkou et al. based their conclusions on a single run, whereas our reanalysis incorporates replicate means and variances, leading to substantive differences—for example, a reduction in reported peaks from 514 to 127. By recalibrating the analysis, we provide a more rigorous account of this landscape and highlight how methodological choices affect conclusions.
We also agree that some nucleotide-level epistasis reflects the structure of the genetic code (i.e., codon degeneracy and context-dependence of amino acid substitutions). In the revised manuscript, we explicitly separate epistasis attributable to codon structure from epistasis arising among codons. For example, synonymous mutations that alter epistasis within codons are treated separately from those affecting interactions across codons, and this distinction is now clearly indicated in the Results.
Reviewer #3 (Recommendations for the authors):
(1) The analysis of peak density and accessibility in the paragraph starting on line 96 seems a bit out of context. Its connection with the various forms of epistasis treated in the rest of the paper is unclear.
(2) As mentioned in the Public Review, the term 'sign epistasis' has been used in a non-standard way. My suggestion would be to use a different term. Even a slightly modified term, such as "strong sign epistasis", should help to avoid any confusion.
(3) mentioned in the public review that it is not clear whether the synonymous mutations that change the type of epistasis also tend to be non-neutral. This issue could be addressed by computing, for example, the fitness effects of all synonymous mutations for backgrounds and mutation pairs where a switch in epistasis occurs, and comparing it with fitness effects where no such switch occurs.
(4) Do the authors have any proposal for why synonymous mutations seem to cause more frequent changes in epistasis in low-fitness backgrounds? Related to this, is there any systematic difference between the types of switch caused by synonymous mutations in the low- versus high-fitness backgrounds?
(5) It is unclear exactly how the pivot points were determined, especially since the data for many mutations is noisy. The protocol should be provided in the Methods section.
(6) Line 303: possible typo, "accurate" --> "inaccurate".
(7) The value of Delta used for the "phenotypic DFE" has not been mentioned in the main text (including Methods).
We agree that the connection needed to be clearer. In the revised manuscript, we (i) relocate and retitle this material as a brief “Landscape overview” preceding the epistasis analyses, (ii) explicitly link multi-peakedness and path accessibility to epistasis (e.g., multi-peak structure implies the presence of sign/reciprocal-sign epistasis; accessibility is shaped by background-dependent effects), and (iii) move derivations to the Supplement. We also recomputed peak density and accessibility using replicate-averaged fitness with replicate SDs, so the overview and downstream epistasis sections now use a single, error-aware landscape (updated in Figs. 1–3, with cross-references in the text).
We have aligned our terminology and now state definitions upfront.
After replacing fixed cutoffs with replicate-based error criteria, switches are more frequent in high-fitness backgrounds (Fig. 3). Mechanistically, near the lower fitness bound, deleterious effects are masked (global nonlinearity), reducing apparent switching. Functional/high-fitness backgrounds allow both beneficial and deleterious outcomes, so background-dependent (higher-order) interactions manifest more readily. Switch types also vary by background fitness: high-fitness backgrounds show more sign/strong-sign switches, whereas low-fitness backgrounds show mostly magnitude reclassifications (Fig. 3C; Supplement Fig. Sx).
Finally, we corrected a typo by replacing “accurate” with “inaccurate” and now define Δ (equal to 0.05) in the main text (in Results and Figure 8 caption).