Sacrament day
Marks the religious setting: the sermon is delivered during a sacred ritual, intensifying its emotional impact. Parris uses the solemnity of the sacrament to lend authority and urgency to his message.
Sacrament day
Marks the religious setting: the sermon is delivered during a sacred ritual, intensifying its emotional impact. Parris uses the solemnity of the sacrament to lend authority and urgency to his message.
Such incurr the hottest of Gods wrath, as follows. 22. v. Now if we would not be Devils we must give our selves wholly up to Christ: & not suffer the predominancy of one lust, & particularly that lust of covetousness, which is made so light of, & which so sadly prevails in these perilous times
Links sin (especially greed) directly to spiritual damnation. Greed is portrayed as a moral failing that can turn believers into “Devils,” reflecting Puritan values and fear of moral corruption.
We are either Saints, or Devils, the scripture gives us no medium.
Reinforces the sermon’s binary opposition between good and evil. Fear-based rhetoric: the congregation is warned that there is no middle ground between virtue and sin. Encourages self-reflection and vigilance.
One sinner destroys much good; how much more one Devil. Pray we also that not one true Saint may suffer as a Devil, either in name, or body.
Shows fear-based moral reasoning: one evil person (or “Devil”) can ruin the spiritual integrity of the whole church. Reinforces urgency for communal purity.
To be much in prayer that God would deliver our Churches from Devils. That God would not suffer Devils in the guise of Saints to associate with us.
Encourages spiritual vigilance and prayer as protection against evil. Highlights the Puritan belief that unseen spiritual threats can infiltrate the community.
To be deeply humbled for the appearances of Devils among our Churches. If the Church of Corinth were called to mourn because of one incestuous person among them. 1 Cor. 5 initio How much more may N-E Churches mourn that such as work witchcraft, or are vehemently suspected so to do should be found among them.
Parris compares Salem’s situation to biblical examples (Corinth) to emphasize moral accountability. He heightens fear by linking witchcraft suspicions to spiritual corruption.
Examine we our selves well, what we are: what we Church-members are
Call for self-reflection. Connects moral and spiritual vigilance to everyday behavior and participation in the church.
too often there are Devils found among the Saints.
This is the central theme of the sermon. Moral binary of good vs evil within the church. Shows the coexistence of virtuous and corrupt individuals.
For a little pelf, men sell Christ to his Enemies, & their Souls to the Devil.
Moral lesson linking greed/corruption to spiritual damnation. Suggests that small sins can lead to a devil-like state.
a collection of n soft-ware agents. The minimum size of n required toauthentically represent an actual group of real peopleusing software is a very relevant question, but we will notgo into further detail here
Oh, okay!
In summary, based on these observations, we believethat artificial societies with their representations of individ-uals, social networks, and the situated environment are themost promising paradigm to support policymaking when itcomes to modeling human behavior in a social contact.
In summary? You didn't compare these to any alternative!
fled
fuggire
its own accord
di sua spontanea volontà
swift
rapidi
sent for him.
convocare, mandare a chiamare
sprinkled
spruzzare, schizzare
maiden
fanciulla
[https://dictionary.cambridge.org/dictionary/english-italian/maiden]
calabash
zucca a fiasco (che, una volta svuotata e lasciata asciugare, indurisce e può essere utilizzata come contenitore)
quiet
zittire
sorcerer’s
stregone
break into
sforzare l'ingresso, irrompere in casa
staff
bastone
rescue
salvare
vines
vite
ferns
felce
tangled
aggrovigliate
roughly
aspramente
dwarf
gnomo
Ka wai ola a Kāne
La parola kānaka maoli per indicare l'acqua è wai, mentre quella hawaiana per indicare la ricchezza è waiwai, a indicare che l'acqua è fonte di benessere e ricchezza
restore
ripristinare
road-side
ciglio della strada
drop down dead
morire improvvisamente
dig
scavare
yam-hills
La coltivazione dell'igname in Giamaica utilizza il metodo dello “stick” (bastone), con materiale di piantagione chiamato “yam heads” (teste di igname) sepolto in singoli cumuli di terra chiamati “yam hills” (collinette di igname).
Br’er
abbreviazione di brother, fratello
Bredder
vedere Br'er
mawning
nel creolo Giamaicano è "morning"
Hog
maiale
Cho
nel creolo giamaicano, termine utilizzato per rappresentare frustrazione o scocciatura nei confronti di qualcuno o qualcosa.
came down
scendere
sit down upon
su cui siede
deh
in creolo giamaicano, il luogo (su cui siede)
vex
annoiato, infastidito
fool
ingannare
hook
l'amo
drifted down
immergersi
ill fortune
sfortuna
crept into
strisciare, trascinarsi
tangled
aggrovigliati
filled out
riempire, riempirsi
mighty
forte, potente, rumoroso
[https://dictionary.cambridge.org/it/dizionario/inglese-italiano/mighty]
banged on
battere, colpire, picchiare
slaked
slake the thirst: placare la sete
crawled over
strisciare
clanked
sferragliare
tinkled
tintinnare
glisten
scintillare
drowsy
assonnato
uttered
proferire
furs
pelliccia
untangle
districarla, liberarla
tumble
rovesciata sul pavimento
whale oil lamp
lampada all'olio di balena
snowhouse
igloo
rib cage
cassa toracica
grimy
lercio
sobbing
singhiozzando
Panting
affannarsi
scrabbled
avanzare a fatica
dove
past simple di dive, immergersi
depths
profondità, abissi
shoreline
bagnasciuga
snatch him down
atterrare
flailed out
sbracciarsi
steam
vapore
zigged
andare a zigzag
paddling
pagaiare
oar
remo
prow
prua
knocked her off
buttarla giù
blazed
risplendere
bucked
sobbalzare, impennarsi
shook
oscillare
froth
spuma, schiuma che creano le onde quando si infrangono
sea was stirred
mare mosso, agitato
inlet
baia, piccola insenatura
had drifted
andare alla deriva
plucked out
strappare via
flesh
le sue carni [https://www.wordreference.com/enit/flesh]
coal of fire
un carbone ardente
[https://www.collinsdictionary.com/it/dizionario/inglese-italiano/coal]
scorched
bruciato
U′guku
Il nome nella mitologia Cherokee per il gufo che ulula, spesso identificato con il gufo barrato (Syrnium nebulosum), un uccello distintivo noto per i suoi richiami profondi e risonanti.
Tskĭlĭ
Horned Owl, o Tskĭlĭ in Cherokee, è un tipo di gufo caratterizzato dalle “corna” di piume sopra la testa.
Uksu′hĭ snake, the black racer
In Cherokee, Uksu′hĭ (a volte scritto Uksuhi) è il nome per il serpente Black Racer (Coluber constrictor), un serpente comune in Nord America.
Screech-owl
gufo
tusti bowl
recipiente intrecciato
spun
tessere
Kănăne′skĭ Amai′yĕhĭ
Kănăne′skĭ Amai′yĕhĭ, il ragno d'acqua nella mitologia Cherokee.
[https://www.cherokeepins.org/post/cherokee-stories-and-the-birth-of-a-new-world-the-first-fire]
Gûle′gĭ
"Blacksnake the climber" si riferisce a diversi serpenti di colore nero, in particolare al serpente nero dei ratti (black rat snake), noto per le sue eccezionali capacità di arrampicata grazie alle sue scaglie ventrali specializzate. Gûle′gĭ nella mitologia Cherokjee.
blast
folata (d'aria)
[https://dictionary.cambridge.org/it/dizionario/inglese-italiano/blast]
Wa′huhu′
Nella mitologia Cherokee "il gufo".
[https://www.cherokeepins.org/post/cherokee-stories-and-the-birth-of-a-new-world-the-first-fire]
had scorched
si era bruciato
The Raven
corvo maggiore
sycamore tree
platano
Ani′-Hyûñ′tĭkwălâ′skĭ
Ani′-Hyûñ′tĭkwălâ′skĭ è il nome, nella mitologia Cherokee, per "The Thunders," il termine plurale per indicare "coloro che hanno dato il fuoco".
