Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public Review):
Faiz et al. investigate small molecule-driven direct lineage reprogramming of mouse postnatal mouse astrocytes to oligodendrocyte lineage cells (OLCs). They use a combination of in vitro, in vivo, and computational approaches to confirm lineage conversion and to examine the key underlying transcription factors and signaling pathways. Lentiviral delivery of transcription factors previously reported to be essential in OLC fate determination-Sox10, Olig2, and Nkx2.2-to astrocytes allows for lineage tracing. They found that these transcription factors are sufficient in reprogramming astrocytes to iOLCs, but that the OLCs range in maturity level depending on which factor they are transfected with. They followed up with scRNA-seq analysis of transfected and control cultures 14DPT, confirming that TF-induced astrocytes take on canonical OLC gene signatures. By performing astrocyte lineage fate mapping, they further confirmed that TF-induced astrocytes give rise to iOLCs. Finally, they examined the distinct genetic drivers of this fate conversion using scRNA-seq and deep learning models of Sox10- astrocytes at multiple time points throughout the reprogramming. These findings are certainly relevant to diseases characterized by the perturbation of OLC maturation and/or myelination, such as Multiple Sclerosis and Alzheimer's Disease. Their application of such a wide array of experimental approaches gives more weight to their findings and allows for the identification of additional genetic drivers of astrocyte to iOLC conversion that could be explored in future studies. Overall, I find this manuscript thoughtfully constructed and only have a few questions to be addressed.
(1) The authors suggest that Sox10- and Olig2- transduced astrocytes result in distinct subpopulations iOLCs. Considering it was discussed in the introduction that these TFs cyclically regulate one another throughout differentiation, could they speculate as to why such varying iOLCs resulted from the induction of these two TFs?
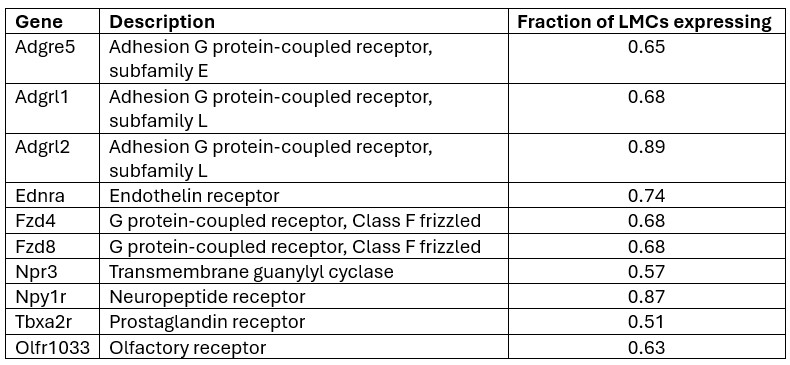
We thank the Reviewer for the opportunity to speculate. We hypothesize that Sox10 and Olig2 may induce different OLCs as a result of differential activation of downstream genes within the gene regulatory network, which are important for OPC, committed OLC and mature OL identity [1]. In support of this, we found different expression levels of genes involved in downstream OLC specification networks [1], including Sox6, Tcfl2 and Myrf, at D14 (Author response image 1), following further analysis of our RNA-seq data.
Author response image 1.
Expression of OLC regulatory network genes in Sox10- and Olig2- cultures. Violin plots show gene expression levels (log-normalized) of downstream OLC regulatory genes (Sox6, Zeb2, Tcf7l2, Myrf, Zfp488, Nfatc2, Hes5, Id2) between Sox10 and Olig2 treated OLCs at 14 days post transduction. Analysis was performed on oligodendrocyte progenitor and mature oligodendrocyte clusters (from Manuscript Figure 1D, clusters 3 and 8).
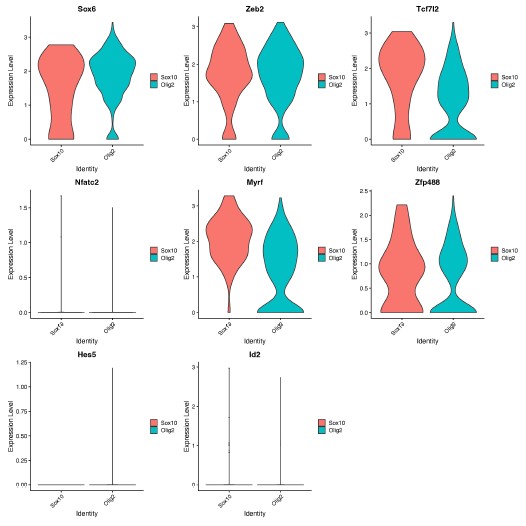
(2) In Figure 1B it appears that the Sox10- MBP+ tdTomato+ cells decreases from D12 to D14. Does this make sense considering MBP is a marker of more mature OLCs?
