7,847 Matching Annotations
- Sep 2025
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
bmjopen.bmj.com bmjopen.bmj.com
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
pmc.ncbi.nlm.nih.gov pmc.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
pmc.ncbi.nlm.nih.gov pmc.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
bmjopen.bmj.com bmjopen.bmj.com
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
www.medrxiv.org www.medrxiv.org
-
LivingReviews:Protocol
-
-
wellcomeopenresearch.org wellcomeopenresearch.org
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
-
Tags
Annotators
URL
-
-
papers.ssrn.com papers.ssrn.com
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
pmc.ncbi.nlm.nih.gov pmc.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
-
Tags
Annotators
URL
-
-
bmjopen.bmj.com bmjopen.bmj.com
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
bmjopen.bmj.com bmjopen.bmj.com
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
systematicreviewsjournal.biomedcentral.com systematicreviewsjournal.biomedcentral.com
-
LivingReviews:Protocol
-
-
www.medrxiv.org www.medrxiv.org
-
LivingReviews:Protocol
-
-
www.sciencedirect.com www.sciencedirect.com
-
LivingReviews:Protocol
Dos? Revisar
-
-
www.sciencedirect.com www.sciencedirect.com
-
www.sciencedirect.com www.sciencedirect.com
-
LivingReviews:Protocol
-
-
www.sciencedirect.com www.sciencedirect.com
-
LivingReviews:Protocol
-
-
www.sciencedirect.com www.sciencedirect.com
-
LivingReviews:Protocol
-
-
www.sciencedirect.com www.sciencedirect.com
-
LivingReviews:Protocol
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
Tags
Annotators
URL
-
-
pmc.ncbi.nlm.nih.gov pmc.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
Child[MeSH]
Tags
Annotators
URL
-
-
pmc.ncbi.nlm.nih.gov pmc.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
Child[MeSH]
Pedriatrics
Tags
Annotators
URL
-
-
pmc.ncbi.nlm.nih.gov pmc.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
Pediatrics
Child[MeSH]
Tags
Annotators
URL
-
-
pmc.ncbi.nlm.nih.gov pmc.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
Pediatrics
Tags
Annotators
URL
-
-
onlinelibrary.wiley.com onlinelibrary.wiley.com
-
Pediatrics
-
-
pmc.ncbi.nlm.nih.gov pmc.ncbi.nlm.nih.gov
-
Pediatrics
-
-
pmc.ncbi.nlm.nih.gov pmc.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
-
-
pmc.ncbi.nlm.nih.gov pmc.ncbi.nlm.nih.gov
-
LivingReviews:Protocol
-
-
pmc.ncbi.nlm.nih.gov pmc.ncbi.nlm.nih.gov
-
Pediatrics
Child[MeSH]
-
-
pmc.ncbi.nlm.nih.gov pmc.ncbi.nlm.nih.gov
-
Updates https://www.bmj.com/content/377/bmj.o1205 2022
Pedriatrics
Tags
Annotators
URL
-
-
pmc.ncbi.nlm.nih.gov pmc.ncbi.nlm.nih.gov
-
Infant[MeSH]
Pedriatrics
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
As part of the living systematic
-
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
living systematic review
-
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
Four RCTs and 19 NRSs
Sample
-
More detail in https://github.com/lmichan/LivingReviews
https://www.zotero.org/groups/6122210/livingreviews/items/V2GPNQGR/item-list
LivingReviewsNeurosciences🧠
Tags
Annotators
URL
-
-
www.sciencedirect.com www.sciencedirect.com
-
www.sciencedirect.com www.sciencedirect.com
-
www.sciencedirect.com www.sciencedirect.com
-
www.sciencedirect.com www.sciencedirect.com
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
3 new RCTs
Sample 23
-
-
www.sciencedirect.com www.sciencedirect.com
-
www.sciencedirect.com www.sciencedirect.com
-
pmc.ncbi.nlm.nih.gov pmc.ncbi.nlm.nih.gov
-
www.sciencedirect.com www.sciencedirect.com
-
www.sciencedirect.com www.sciencedirect.com
-
www.sciencedirect.com www.sciencedirect.com
-
www.sciencedirect.com www.sciencedirect.com
-
70
Sample
-
More detail in https://github.com/lmichan/LivingReviews
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
More detail in https://github.com/lmichan/LivingReviews
Updates 4