[https://www.cherokeepins.org/post/cherokee-stories-and-the-birth-of-a-new-world-the-first-fire]
Gălûñ′lătĭ
Gălûñ′lătĭ, secondo la mitologia Cherokee, è un mondo creato prima dell'esistenza della Terra.
[https://www.cherokeepins.org/post/cherokee-stories-and-the-birth-of-a-new-world-the-first-fire]
Kaatskill mountains
The Catskill Mountains, or the Catskills, are a group of mountains in southeastern New York and are part of the larger Appalachian Mountain range.
that stood in the shade of
"to stand in the shade of sth" posizionarsi nell'aria d'ombra creata da un grosso oggetto, come un albero.
eLife Assessment
This Review Article provides a thorough overview of whole-brain activity changes induced by brain stimulation and summarizes the current state of the field. However, it lacks integration across spatial and mechanistic scales, which limits the reader's ability to understand how the different findings relate to one another. In addition, several key concepts are not explained in sufficient depth for non-expert readers. The manuscript would benefit from the development of a cohesive conceptual framework to more clearly synthesize the existing literature.
Reviewer #1 (Public review):
Summary:
This paper is a comprehensive review of perturbation studies and the state-dependence of the brain's response to perturbation at the circuit, mesoscale, and macroscale levels.
Strengths:
The strengths of the paper are the thorough description of many perturbation studies at different levels of organization, and the integration of both experimental and modeling studies. The review clearly communicates the need to consider (1) brain or local-population state, and (2) multiple levels of organization, in order to understand perturbation responses. Another major strength is the ability for the reader to reproduce figures using the EBRAINS platform.
Weaknesses:
Two major points of improvement should be resolved with the review, in order to make it useful for a broad audience.
The first is that the review does not include a significant integration across scales, and as a result, reads like three separate (though comprehensive) reviews. Currently, the only integration across the scales is in the brief conclusion paragraph. I would recommend adding an additional section, in which the overarching picture is discussed. (i.e. a unifying view of state dependence, and what is learned by considering across scales). This need not be too long, but it should be longer than a single conclusion paragraph.
The second major weakness is that there is a lack of clarity on many points throughout, which is needed for the reader to fully understand the results described.
Reviewer #2 (Public review):
Summary:
In this review article, the authors discuss the whole-brain activity changes induced by brain stimulation. They review the literature on how these activity changes depend on the cognitive state of the brain and divide the results by the scale of the change being induced, from microscale changes across small groups of neurons, up to macroscale changes across the entire brain. Finally, they describe attempts to model these changes using computational models.
Strengths:
The review provides an overview of the results within this subfield of neuroscience, and the authors are able to discuss a lot of prior results. The framing of the changes in neuronal activity in terms of computational changes is also a helpful approach.
Weaknesses:
However, the authors are not able to contextualize these results within a single framework, i.e. explaining from first principles how different aspects of stimulus-induced changes interact to generate functional changes in the brain, and how different changes - at distinct spatiotemporal scales - combine to form larger effects. This is a significant weakness in generating a review of the literature, since the authors do not provide a cohesive conceptual framework on which to frame the results. Similarly, the authors do not explain how their different computational models fit together, and how one can get a singular computational understanding of the distinct mechanisms of brain activity changes due to stimulation under different brain states, by combining the results derived from each separate model.
Major Comments:
(1) The authors have written this review as if it were intended for an audience who is already familiar with the topics. For example, they introduce concepts like complexity, spiral vs planar waves, without much explanation.
(2) Regarding complexity, the authors present a quantification termed PCI. However, in the associated box, they state that PCI could be implemented in a number of different ways, using analogous metrics (which are, nonetheless, not identical). Yet the authors simply claim that all these metrics are sufficiently similar to be grouped together as "PCI". The authors do not provide much intuition about this, and they also don't present any other potential quantifications. This makes any interpretation of their results strongly dependent on your understanding of the concept of PCI. It would be helpful to present some other, analogous metric to demonstrate that the results that the authors are focusing on are not somehow tied to the specific computational structure of the PCI metric.
(3) The authors divide the review into sections organized by the spatial extent of the effects that they are exploring (e.g. from microscale to macroscale). However, they don't bring together these insights into a cohesive structure - for example, by providing potential explanations of the macroscale effects by using the microscale changes.
(4) The authors completely ignore any aspect of cell-type specificity in their review, despite the known importance of specific cell types at the microcircuit scale. This makes it difficult to map their results onto the true biological system.
(5) The authors introduce several different computational models, such as the Hopf model, the AdEx model, and the MPR model. However, they do not provide the reader with a conceptual understanding of the structure of each of these models (except through potentially more complex terminology, e.g. the Hopf model is a "phenomenological Stuart-Landau nonlinear oscillator"). Additionally, though they present the results of each simulation, they don't provide the reader with intuition about how these models compare against each other, and how best to interpret results derived from each model.
(6) In several cases, the authors make statements that they appear to believe to be completely straightforward (and require no justification), but that do not appear so to the reader. For example, they mention: "In wakefulness and REM sleep, ..., the membrane potential is depolarized and close to the spike threshold, which explains why neurons respond more reliably and with less response variability compared with slow-wave sleep". However, this statement is not obvious to the reader and requires explanation (for example, in a system that is close to balance, bringing cells closer to the firing threshold can result in increased response jitter).
Author response:
We would like to thank the editors and reviewers for excellent suggestions to improve the paper. We will carefully address these suggestions in a revised version.
As one of the community and public health nursing Specialists competences is the community empowerment [3], there is a great management opportunity for primary health care leaders to ensure that these specialists are allocated to public health units
No, only people with accounts that I have authorized can post here. The vast majority of the edits are made by me, but a small number may be made by graduate or undergraduate research assistants. Check the “history” tab on any given page to see who changed what; any changes made by “Caleb McDaniel” or “wcaleb” are attributable to me.
Open access to thoughts, etc. but restricted when it comes to collaboration, as it is not a collaborative project / looking for information or input from its viewers
In that time, the notebook has seen six projects go from conception to publication, and a seventh founder on a null result (see #tribolium). Several more projects continue to unfold.
We have open access to information pertaining to projects (unreleased/published) - this is a newer process after changing mediums for some (sharing information before releasing official versions)
Welcome to my open lab notebook. This is the active, permanent record of my scientific research, standing in place of the traditional paper bound lab notebook. The notebook is primarily a tool for me to do science, not communicate it. I write my entries with the hope that they are intelligible to my future self; and maybe my collaborators and experts in my field. Only the occasional entry will be written for a more general audience. […] In these pages you will find not only thoughts and ideas, but references to the literature I read, the codes or manuscripts I write, derivations I scribble and graphs I create and mistakes I make.
On 24 November 2023, the EU and Canada launched a Digital Partnership to reinforce cooperation on digital issues. The partnership reflects a shared vision for a positive and human-centric digital economy and society. The EU and Canada agreed to work together in crucial areas such as AI, secure international connectivity, cyber security, online platforms, digital identity and digital skills. This Digital Partnership and its importance were highlighted in the New EU-Canada Strategic Partnership of the Future, adopted at the Canada-EU Summit on 23 June 2025.
Digital Partnership started in 2023. June 2025 incorporated in strategic partnership. Now a first physical meeting at high level.
EU and Canada are committed to working together on secure international connectivity, for example in 5G and subsea cables, and have agreed to explore new cable routes to strengthen global network resilience, including in the Arctic region. The EU and Canada will deepen the collaboration in priority topics such as quantum technologies, semiconductors, and high-performance computing. They also reaffirmed their commitment to resilient semiconductor supply chains and secure and sovereign cloud infrastructure and data centres.
Covering deeper layers of the stack. So it's a digital sovereignty / autonomy pact.
explore cooperation on enhancing information integrity online, including on the challenges posed by generative AI and the risks stemming from foreign information manipulation and interference.