Thank you for this comment. To address this, we compared the number of MBP+tdTomato+ Sox10 cells across reprogramming timepoints. We saw no difference between the number of MBP+tdTomato+ OLs at D12 and D14 (Author response image 2, p = 0.2314). However, we do see a [nonsignificant] decrease in MBP+tdTomato+ Sox10 cells from D12 to D22 (Manuscript Supplementary Figure 3B, Author response image 2, p= 0.0543), which suggests that culture conditions are not optimal for longer-term cell survival [2], [3], [4].
Author response image 2.
Comparison of Sox10- induced MBP+tdTomato+ iOLCs over time. Quantification of MBP<sup>+</sup>tdTomato<sup>+</sup> iOLs in Sox10 cultures at D8 (n=5), D10 (n=5), D12 (n=5), D14 (n=7) and D22 (n=3) post transduction. Data are presented as mean ± SEM, each data point represents one individual cell culture experiment, Brown-Forsythe and Welch ANOVA on transformed percentages with Dunnett’s T3 multiple comparisons test (*= p<0.05).
(3) Previous studies have shown that MBP expression and myelination in vitro occurs at the earliest around 4-6 weeks of culturing. When assessing whether further maturation would increase MBP positivity, authors only cultured cells up to 22 DPT and saw no significant increase. Has a lengthier culture timeline been attempted?
We agree with the Reviewer that previous studies of pluripotent stem cell derived (hESCs or iPSCs) have shown MBP+ OLCs in vitro around 4-6 weeks [5], [6], [7]. However, studies of neural stem cells [8] or fibroblasts [9] conversion show OLC appearance after 7 and 24 days, respectively, demonstrating that OLCs can be generated in vitro within 1-3 weeks of plating. Moreover, as noted above in response to #2, we see fewer MBP+ cells at 22DPT, suggesting that extended time in culture may require additional factors for support. Therefore, we did not attempt longer timepoints.
(4) Figure S4D is described as "examples of tdTomatonegzsGreen+OLCmarker+ cells that arose from a tdTomatoneg cell with an astrocyte morphology." The zsGreen+ tdTomato- cell is not convincingly of "astrocyte morphology"; it could be a bipolar OLC. To strengthen the conclusions and remove this subjectivity, more extensive characterizations of astrocyte versus OLC morphology in the introduction or results are warranted. This would make this observation more convincing since there is clearly an overlap in the characteristics of these cell types.
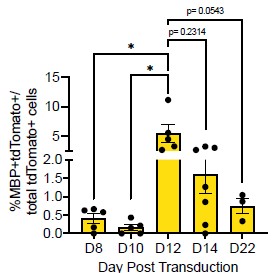
We thank the reviewer for this excellent suggestion. To assess astrocyte morphology, we measured the cell size, nucleus size, number of branches and branch thickness of 70 Aldh1l1+tdTomato+ astrocytes in tamoxifen-labelled Aldh1l1-CreERT2;Ai14 cultures (new Supplemental Table 1). To assess OPC morphology, we performed IHC for PDGFRa in iOLC cultures and measured the same parameters in 70 PDGFRa+ OPCs (new Supplemental Table 1). We found that astrocytes were characterized by larger branch thickness, cell length and nucleus size, while OPCs showed a larger number of branches (new Supplemental Figure 1, and Author response image 3 below). Based on this framework, the AAV9-GFAP::zsGreen<sup>pos</sup>Aldh1l1-tdTomato<sup>neg</sup> and AAV9-GFAP::zsGreen<sup>pos</sup>Aldh1l1-tdTomato<sup>pos</sup>starting cells tracked fall within the bounds of ‘astrocytes’. We have revised the manuscript to include this more rigorous characterization (Line 119-124, Page 4; Line 307-312, Page 9; Line 323-326, Page 9). We also demonstrate (below) that the GFAP::zsGreen<sup>pos</sup> Aldh1l1-tdTomato<sup>pos</sup> and GFAP::zsGreen<sup>pos</sup>Aldh1l1-tdTomato<sup>neg</sup> starting cell depicted in Figure 2G and Supplemental Figure 5D is consistent with astrocyte morphology (Author response image 3).
Author response image 3.