2007 2010 2013 2017
-
12
Sample
-
living systematic review
https://www.zotero.org/groups/6122210/livingreviews/items/GV8IES9W/item-list
Tags
Annotators
URL
-
-
pmc.ncbi.nlm.nih.gov pmc.ncbi.nlm.nih.gov
-
www.sciencedirect.com www.sciencedirect.com
-
Update
-
-
bmcmedresmethodol.biomedcentral.com bmcmedresmethodol.biomedcentral.com
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
More detail in https://github.com/lmichan/LivingReviews
-
-
pmc.ncbi.nlm.nih.gov pmc.ncbi.nlm.nih.gov
-
review as a living systematic review
Tags
Annotators
URL
-
-
www.cochranelibrary.com www.cochranelibrary.com
-
This was previously a living systematic review
Revisar
Tags
Annotators
URL
-
-
open-research-europe.ec.europa.eu open-research-europe.ec.europa.eu
-
www.cochranelibrary.com www.cochranelibrary.com
-
wellcomeopenresearch.org wellcomeopenresearch.org
Tags
Annotators
URL
-
-
www.ahajournals.org www.ahajournals.org
-
www.cambridge.org www.cambridge.org
Tags
Annotators
URL
-
-
preprints.jmir.org preprints.jmir.org
Tags
Annotators
URL
-
-
www.taylorfrancis.com www.taylorfrancis.com
-
open-research-europe.ec.europa.eu open-research-europe.ec.europa.eu
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
jamanetwork.com jamanetwork.com
-
www.jclinepi.com www.jclinepi.com
-
Revisar
-
-
effectivehealthcare.ahrq.gov effectivehealthcare.ahrq.gov
-
pmc.ncbi.nlm.nih.gov pmc.ncbi.nlm.nih.gov
-
Updates https://doi.org/10.1136/bmj.p688 2023
Revisar
-
-
www.ahajournals.org www.ahajournals.org
-
pubs.rsna.org pubs.rsna.org
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
academic.oup.com academic.oup.com
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
Updates https://doi.org/10.1186/s13643-021-01693-7 2021
Revisar
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
Update 2021
Revisar
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
www.sciencedirect.com www.sciencedirect.com
-
www.bmj.com www.bmj.com
-
www.bmj.com www.bmj.com
Tags
Annotators
URL
-
www.bmj.com www.bmj.com
-
Updates
Tags
Annotators
URL
-
-
www.bmj.com www.bmj.com
-
www.bmj.com www.bmj.com
Tags
Annotators
URL
-
pmc.ncbi.nlm.nih.gov pmc.ncbi.nlm.nih.gov
-
Updates https://doi.org/10.1136/bmj.p2929 2024
Tags
Annotators
URL
-
-
f1000research.com f1000research.com
-
www.sciencedirect.com www.sciencedirect.com
- Aug 2025
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
iving systematic review
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
-
living systematic review
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
living systematic review
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
living systematic review
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
living systematic review
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
En Acceso Cerrado?
Tags
Annotators
URL
-
-
www.mdpi.com www.mdpi.com
-
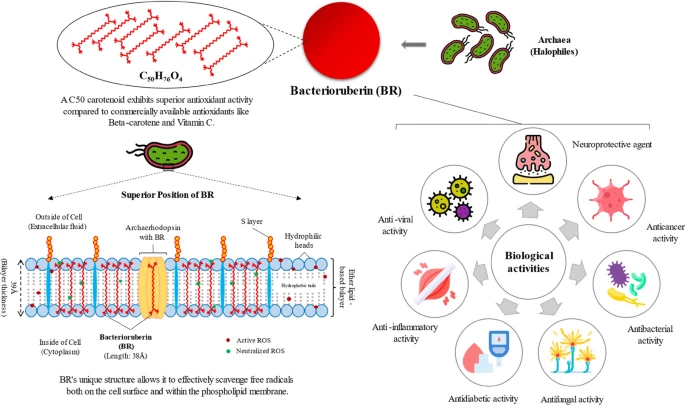
Besides, potential biotechnological uses of haloarchaeal pigments are poorly explored. This work summarises what it has been described so far about carotenoids from haloarchaea and their production at mid- and large-scale, paying special attention to the most recent findings on the potential uses of haloarchaeal pigments in biomedicine.
Además, los posibles usos biotecnológicos de los pigmentos haloarqueales están poco explorados. Este trabajo resume lo descrito hasta la fecha sobre los carotenoides de las haloarqueas y su producción a mediana y gran escala, con especial atención a los hallazgos más recientes sobre los posibles usos de los pigmentos haloarqueales en biomedicina.
-
-
pmc.ncbi.nlm.nih.gov pmc.ncbi.nlm.nih.gov
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-
-
link.springer.com link.springer.com
-
www.nature.com www.nature.com
Tags
Annotators
URL
-
-
www.microbiologyresearch.org www.microbiologyresearch.org
-
pubmed.ncbi.nlm.nih.gov pubmed.ncbi.nlm.nih.gov
Tags
Annotators
URL
-