DSA aspects
the cooperation on digital credentials and trust services, including on technical interoperability and solutions based on digital identity wallets, the EU and Canada signed a Memorandum of Understanding on Digital Credentials and Trust Services.
Relevant to EU wallett efforts and to trusted data transactions. This is a 2nd MoU
the partners will share best practices to accelerate AI adoption in strategic sectors such as healthcare, manufacturing, energy, culture, science and public services, and support SMEs. They committed to work together on large AI infrastructures and support industry and academia's access to AI compute capacity. They will also explore scientific cooperation on fundamental AI research, and the development of advanced AI models for the public good, including in areas such as extreme weather monitoring and climate change. In addition, the EU and Canada will set up a structured dialogue on data spaces, of particular relevance to the development of large AI models.
Elements in the MoU: - share good practices to support adoption - collab on large AI infrastructure (Apply AI strat EU, HPC network) - collab on access to HPC (in line w AI factories in EU) - explore coop in fundamental ai research (weak) - development of AI for public good (in line w EU AI goals) - structured dialogue on data spaces (as data source for AI models) Only the last one is not immediately obviously connected to existing EU efforts and actions.
the first Digital Partnership Council held today in Montreal, Canada
Canada and EU have a shared Digital Partnership Council. First meeting #2025/12/08
Unless otherwise noted (i.e., on a draft manuscript), notes posted here is made available under the Creative Commons NonCommercial-Attribution-ShareAlike license. This means you are free to make use of it, change it, use it for any non-commercial purposes, as long as you acknowledge the source. Journal manuscripts under development here are often NOT covered by this Creative Commons license, because they will eventually be subject to whatever license the target journal requires. Thus, drafts are readable in their posted form, but all rights are reserved beyond viewing (and, of course, having your own ideas with respect to the material).
This is the prime reason why digital humanities was taken over so soon by multiple disciples. The main purpose was to connect other intellects and even those who are just curious - the information can be added/edited by anyone who has something to add.
But by far the more important aspect of open science is an open process, and reproducible results. To that end, I have been exploring the use of wikis and blogs to record interim thinking on research topics, and this is the second iteration of an online "lab notebook" that goes beyond occasional blog posting. My first digital lab notebook was a local installation of the Instiki wiki, synchronized with Dropbox. This was useful for doing my own work wherever I happened to be, but was not truly "open" in the sense of public access. I have been migrating some of those reading notes, and topical notebooks to this current iteration, and that process is ongoing. My first "online open notebook" was hosted by Wikispaces, but I found that the lack of offline access was difficult for me, given travel and limited internet access where I live and work.
This author is presenting academic blogging as a way to connect with others (whether this be students or other teachers, etc.)
My commitment to open science began with open access, and attempts to ensure that my written output was -- to the extent possible -- available online in freely downloadable format. This is always a work in progress, because older publications are often unavailable given paywalls or commercial licenses by academic publishers. To the extent possible, I will always make versions of publications available online, and I will attempt to choose journals with permissive preprint/postprint policies. I am slowly attempting to reconstruct PDF versions of older conference papers, many of which I have only in print files, which will need to be scanned.
The author is aware that with the changing of mediums - it has become a different playing field for the world of information. They have shown that there is an understanding for the need for information especially in this day-in-age where we are more used to digital platforms and halted by subscriptions.
Cohesion refers to the flow from sentence to sentence. For example, compare these passages: Version A (That I Rewrote): Granovetter begins by looking at balance theory. If an actor, A, is strongly tied to both B and C, it is extremely likely that B and C are, sooner or later, going to be tied to each other, according to balance theory (1973:1363).[10] Bridge ties between cliques are always weak ties, Granovetter argues (1973:1364). Weak ties may not necessarily be bridges, but Granovetter argues that bridges will be weak. If two actors share a strong tie, they will draw in their other strong relations and will eventually form a clique. Only weak ties that do not have the strength to draw together all the “friends of friends” can connect people in different cliques. Version B (The Original By Giuffre): Granovetter begins by looking at balance theory. In brief, balance theory tells us that if an actor, A, is strongly tied to both B and C, it is extremely likely that B and C are, sooner or later, going to be tied to each other (1973:1363). Granovetter argues that because of this, bridge ties between cliques are always weak ties (1973:1364). Weak ties may not necessarily be bridges, but Granovetter argues that bridges will be weak. This is because if two actors share a strong tie, they will draw in their other strong relations and will eventually form a clique. The only way, therefore, that people in different cliques can be connected is through weak ties that do not have the strength to draw together all the “friends of friends.” [11] Version A has the exact same information as version B, but it is harder to read because it is less cohesive.
The comparison between Version A and Version B shows me how cohesion works. I realize my writing feels choppy sometimes because I don’t connect old and new information clearly.
In academic writing, readers expect each paragraph to have a sentence or two that captures its main point. They’re often called “topic sentences,” though many writing instructors prefer to call them “key sentences.” There are at least two downsides of the phrase “topic sentence.” First, it makes it seem like the paramount job of that sentence is simply to announce the topic of the paragraph. Second, it makes it seem like the topic sentence must always be a single grammatical sentence. Calling it a “key sentence” reminds us that it expresses the central idea of the paragraph.
The examples of Version A vs. Version B help me understand why key sentences matter. They don’t just introduce topics—they connect each paragraph to the argument.
Effective paragraphs are the fundamental units of academic writing; consequently, the thoughtful, multifaceted arguments that your professors expect depend on them. Without good paragraphs, you simply cannot clearly convey sequential points and their relationships to one another
This section reminds me how important paragraph organization is. I used to think content and style were separate, but the author shows they work together.
oes the Bystander Effect Always Occur?
Are there any other variables that causes this effect?
sibility could explain why you might not put in as much effort on a group project as you would on an individual project (because your classmates are also responsible for doing the work). It can also explain why sharing chores with roomm
It is very common in many projects
ent to go get help while the participant was still having the seizure, and everyone reported it before the experimental session ended). However, when the participants believed that they were in groups of six—that is, when they thought there were four other people who could also report the seizure—they were less likely to get help: only 31% of participants reported the emergency while the seizure was happening, and only 62% reported it by the end of the experiment. In another condition, in which participants were in groups o
Maybe people's afraid of other consequences that might occur, so they choose to hope the job will be done by other people around
tervene and help others? Psychologists have found that people are sometimes less likely to help out when there are others present, a phenomenon known as the bystander effect. One reason the bystander effect occurs is due to diffusion o
Connect to the text, little people want to help aliens out of many watch. This action might be caused by the bystander effect.
threat (the host nation failing to provide needed support for refugees)
Connect to text, in the novel there is a part about the aliens are hard to be alive on Earth, and countries might fail.
o how can we reduce perceptions of different types of threats associated with refugees and immigrants? Here are some suggestions: To understand the real effects of migration on us and our cou
Can people at last manage to solve these nationalist problems ?
dy 4 used a more representative sample, consisting of 111 women and 140 men (mean age of 50 years). Compared to college students in previous samples, these participants reported perceiving even stronger threats and experiencing more hostility toward refugees. And the results again showed support for the six threat types. Every threat type was correlated with unfavorable views of migr
Why old people are more controversal?
efugees, for instance, pose an economic threat because they need jobs, low-cost housing, access to health care, etc. In addition, they pose a health threat because some refugees come f
Linking with some personal experience, I've meet some English people whom said that many for their people don't like refugee because they have access to benefits but don't contribute to the society as much.
you are among those who hate refugees, do you know why you feel this way? Is it a vague feeling of hostility or does it
On Chinese internet, many people are preventing refugees to enter the boarder, even though China isn't a immigrant country and non of them experience negative side of it.
With the recent mass shooting in Germany, some people are again asking w
This remained me about the recent Germany party of AFD that also trys to keep refugee and immigrants out
They’re not afraid to make mistakes (own them even), and they know that struggle can be a rewarding part of the process. By equal measure, mature learners celebrate their strengths and use them strategically.
The author explains that reflection, looking back at past experiences and noticing how your thinking has evolved is important in this process.
9 percent, up from 7 percent last year
It's ironical linking to the text about "harder college application for children", because the aliens represents refugees.