Morphological characterization of astrocytes, oligodendrocyte lineage cells, and starting cells. Quantification of the (A) cell length, (B) nucleus size, (C) number of branches, and (D) branch thickness iAldh1l1+tdTomato+ and PDGFRα+ OPCs (n= 70 per cell type, data are presented as mean ± SEM). Orange line indicates parameter value for GFAP::zsGreen<sup>pos</sup>Aldh1l1-tdTomato<sup>pos</sup> starting cell in Figure 2G. Green line indicates parameter value for GFAP::zsGreen<sup>pos</sup> Aldh1l1-tdTomato<sup>neg</sup> starting cell in Supplemental Figure 5D.
Reviewer #2 (Public Review):
The study by Bajohr investigates the important question of whether astrocytes can generate oligodendrocytes by direct lineage conversion (DLR). The authors ectopically express three transcription factors - Sox10, Olig2 and Nkx6.2 - in cultured postnatal mouse astrocytes and use a combination of Aldh1|1-astrocyte fate mapping and live cell imaging to demonstrate that Sox10 converts astrocytes to MBP+ oligodendrocytes, whereas Olig2 expression converts astrocytes to PDFRalpha+ oligodendrocyte progenitor cells. Nkx6.2 does not induce lineage conversion. The authors use single-cell RNAseq over 14 days post-transduction to uncover molecular signatures of newly generated iOLs.
The potential to convert astrocytes to oligodendrocytes has been previously analyzed and demonstrated. Despite the extensive molecular characterization of the direct astrocyteoligodendrocyte lineage conversion, the paper by Bajohr et al. does not represent significant progress. The entire study is performed in cultured cells, and it is not demonstrated whether this lineage conversion can be induced in astrocytes in vivo, particularly at which developmental stage (postnatal, adult?) and in which brain region. The authors also state that generating oligodendrocytes from astrocytes could be relevant for oligodendrocyte regeneration and myelin repair, but they don't demonstrate that lineage conversion can be induced under pathological conditions, particularly after white matter demyelination. Specific issues are outlined below.
We thank the reviewer for this summary. We agree that there are a handful of reports of astrocytelike cells to OLC conversion [10], [11]. However, our study is the first study to confirm bonafide astrocyte to OLC conversion, which is important given the recent controversy in the field of in vivo astrocyte to neuron reprogramming [12]. In addition, the extensive characterization of the molecular timeline of reprogramming, highlights that although conversion of astrocytes is possible by ectopic expression of any of the three factors, the subtypes of astrocytes converted and maturity of OLCs produced may vary depending on the choice of TF delivered. Our findings will inform future in vivo studies of iOLC generation that aim to understand the impact of brain region, age, pathology, and sex, which are especially important given the diversity of astrocyte responses to disease [13], [14], [15].
(1) The authors perform an extensive characterization of Sox10-mediated DLR by scRNAseq and demonstrate a clear trajectory of lineage conversion from astrocytes to terminally differentiated MBP+ iOLCs. A similar type of analysis should be performed after Olig2 transduction, to determine whether transcriptomics of olig2 conversion overlaps with any phase of sox10 conversion.
We thank the Reviewer for this excellent comment. We chose to include an in-depth analysis of Sox10 in the manuscript, as Sox10-transduced cultures showed a higher percentage of mature iOLCs compared to Olig2 in our studies. We have added this specific rationale to the manuscript (Line 329-330-Page 9).
Nonetheless, we also agree that understanding the underpinnings of Olig2-mediated conversion is important. Therefore, we used Cell Oracle [16] to understand the regulation of cell identity by Olig2. in silico overexpression of Olig2 in our control time course dataset (D0, D3, D8 and D14) showed cell movement from cluster 1, characterized by astrocyte genes [Mmd2[17], Entpd2[18], H2-D1[19]], towards cluster 5, characterized by OPC genes [Pdgfra[20], Myt1[21]] validating astrocyte to OLC conversion by Olig2 (Author response image 4).
We hypothesize that reprogramming via Sox10 and Olig2 take different conversion paths to oligodendrocytes for the following reasons.
(1) Differential astrocyte gene expression at D14 when cells are exposed to Sox10 and Olig2 (Manuscript Figure 1D-E [Sox10 characterized by Lcn2[19], C3[19]; Olig2 characterized by Slc6a11[22], Slc1a2[23]].
(2) Differential expression of key OLC gene regulatory network genes at D14 between cells treated with Sox10 and Olig2 (Author response image 1).