Europe remains among the regions with the highest number of displaced people globally, hosting 13.2 million refugees.
It is very common to see refugees will visiting Europe
2025, more than 117 million people were forcibly displaced worldwide as a result of persecution, conflict, violence or human rights violations. This includes:
Do this include all the descendents of the refugees?
eLife Assessment
This potentially valuable cross-sectional longitudinal study leverages high-definition transcranial direct current stimulation to the left dorsolateral prefrontal cortex to examine its effect on procrastination behavior over an extended time span. Support for the conclusions is incomplete owing to missing information about the analyses, the nature of the procrastination tasks, and the derived dependent measures.
Reviewer #1 (Public review):
Summary:
The authors report the results of a tDCS brain stimulation study (verum vs sham stimulation of left DLPFC; between-subjects) in 46 participants, using an intense stimulation protocol over 2 weeks, combined with an experience-sampling approach, plus follow-up measures after 6 months.
Strengths:
The authors are studying a relevant and interesting research question using an intriguing design, following participants quite intensely over time and even at a follow-up time point. The use of an experience-sampling approach is another strength of the work.
Weaknesses:
There are quite a few weaknesses, some related to the actual study and some more strongly related to the reporting about the study in the manuscript. The concerns are listed roughly in the order in which they appear in the manuscript.
(1) In the introduction, the authors present procrastination nearly as if it were the most relevant and problematic issue there is in psychology. Surely, procrastination is a relevant and study-worthy topic, but that is also true if it is presented in more modest (and appropriate) terms. The manuscript mentions that procrastination is a main cause of psychopathology and bodily disease. These claims could possibly be described as 'sensationalized'. Also, the studies to support these claims seem to report associations, not causal mechanisms, as is implied in the manuscript.
(2) It is laudable that the study was pre-registered; however, the cited OSF repository cannot be accessed and therefore, the OSF materials cannot be used to (a) check the preregistration or to (b) fill in the gaps and uncertainties about the exact analyses the authors conducted (this is important because the description of the analyses is insufficiently detailed and it is often unclear how they analyzed the data).
(3) Related to the previous point: I find it impossible to check the analyses with respect to their appropriateness because too little detail and/or explanation is given. Therefore, I find it impossible to evaluate whether the conclusions are valid and warranted.
(4) Why is a medium effect size chosen for the a priori power analysis? Is it reasonable to assume a medium effect size? This should be discussed/motivated. Related: 18 participants for a medium effect size in a between-subjects design strikes me as implausibly low; even for a within-subjects design, it would appear low (but perhaps I am just not fully understanding the details of the power analysis).
(5) It remains somewhat ambiguous whether the sham group had the same number of stimulation sessions as the verum stimulation group; please clarify: Did both groups come in the same number of times into the lab? I.e., were all procedures identical except whether the stimulation was verum or sham?
(6) The TDM analysis and hyperbolic discounting approach were unclear to me; this needs to be described in more detail, otherwise it cannot be evaluated.
(7) Coming back to the point about the statistical analyses not being described in enough detail: One important example of this is the inclusion of random slopes in their mixed-effects model which is unclear. This is highly relevant as omission of random slopes has been repeatedly shown that it can lead to extremely inflated Type 1 errors (e.g., inflating Type 1 errors by a factor of then, e.g., a significant p value of .05 might be obtained when the true p value is .5). Thus, if indeed random slopes have been omitted, then it is possible that significant effects are significant only due to inflated Type 1 error. Without more information about the models, this cannot be ruled out.
(8) Related to the previous point: The authors report, for example, on the first results page, line 420, an F-test as F(1, 269). This means the test has 269 residual degrees of freedom despite a sample size of about 50 participants. This likely suggests that relevant random slopes for this test were omitted, meaning that this statistical test likely suffers from inflated Type 1 error, and the reported p-value < .001 might be severely inflated. If that is the case, each observation was treated as independent instead of accounting for the nestedness of data within participants. The authors should check this carefully for this and all other statistical tests using mixed-effects models.
(9) Many of the statistical procedures seem quite complex and hard to follow. If the results are indeed so robust as they are presented to be, would it make sense to use simpler analysis approaches (perhaps in addition to the complex ones) that are easier for the average reader to understand and comprehend?
(10) As was noted by an earlier reviewer, the paper reports nearly exclusively about the role of the left DLPFC, while there is also work that demonstrates the role of the right DLPFC in self-control. A more balanced presentation of the relevant scientific literature would be desirable.
(11) Active stimulation reduced procrastination, reduced task aversiveness, and increased the outcome value. If I am not mistaken, the authors claim based on these results that the brain stimulation effect operates via self-control, but - unless I missed it - the authors do not have any direct evidence (such as measures or specific task measures) that actually capture self-control. Thus, that self-control is involved seems speculation, but there is no empirical evidence for this; or am I mistaken about this? If that is indeed correct, I think it needs to be made explicit that it is an untested assumption (which might be very plausible, but it is still in the current study not empirically tested) that self-control plays any role in the reported results.
(12) Figures 3F and 3H show that procrastination rates in the active modulation group go to 0 in all participants by sessions 6 and 7. This seems surprising and, to be honest, rather unlikely that there is absolutely no individual variation in this group anymore. In any case, this is quite extraordinary and should be explicitly discussed, if this is indeed correct: What might be the reasons that this is such an extreme pattern? Just a random fluctuation? Are the results robust if these extreme cells are ignored? The authors remove other cells in their design due to unusual patterns, so perhaps the same should be done here, at least as a robustness check.
(13) The supplemental materials, unfortunately, do not give more information, which would be needed to understand the analyses the authors actually conducted. I had hoped I would find the missing information there, but it's not there.
In sum, the reported/cited/discussed literature gives the impression of being incomplete/selectively reported; the analyses are not reported sufficiently transparently/fully to evaluate whether they are appropriate and thus whether the results are trustworthy or not. At least some of the patterns in the results seem highly unlikely (0 procrastination in the verum group in the last 2 observation periods), and the sample size seems very small for a between-subjects design.
Reviewer #2 (Public review):
Summary:
Chen and colleagues conducted a cross-sectional longitudinal study, administering high-definition transcranial direct stimulation targeting the left DLPFC to examine the effect of HD-tDCS on real-world procrastination behavior. They find that seven sessions of active neuromodulation to the left DLPFC elicited greater modulation of procrastination measures (e.g., task-execution willingness, procrastination rates, task aversiveness, outcome value) relative to sham. They report that tDCS effects on task-execution willingness and procrastination are mediated by task outcome value and claim that this neuromodulatory intervention reduces procrastination rates quantified by their task. Although the study addresses an interesting question regarding the role of DLPFC on procrastination, concerns about the validity of the procrastination moderate enthusiasm for the study and limit the interpretability of the mechanism underlying the reported findings.
Strengths:
(1) This is a well-designed protocol with rigorous administration of high-definition transcranial direct current stimulation across multiple sessions. The approach is solid and aims to address an important question regarding the putative role of DLPFC in modulating chronic procrastination behavior.
(2) The quantification of task aversiveness through AUC metrics is a clever approach to account for the temporal dynamics of task aversiveness, which is notoriously difficult to quantify.
Weaknesses:
(1) The lack of specificity surrounding the "real-world measures" of procrastination is problematic and undermines the strength of the evidence surrounding the DLPFC effects on procrastination behavior. It would be helpful to detail what "real-world tasks" individuals reported, which would inform the efficacy of the intervention on procrastination performance across the diversity of tasks. It is also unclear when and how tasks were reported using the ESM procedure. Providing greater detail of these measures overall would enhance the paper's impact.
(2) Additionally, it is unclear whether the reported effects could be due to differential reporting of tasks (e.g., it could be that participants learned across sessions to report more achievable or less aversive task goals, rather than stimulation of DLPFC reducing procrastination per se). It would be helpful to demonstrate whether these self-reported tasks are consistent across sessions and similar in difficulty within each participant, which would strengthen the claims regarding the intervention.
(3) It would be helpful to show evidence that the procrastination measures are valid and consistent, and detail how each of these measures was quantified and differed across sessions and by intervention. For instance, while the AUC metric is an innovative way to quantify the temporal dynamics of task-aversiveness, it was unclear how the timepoints were collected relative to the task deadline. It would be helpful to include greater detail on how these self-reported tasks and deadlines were determined and collected, which would clarify how these procrastination measures were quantified and varied across time.
(4) There are strong claims about the multi-session neuromodulation alleviating chronic procrastination, which should be moderated, given the concerns regarding how procrastination was quantified. It would also be helpful to clarify whether DLPFC stimulation modulates subjective measures of procrastination, or alternatively, whether these effects could be driven by improved working memory or attention to the reported tasks. In general, more work is needed to clarify whether the targeted mechanisms are specific to procrastination and/or to rule out alternative explanations.
Reviewer #3 (Public review):
This manuscript explores whether high-definition transcranial direct current stimulation (HD-tDCS) of the left DLPFC can reduce real-world procrastination, as predicted by the Temporal Decision Model (TDM). The research question is interesting, and the topic - neuromodulation of self-regulatory behavior - is timely.
However, the study also suffers from a limited sample size, and sometimes it was difficult to follow the statistics.
The preregistration and ecological design (ESM) are commendable, but I was not able the find the preregistration, as reported in the paper.
Overall, the paper requires substantial clarification and tightening.
Author response:
Reviewer #1:
(1) We fully thank you to point out the risks of sensationalizing ramification of procrastination on psychopathology, and would rewrite the Introduction section by adding balanced evidence and overall toning down such inappropriate claims meanwhile.
(2) Thank you to raise this crucial question. We are sorry for this fundamental technical issue to preregistration. This occurs from a seriously technical hurdle. The OSF has banned my OSF account, as it claimed to detect “suspicious user’s activities” in my account. This causes no accesses to all materials that already deposited in this OSF account, including this preregistration. We have contacted OSF team, but received no valid technical solution. We reckon that this may be mistaken by my affiliation changes to Third Military Medical University of People’s Liberation Army (PLA). To tackle with this technical issue, we shall upload preregistration in a new repository soon.
(3) This is a back-to-back study to conceptually probe into whether strengthening left DLPFC can mitigate procrastination via reducing task aversiveness or weighting outcome value. Thus, the current study selected a medium effect size in aprior by following the previous one (Xu et al., 2023). This effect size is calculated by the new tool called “Power Contours” (Baker et al., 2021), which weights statistical power by increasing within-subject repeated measures. As you kindly pointed out, we shall clarify effect size calculation in the revised manuscript.
(4) Yes, both groups come in the same number of times into the lab for tDCS stimulation, except to the type (active vs sham).
(5) We shall add full details for clarifying TDM and hyperbolic discounting modeling.
(6) Thank you to raise this very crucial statistical question. We shall double-check whether multiple sessions are modeled as random slopes, and would like to reanalysis it in case which those random slopes are omitted.
(7) Thank you. We have no intentions of confusing you by adding those complicated statistics, but indeed enrich understanding of how we can interpret those findings.
(8) Yes, as mentioned above, we shall add balanced evidence to clarify both left and right DLPFC may function to self-control capability in the Introduction section.
(9) Yes, this is a conceptual hypothesis --- actively stimulating left DLPFC could improve self-control functions. Thank you for this very nuanced but crucial insight, and we could explicitly clarify the nature of our conclusions.
(10) Yes, we ensure that all the participants successfully completed their tasks before deadline at session 6 and 7, and the procrastination rates have been all decreased to 0. Personally speaking, this is somewhat surprise to us as well, but we affirmed this case. For a portion of participants included in the active group, we have received written letters of thanks from them. Thus, this is surprise but exciting finding. Furthermore, thank you for this helpful suggestion, and we would like to do this robustness check by iteratively removing each session, to obviate the statistical biases from an extreme pattern.
(11) Yep, we fully agree with you to add full details in the main text rather in Supplemental materials, and would like to do so in the first round of revision.
Reviewer #2:
(1) Thank you for this very crucial suggestion. We are sorry for this case that much details are omitted to comply with editorial requirement at Nature Human Behaviour (last submission). We do apologize to confuse you as those ambiguous descriptions, and would like to clearly clarify how we measure participants’ procrastination in the real-world tasks. In brief, we asked participant to report a real task that would really happen in the tomorrow and its deadline is also no more than tomorrow. When tomorrow comes, we used ESM to require participant reporting real task completion rate (0-100%) at five time points before the deadline. The five time points are determined by a hyperbolic discounting model (see how and why we set those five time points in the full author’s response letter later). When participant reports the real task completion rate (0-100%) at a given time point, she/he is required to provide a photo to prove its authenticity. The dependent variable --- real-world procrastination rates --- is thus calculated as 100% subtracts the task completion rate (0-100%) when the deadline meets. That is to say, if participant reports task has been fully completed before or when deadline meets, his/her real-world procrastination rate is 100% - 100% = 0%; if reporting task has been completed 60% when deadline meets, the real-world procrastination rate is determined as 100% - 60% = 40%. Do not worry for spurious reporting, we asked all the participants to provide photo verifying the real task completion rate. This is merely a short instance. We shall show the full details in the formal author response letter later.
(2) This is a very meaningful point. We agree with you for this case that participants may learn how to complete this experiment task swiftly rather benefit from neuromodulation. This speculation makes sense, but is compromised by experimental control and empirical observations. Firstly, we do not say “You must complete this task” or “The task completion is associated with bonus/rewards you may get” for participants, which indicates no motivations to do so. Then, the measures to task completion rate are not yet fully based on self-reporting, and we mandate them to provide photos for verification. Thus, this controls the marked risks of spurious reporting. Lastly, all the participants, including ones in either active or sham group, received all the same treatments, excepting “real simulation” and “sham simulation” protocol. Results demonstrated the significant amelioration in the active group rather sham one, indicating no significant “placebo” or “task learning” side effect.
(3) Thank you. As you kindly suggested, we would like to add huge details for those measures in the revised manuscript. While this is a great idea, we did not collect procrastination scores from scales after neuromodulation, and would like to warrant this point into the Limitation section.
(4) Yep, this is a conceptual hypothesis --- actively stimulating left DLPFC could improve self-control functions. We cannot rule out possibilities of amplifying working memory, attention or other cognitive components from this neuromodulation protocol. We fully agree with you for this helpful recommendation --- we would like tone down those claims regarding the roles of DLPFC on self-control, and explicitly warrant that this mechanism may be specialized to the procrastination.
Reviewer #3:
(1) Thank you for taking valuable time to review our manuscript. Yep, limited sample size should warrant cautions to draw a solid conclusion. We would like to claim it into the limitation section. Also, we have streamlined and tightened statistic section by removing complicated and redundancy statistical models.
(2) As mentioned above, we are sorry for this fundamental technical issue to preregistration. This occurs from a seriously technical hurdle. The OSF has banned my OSF account, as it claimed to detect “suspicious user’s activities” in my account. This causes no accesses to all materials that already deposited in this OSF account, including this preregistration. We have contacted OSF team, but received no valid technical solution. We reckon that this may be mistaken by my affiliation changes to Third Military Medical University of People’s Liberation Army (PLA). To tackle with this technical issue, we shall upload preregistration in a new repository soon.
(3) Yep, thank you for this very helpful suggestion. As you kindly indicated, we would like to clarify measures, analyses, methods, and protocols, as well as tighten the whole manuscript.