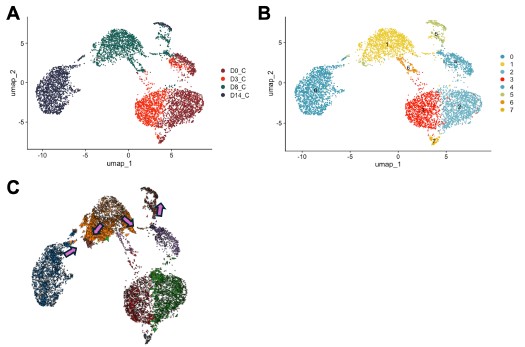
Author response image 4.
in silico modeling of Olig2 reprogramming (A) UMAP clustering of Cre control treated cells from 0, 3, 8, and 14 days post transduction (DPT). (B) UMAP clustering from (A) overlayed with timepoint and treatment group. (C) Cell Oracle modeling of predicted cell trajectories following Olig2 knock in (KI), overlaid onto UMAP plot. Arrows indicate cell movement prediction with Olig2 KI perturbation.
(2) A complete immunohistochemical characterization of the cultures should be performed at different time points after Sox10 and Olig2 transduction to confirm OL lineage cell phenotypes.
We performed a complete immunohistochemical characterization of Ai14 cultures transduced with GFAP::Sox10-Cre and GFAP::Olig2-Cre. This system allows permanent labelling and therefore, enabled the tracking of transduced cells through the process or DLR, which we believe is the most appropriate way to characterize iOLC conversion efficiencies. We then confirmed the conversion of Aldh1l1+ astrocytes in Aldh1l1-CreERT2;Ai14 cultures transduced with GFAP::Sox10-zsGreen and GFAP::Olig2-zsGreen. In this system, GFAP drives the expression of zsGreen, and therefore, may not faithfully track all cells and lead to an underestimate of the numbers of converted cells. For example, iOLCs from Aldh1l1<sup>neg</sup> astrocytes or iOLCs that have lost zsGreen expression following conversion. Therefore we use this system only to confirm astrocyte origin.
Nonetheless, we appreciate this comment and recognize that there may be differences in conversion efficiencies when analyzing Aldh1l1+ astrocytes versus all transduced cells. Therefore, we have softened the language in the manuscript (see below) regarding Olig2 and Sox10 generating different OLC phenotypes and now claim iOLC generation from both Sox10 and Olig2. We thank the Reviewer for this comment, and believe it has strengthened the discussion.
Line 240, Page 7
Line 261-263, Page 8
Line 304-307, Page 8/9
Line 413-414, Page 11
References
(1) E. Sock and M. Wegner, “Using the lineage determinants Olig2 and Sox10 to explore transcriptional regulation of oligodendrocyte development,” Dev Neurobiol, vol. 81, no. 7, pp. 892–901, Oct. 2021, doi: 10.1002/dneu.22849.
(2) B. A. Barres, M. D. Jacobson, R. Schmid, M. Sendtner, and M. C. Raff, “Does oligodendrocyte survival depend on axons?,” Current Biology, vol. 3, no. 8, pp. 489–497, Aug. 1993, doi: 10.1016/0960-9822(93)90039-Q.
(3) A.-N. Cho et al., “Aligned Brain Extracellular Matrix Promotes Differentiation and Myelination of Human-Induced Pluripotent Stem Cell-Derived Oligodendrocytes,” ACS Appl. Mater. Interfaces, vol. 11, no. 17, pp. 15344–15353, May 2019, doi: 10.1021/acsami.9b03242.
(4) E. G. Hughes and M. E. Stockton, “Premyelinating Oligodendrocytes: Mechanisms Underlying Cell Survival and Integration,” Front. Cell Dev. Biol., vol. 9, Jul. 2021, doi: 10.3389/fcell.2021.714169.
(5) M. Ehrlich et al., “Rapid and efficient generation of oligodendrocytes from human induced pluripotent stem cells using transcription factors,” Proc Natl Acad Sci U S A, vol. 114, no. 11, pp. E2243–E2252, Mar. 2017, doi: 10.1073/pnas.1614412114.
(6) Y. Liu, P. Jiang, and W. Deng, “OLIG gene targeting in human pluripotent stem cells for motor neuron and oligodendrocyte differentiation,” Nat Protoc, vol. 6, no. 5, pp. 640–655, May 2011, doi: 10.1038/nprot.2011.310.
(7) S. A. Goldman and N. J. Kuypers, “How to make an oligodendrocyte,” Development, vol. 142, no. 23, pp. 3983–3995, Dec. 2015, doi: 10.1242/dev.126409.
(8) M. Faiz, N. Sachewsky, S. Gascón, K. W. A. Bang, C. M. Morshead, and A. Nagy, “Adult Neural Stem Cells from the Subventricular Zone Give Rise to Reactive Astrocytes in the Cortex after Stroke,” Cell Stem Cell, vol. 17, no. 5, pp. 624–634, Nov. 2015, doi:10.1016/j.stem.2015.08.002.