References
Baker, D. H., Vilidaite, G., Lygo, F. A., Smith, A. K., Flack, T. R., Gouws, A. D., & Andrews, T. J. (2021). Power contours: Optimising sample size and precision in experimental psychology and human neuroscience. Psychological methods, 26(3), 295–314. https://doi.org/10.1037/met0000337
Xu, T., Zhang, S., Zhou, F., & Feng, T. (2023). Stimulation of left dorsolateral prefrontal cortex enhances willingness for task completion by amplifying task outcome value. Journal of experimental psychology. General, 152(4), 1122-1133. https://doi.org/10.1037/xge0001312
Again, we wholeheartedly appreciate all of those very helpful and insightful comments, with each one to contribute substantially for the quality of this manuscript. Notably, those response we presented above are merely provisional and initial. We shall revise our manuscript following those suggestions, one-by-one, along with a full-length response letter.
eLife Assessment
In this Review Article, the authors survey the literature describing how correlated dynamical states relate to various cognitive states, including anesthesia and sleep. While the topic is significant and the coverage broad, the manuscript does not yet provide a synthesis that connects the many available findings or highlights converging themes across studies. Additionally, many of the disparate concepts are not introduced at the level of first principles. As a result, the Review remains difficult to access for readers outside the immediate subfield. Developing a clearer integrative perspective would help make the article informative to a wider audience.
Reviewer #1 (Public review):
Summary:
In the paper, the authors review literature on synchronous activity, its relationship to brain state, and the multi-scale mechanisms underlying it.
Strengths:
The overall strength of the paper is the wide range of information reviewed, and the diversity of perspectives/approaches it brings together.
Weaknesses:
However, this strength is also the source of its major weaknesses - namely, that the overall structure lacks clarity, and there are inconsistencies throughout. Overall, in the opinion of this reviewer, the manuscript reads as disorganized and incomplete. Major and minor points are delineated below.
Major points:
(1) Most of the text in many figures was too small to read.
(2) Terminology is inconsistent throughout the manuscript. What is the difference between slow oscillations and delta waves? Sometimes the term slow waves is used instead. For sleep state, sometimes the term SWS is used, sometimes non-REM. Similarly, "spindle activity" is not defined, but simply stated as if the reader knows. This brings up two issues: (a) the manuscript should be clearer and more consistent about its terminology, and (b) it's unclear who is the intended readership of the review - is it a pedagogical review for people outside the field of sleep and slow oscillations, or is it meant to be a consensus statement for readers who are already in the field in which a pressing concern has been addressed? It seems part way between these two, and as a result, is ineffective at either goal.
(3) I suggest the authors look again at the overall structure and flow of the review... many sections feel redundant, and it's unclear how they fit together into a single review.
(4) There are many speculative statements in the review that are not justified or explained sufficiently for the reader. For example: "While highly regular slow waves in vivo suggest a single mechanism of generation, namely local cortical circuits, irregular cycles are compatible with a larger role of subcortical nuclei, ..."; "The involvement of different cortical areas and subcortical nuclei can form the basis of these different roles in memory.". For these statements, I assume the relationship between slow wave statistics, subcortical nuclei, and memory either has been written about before, and then should be cited and summarized, or is a novel claim of the authors, which then should be explained and defended rather than stated. There are other similar examples, and I suggest the authors go through the manuscript and make sure that it's clear what is a novel claim of the authors vs a cited claim, and make sure that both are sufficiently justified for the reader.
(5) An especially notable example can be found in the section on the role of the thalamus, where the authors state that they "hold that slow oscillations are fundamentally cortical". However, this section is far too short, and very little evidence is provided to back up this claim. Please review the ways in which the thalamus modulates, and, e.g., ways in which up-down is similar/different without the thalamus.
Reviewer #2 (Public review):
Summary:
In this review article, the authors discuss the correlated dynamical states associated with distinct cognitive states, including those associated with anesthesia and sleep. They present evidence that these states are primarily cortically generated, and demonstrate the properties of these dynamical states at different levels, from the microscale dynamics in individual neurons to the macroscale dynamics across the brain.
Strengths:
Multiple groups have been adding to this field over the past decades, and therefore, a review of this literature is very helpful. This review collates a large amount of the literature within this field into a single document, which should make it a valuable resource within this area of neuroscience.
Weaknesses:
Unfortunately, this review does not seem to be a balanced viewpoint of the field in question. Although there are a lot of authors in the review, it feels as if they are from a common school of thought. The authors provide only a single perspective on these dynamical states, focusing on the perspective of wave-like electrical dynamics across the cortex. Their perspective is embedded in methods such as EEG and LFP recordings. This makes the work hard to interpret outside of the field in which the authors reside. Indeed, the review seems intended for a more specialized audience.
In addition, the article reads more like a catalog of prior studies as opposed to a true synthesis across the large volume of data in this field that highlights links across multiple sources. Hence, it does not seem to provide a novel way of understanding the dynamics involved in cognitive state transitions.
We have included more details on these general comments below:
Major Comments:
(1) The authors have written this review as if it were intended for an audience who is already familiar with these topics. They do not define many of the terms that they introduce within the review, including concepts like complexity, metastability, and oscillations that are fundamental to the concepts that the authors are introducing. Though these may seem like first principles concepts to the authors, they often introduce assumptions that may be unfamiliar to the general reader. For example, are slow wave oscillations periodic? A naïve reader may assume that oscillations - characterized by their frequency - should be somewhat periodic, but that is often not the case. For a journal with a general biological science readership, it would be particularly helpful for each of these terms to be formally defined and characterized.
(2) It would be helpful for the authors to reframe their work in different perspectives and to incorporate all the literature on the dynamics of cortical brain states, and not simply the work that is most familiar to them. As one example, the authors do not discuss cell-type-specific changes in brain state during anesthesia and in altered states of consciousness (including dissociative states and hallucinatory states). There is recent work in this vein (Suzuki and Larkum, 2020; Vesuna et al, 2020; Bharioke, Munz et al, 2023), and yet the authors do not discuss these papers.
(3) Given the authors' clear, extensive knowledge of their field, it would also be extremely helpful for the authors to reframe fundamental concepts in terms of neuronal population activity, trajectory analyses, etc. This would enable a more general audience to better understand their work.
(4) The authors have one section focused on thalamic contributions to cortical wave-like activity. This is a cursory treatment of a subject that is quite controversial in the field. It would be helpful if the authors could provide a more balanced consideration of all the evidence regarding potential thalamocortical interactions and their role in wave-like activity.
(5) The authors present many computational models and describe the results of simulations with these different models. However, this doesn't provide the reader with intuition about what each model adds or removes from the true biological picture. It would be helpful for the authors to provide some intuition about the assumptions and constraints that underlie each model.
(6) The authors state that "The main mechanism [of slow oscillatory dynamics] consists of a combination of two ingredients: the recurrent connectivity, which maintains the excitability in the network, and adaptation, an activity-dependent fatigue variable that provides inhibitory feedback". They make this statement as a fact, yet they don't provide much justification for it. Additionally, it's not clear that any other possible combination of ingredients would be able to produce slow oscillatory dynamics.
(7) The authors often define one concept in terms of other equally complex concepts. For example: "EIA (excitatory-inhibitory with adaptation) cortical circuits then display the typical slow-fast dynamics of relaxation oscillators". The reader would need an explanation of slow-fast dynamics and relaxation oscillators to understand this line, neither of which is provided in the text.
(8) When discussing sleep, the authors do not discuss REM sleep, focusing on slow-wave non-REM sleep. It would be helpful if the authors could at least frame the full sleep cycle and discuss why they are focusing on one part of it.
(9) The authors introduce the concept of sleep spindles without any explanation.
eLife Assessment
This important work combines theoretical analysis with precise experimental perturbation to demonstrate a previously unappreciated quantitative characteristic of the Wnt signaling pathway, which is anti-resonance, or a suppression of pathway output at intermediate activation frequencies. This effect is demonstrated experimentally with compelling evidence from optogenetic stimulation in multiple cell types, alongside modeling results that corroborate the phenomenon. While the demonstration of this phenomenon has yet to be extended to fully physiological situations, its clear existence within optogenetically stimulated systems shows that it is likely a significant factor that contributes to the behavior of this central signaling pathway.