(9) F. J. Najm et al., “Transcription factor–mediated reprogramming of fibroblasts to expandable, myelinogenic oligodendrocyte progenitor cells,” Nat Biotechnol, vol. 31, no. 5, pp. 426–433, May 2013, doi: 10.1038/nbt.2561.
(10) A. Mokhtarzadeh Khanghahi, L. Satarian, W. Deng, H. Baharvand, and M. Javan, “In vivo conversion of astrocytes into oligodendrocyte lineage cells with transcription factor Sox10; Promise for myelin repair in multiple sclerosis,” PLoS One, vol. 13, no. 9, p. e0203785, Sep. 2018, doi: 10.1371/journal.pone.0203785.
(11) S. Farhangi, S. Dehghan, M. Totonchi, and M. Javan, “In vivo conversion of astrocytes to oligodendrocyte lineage cells in adult mice demyelinated brains by Sox2,” Mult Scler Relat Disord, vol. 28, pp. 263–272, Feb. 2019, doi: 10.1016/j.msard.2018.12.041.
(12) L.-L. Wang, C. Serrano, X. Zhong, S. Ma, Y. Zou, and C.-L. Zhang, “Revisiting astrocyte to neuron conversion with lineage tracing in vivo,” Cell, vol. 184, no. 21, pp. 5465-5481.e16, Oct. 2021, doi: 10.1016/j.cell.2021.09.005.
(13) I Matias, J. Morgado, and F. C. A. Gomes, “Astrocyte Heterogeneity: Impact to Brain Aging and Disease,” Front. Aging Neurosci., vol. 11, Mar. 2019, doi: 10.3389/fnagi.2019.00059.
(14) N. Habib et al., “Disease-associated astrocytes in Alzheimer’s disease and aging,” Nat Neurosci, vol. 23, no. 6, pp. 701–706, Jun. 2020, doi: 10.1038/s41593-020-0624-8.
(15) M. A. Wheeler et al., “MAFG-driven astrocytes promote CNS inflammation,” Nature, vol. 578, no. 7796, pp. 593–599, Feb. 2020, doi: 10.1038/s41586-020-1999-0.
(16) K. Kamimoto, B. Stringa, C. M. Hoffmann, K. Jindal, L. Solnica-Krezel, and S. A. Morris, “Dissecting cell identity via network inference and in silico gene perturbation,” Nature, vol. 614, no. 7949, pp. 742–751, Feb. 2023, doi: 10.1038/s41586-022-05688-9.
(17) P. Kang et al., “Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis,” Neuron, vol. 74, no. 1, pp. 79–94, Apr. 2012, doi:10.1016/j.neuron.2012.01.024.
(18) K. Saito et al., “Microglia sense astrocyte dysfunction and prevent disease progression in an Alexander disease model,” Brain, vol. 147, no. 2, pp. 698–716, Nov. 2023, doi:10.1093/brain/awad358.
(19) S. A. Liddelow et al., “Neurotoxic reactive astrocytes are induced by activated microglia,” Nature, vol. 541, no. 7638, pp. 481–487, Jan. 2017, doi: 10.1038/nature21029.
(20) Q. Zhu et al., “Genetic evidence that Nkx2.2 and Pdgfra are major determinants of the timing of oligodendrocyte differentiation in the developing CNS,” Development, vol. 141, no. 3, pp. 548–555, Feb. 2014, doi: 10.1242/dev.095323.
(21) J. A. Nielsen, J. A. Berndt, L. D. Hudson, and R. C. Armstrong, “Myelin transcription factor 1 (Myt1) modulates the proliferation and differentiation of oligodendrocyte lineage cells,” Mol Cell Neurosci, vol. 25, no. 1, pp. 111–123, Jan. 2004, doi:10.1016/j.mcn.2003.10.001.
(22) J. Liu, X. Feng, Y. Wang, X. Xia, and J. C. Zheng, “Astrocytes: GABAceptive and GABAergic Cells in the Brain,” Front. Cell. Neurosci., vol. 16, Jun. 2022, doi:10.3389/fncel.2022.892497.
(23) A. Sharma et al., “Divergent roles of astrocytic versus neuronal EAAT2 deficiency on cognition and overlap with aging and Alzheimer’s molecular signatures,” Proceedings of the National Academy of Sciences, vol. 116, no. 43, pp. 21800–21811, Oct. 2019, doi:10.1073/pnas.1903566116