Reviewer #1 (Public review):
Summary:
This report demonstrates that the gene expression output of the Wnt pathway, when controlled precisely by a synthetic light-based input, depends substantially on the frequency of stimulation. The particular frequency-dependent trend that is observed - anti-resonance, a suppression of target gene expression at intermediate frequencies given a constant duty cycle - is a novel aspect that has not been clearly shown before for this or other signaling pathways. The paper provides both clear experimental evidence of the phenomenon with engineered cellular systems and a model-based analysis of how the pairing of rate constants in pathway activation/deactivation could result in such a trend.
Strengths:
This report couples in vitro experimental data with an abstracted mathematical model. Both of these approaches appear to be technically sound and to provide consistent and strong support for the main conclusion. The experimental data are particularly clear, and the demonstration that Brachyury expression is subject to anti-resonance in ESCs is particularly compelling. The modeling approach is reasonably scaled for the system at the level of detail that is needed in this case, and the hidden variable analysis provides some insight into how the anti-resonance works.
In this revised manuscript, the authors have addressed issues in presentation and in discussing the broader relevance of their study to other pathways. Other limitations of the paper, including the fact that the anti-resonance phenomenon has not yet been demonstrated using physiological Wnt ligands and that the model has not been validated using experimental manipulations to establish that the mechanisms of the cell system and the model are the same, were deemed out of the scope of this initial demonstration by both the reviewers and authors. These questions will provide an interesting basis for further studies.
Reviewer #2 (Public review):
Summary:
By combining optogenetics with theoretical modelling the authors identify an anti-resonance behavior in the WnT signaling pathway. This behavior is manifested as a minimal response at a certain stimulation frequency. Using an abstracted hidden variable model, the authors explain their findings by a competition of timescales. Furthermore, they experimentally show that this anti-resonance influences the cell fate decision involved in human gastrulation.
Strengths:
- This interdisciplinary study combines precise optogenetic manipulation with advanced modelling.<br /> - The results are directly tested in two different systems: HEK293T cells and H9 human embryonic stem cells.<br /> - The model is implemented based on previous literature and has two levels of detail: i) a detailed biochemical model and ii) an abstract model with a hidden parameter
Weaknesses:
- While the experiments provide both single-cell data and population data, the model only considers population data.<br /> - Although the model captures the experimental data for TopFlash very well, the beta-Cat curves (Fig 2B) are only described qualitatively. This discrepancy is not discussed.
Overall Assessment:
The authors convincingly identified an anti-resonance behavior in a signaling pathway that is involved in cell fate decisions. The focus on a dynamic signal and the identification of such a behavior is important. I believe that the model approach of abstracting a complicated pathway with a hidden variable is an important tool to obtain an intuitive understanding of complicated dependencies in biology. Such a combination of precise ontogenetical manipulation with effective models will provide a new perspective on causal dependencies in signaling pathways and should not be limited only to the system that the authors study.
Comments on revisions:
I don't have any more comments for the authors and would like to congratulate them for the nice piece of work!
Author response:
The following is the authors’ response to the original reviews
Public Reviews:
Reviewer #1 (Public review):
Summary:
This report demonstrates that the gene expression output of the Wnt pathway, when controlled precisely by a synthetic light-based input, depends substantially on the frequency of stimulation. The particular frequency-dependent trend that is observed - anti-resonance, a suppression of target gene expression at intermediate frequencies given a constant duty cycle - is a novel aspect that has not been clearly shown before for this or other signaling pathways. The paper provides both clear experimental evidence of the phenomenon with engineered cellular systems and a model-based analysis of how the pairing of rate constants in pathway activation/deactivation could result in such a trend.
Strengths:
This report couples in vitro experimental data with an abstracted mathematical model. Both of these approaches appear to be technically sound and to provide consistent and strong support for the main conclusion. The experimental data are particularly clear, and the demonstration that Brachyury expression is subject to anti-resonance in ESCs is particularly compelling. The modeling approach is reasonably scaled for the system at the level of detail that is needed in this case, and the hidden variable analysis provides some insight into how the anti-resonance works.
Weaknesses:
(1) The anti-resonance phenomenon has not been demonstrated using physiological Wnt ligands; however, I view this as only a minor weakness for an initial report of the phenomenon. The potential significance of the phenomenon for Wnt outweighs the amount of effort it would take to carry the demonstration further - testing different frequencies/duty cycles at the level of ligand stimulus using microfluidics could get quite involved, and would likely take quite some time. Adding some more discussion about how the time scales of ligand-receptor binding could play into the reduced model would further ameliorate this issue.
We thank the reviewer for this comment and the interesting suggestion to test the anti-resonance phenomenon with microfluidics. We agree that combining physiological Wnt ligands with microfluidic stimulation would go beyond the scope of this current study, though it is an interesting extension. One advantage of the optogenetic setup, as mentioned in the discussion, is that the Wnt stimulus can be turned off sharply. This allows us to test the output from perfectly square wave input profiles; in microfluidics, washing the sticky ligand off the cells might “smear” the effective input profile cells respond to.
We show in Supplement Fig. 6, that our reduced model matches the experimental data and that we would expect the antiresonance phenomenon as long as 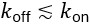 (see Fig. 4). Practically, a smeared input profile implies an effective reduction of 𝑘<sub>off</sub>, which means that the phenomenon would be visible with microfluidics (provided the minimum is deep enough, see Fig. 4). However, this should still be considered with caution, as the antiresonance would then appear because the cells essentially receive a smeared out or continuous pulse in the high frequency limit, rather than cells responding to a square wave in a specific way.
(see Fig. 4). Practically, a smeared input profile implies an effective reduction of 𝑘<sub>off</sub>, which means that the phenomenon would be visible with microfluidics (provided the minimum is deep enough, see Fig. 4). However, this should still be considered with caution, as the antiresonance would then appear because the cells essentially receive a smeared out or continuous pulse in the high frequency limit, rather than cells responding to a square wave in a specific way.
(2) While the model is fully consistent with the data, it has not been validated using experimental manipulations to establish that the mechanisms of the cell system and the model are the same. There may be some ways to make such modifications, for example, using a proteasome inhibitor. An alternative would be to more explicitly mention the need to validate the model's mechanism with experiments.
We thank the reviewer for this valuable and constructive comment. We agree that future experimental perturbations that directly modulate pathway activation and reset kinetics—such as proteasome inhibition, targeted degradation of pathway components, or engineered changes in receptor turnover—would provide an important validation of the model’s mechanistic interpretation. In the present study, our primary goal was to establish the existence and quantitative features of anti-resonance in the Wnt pathway and to identify the minimal set of timescale relationships that can explain it. We view the proposed experimental validations as exciting next steps that extend beyond the scope of the current work, and we are grateful to the reviewer for emphasizing their importance. We now mention this explicitly in the discussion of our manuscript.
(3) I think the manuscript misses an opportunity to discuss the potential of the phenomenon in other pathways. The hedgehog pathway, for example, involves GSK3-mediated partial proteolysis of a transcription factor, which could conceivably be subject to similar behaviors, and there are certainly other examples as well.
We thank the reviewer for pointing out an opportunity to emphasize the possibility of this phenomenon in other pathways. The minimal model indicates that anti-resonance emerges whenever a rapid activating process is paired with a slower deactivating/reset process. Beyond Hedgehog/Gli processing, candidate circuits include: NF-κB (rapid IκBα phosphorylation/degradation vs slower IκBα resynthesis), ERK (fast phosphorylation bursts vs slower transcriptional negative feedback such as DUSPs), Notch (fast γ-secretase NICD release vs slower NICD turnover and feedback), BMP/TGF-β–SMAD (fast R-SMAD phosphorylation vs slower receptor trafficking/SMAD7 feedback), and Hippo/YAP (rapid cytoplasmic sequestration vs slower transcriptional feedback). Each contains the same timescale separation that should create a frequency ‘stop-band,’ predicting suppressed gene expression or fate transitions at intermediate stimulation frequencies. We have updated the manuscript’s discussion to mention the Hedgehog connection with the following added sentence in the discussion: Analogous band-stop filtering should arise in other developmental circuits that couple a fast ‘ON’ step to slower deactivation or negative feedback. In Hedgehog, for example, PKA/CK1/GSK3-mediated partial proteolysis of Gli with slower recovery of full-length Gli creates the same fast-activation/slow-reset motif our hidden-variable model predicts will yield anti-resonance, and Wnt–Hedgehog crosstalk through the shared kinase GSK3 suggests such frequency selectivity could occur in other developmental signaling pathways.
We also added an additional sentence regarding different activation and deactivation timescales in other pathways.
(4) Some aspects of the modeling and hidden variable analysis are not optimally presented in the main text, although when considered together with the Supplemental Data, there are no significant deficiencies.
We have addressed the model choices and analysis now more clearly in the main manuscript and also referred to the Supplemental Data more directly.
Reviewer #2 (Public review):
Summary:
By combining optogenetics with theoretical modelling, the authors identify an anti-resonance behavior in the WnT signaling pathway. This behavior is manifested as a minimal response at a certain stimulation frequency. Using an abstracted hidden variable model, the authors explain their findings by a competition of timescales. Furthermore, they experimentally show that this anti-resonance influences the cell fate decision involved in human gastrulation.
Strengths:
(1) This interdisciplinary study combines precise optogenetic manipulation with advanced modelling.
(2) The results are directly tested in two different systems: HEK293T cells and H9 human embryonic stem cells.
(3) The model is implemented based on previous literature and has two levels of detail: i) a detailed biochemical model and ii) an abstract model with a hidden parameter.
Weaknesses:
(1) While the experiments provide both single-cell data and population data, the model only considers population data.
We thank the reviewer for correctly pointing out that the single-cell measurements would in principle allow us to incorporate the cell-to-cell heterogeneity into the model. In this study, we sought to identify a minimal quantitative model of the Wnt pathway that could explain anti-resonance through competing time scales. We believe that, for our purposes, focusing on population data allowed us to keep the complexity of the model to a minimum to increase its explanatory value. We agree with the reviewer that considering single-cell trajectories is an interesting direction for further work.
(2) Although the model captures the experimental data for TopFlash very well, the beta-Cat curves (Figure 2B) are only described qualitatively. This discrepancy is not discussed.
Indeed, our model fits to mean β-catenin expressions are more qualitative than for TopFlash. The fit for β-catenin was tricky, as expression of β-catenin is typically low and closer to the detectable limits than TopFlash. These experimental constraints mean that the variation between individual signal trajectories is higher for β-catenin compared to the light-off condition than for TopFlash. Therefore, we strove to obtain a qualitative rather than a quantitative fit to the mean expression profile in β-catenin. The current model fit is well within the standard deviation of variation. Given the observed heterogeneity and the fact that we take the parameters from literature (which ensures that the order of magnitude of parameters is in a sensible range), we believe that the model fits are reasonable. We now mention this explicitly in the text.
Overall Assessment:
The authors convincingly identified an anti-resonance behavior in a signaling pathway that is involved in cell fate decisions. The focus on a dynamic signal and the identification of such a behavior is important. I believe that the model approach of abstracting a complicated pathway with a hidden variable is an important tool to obtain an intuitive understanding of complicated dependencies in biology. Such a combination of precise ontogenetic manipulation with effective models will provide a new perspective on causal dependencies in signaling pathways and should not be limited only to the system that the authors study.
We thank both reviewers for the positive assessment of our manuscript.
Recommendations for the authors:
Reviewer #1 (Recommendations for the authors):
There are several points that deserve more discussion, as noted above in the review.
(1) It would be worthwhile to consider whether a relatively simple experiment with a proteasome inhibitor or similar pharmacological manipulation could provide useful validation data for the model.
We address this point above in the weaknesses section from reviewer 1.
(2) The figure legend for S5C should clarify whether the values plotted are at a particular fixed time point, or (more likely) at a certain time following the second pulse, which would be variable.
We have modified the figure caption to clarify that the values plotted are at a fixed time point in the simulation (t\=48 hrs). We chose this timepoint sufficiently long after the second pulse to ensure that there are no residual dynamical effects. We thank the reviewer for noting this.
(3) As noted in the Sci Score document, various aspects of the resource reporter should be improved, such as including RRIDs, etc.
We are sending out our plasmids to AddGene; versions for Python and Matlab are listed in our methods section.
Reviewer #2 (Recommendations for the authors):
I mostly have suggestions to improve the clarity of the presentation.
(1) Not all symbols in the equations given in the main text are explained. This is rather annoying, because either you present them and explain what they are or you don't show them and refer to the supplements. For example, d_0 or c_o or \bar{b} or n or K are not explained.
We have now more clearly presented the parameters in the main text and added signposts to the Methods section.
(2) Overall, it is often not clear what data in the figures are redundant, although the authors referred to them in the text. For example, in Figure 2c, a curve for 24 hours is shown and referred back to Figure 1D. However, in Figure 1D there is no curve for 24 hours. Is the data from Supplementary Figure 1 H and K also in the main text?
We thank the referee for pointing out these redundancies. We have now included the 24hr line in Figure 1D and are now only showing the unsmoothed data, also in the main text of the manuscript. To clarify supplemental figures, we have now removed S1H and S1K since all they showed was the unsmoothed version of the data. The remaining plots in Supplementary Figure 1 are normalized differently from what we show in Figure 1 to demonstrate our choice of normalization is not the reason for the observed optogenetic response.
Next Generation EIF: official kick‑off with Member States The Chair of the independent expert group on next generation EIF, Emilija Stojmenova Duh, presented its recommendations for an EIF blueprint, which will feed into the future EIF revision and proposal by the Board.
EIF proposal presented. Decision q1 2026?
The labelled solutions are: Core Vocabularies – common semantic building blocks that help public administrations describe key concepts (such as persons, organisations or locations) in a consistent way across borders and sectors. DCAT-AP – a common specification that makes data catalogues interoperable and helps make public sector data easier to discover and reuse.
Two things labeled as interoperability solutions now. DCAT-AP and core vocabs. Odd as they're not 'solutions' but DCAT-AP is a standard. Risk of board repeating existing standardisation efforts?
Adam optimizers maintain two moving averages for each model parameter: the first moment (mean) of the gradients and the second moment (uncentered variance) of the gradients. In other words, Adam optimizers store two additional values for each single model parameter in memory.
additional weights that add up the memory.
Companies will not be obliged to use the European Business Wallets. The Regulation places obligations solely on public sector bodies to accept its core functions, while companies remain free to decide whether to adopt the wallets for their commercial operations or interactions with public authorities
wallets are voluntary, but PSBs must accept them. Asymmetry may impact adoption? ('costly to prepare but noone uses them')
In parallel, the Commission will work closely with Member States and the private sector to define the technical standards and requirements for European Business Wallets through ongoing efforts under the European Digital Identity Framework and in large-scale pilot projects funded under the Digital Europe Programme such as the WeBuild consortium.
technical standards and reqs not yet defined.
It is estimated that simplification efforts will save up to €5 billion in administrative costs between now and 2029. Moreover, the European Business Wallets should unlock at least EUR 160 billion in savings for businesses each year.
Savings projects between 2025 and 2029, odd bc it is only now in proposal. May well be 2027 before law is there, implementation another 2 yrs, so no real change until 2029/2030?
The technical architecture and features for the European Business Wallets will build on the one for the EU Digital Identity Wallets.
Business wallets will build on existing personal wallets tech and infra. What about existing entity identifiers (Digidentity e.g.)?
The European Business Wallets are one of the key deliverables of the Digital Package. The Commission proposes a regulation to establish those Wallets as a harmonised digital solution to ease the administrative burden and to enable companies and public sector bodies to identify, to authenticate and to exchange data in a secure and user-friendly way, with full legal effect across the European Union.
EU Business Wallets proposed as regulation in the 2025 work programma. So in first step of legislative process.
yet another rigid bureaucracy for tech governance
eternal tensions :-(
voluntary codes of conduct for information integrity
hasn't worked so far - accountability-washing
Still, even the UN recognizes that effective digital cooperation is not just about new portals or pacts, but about inclusive governance and values. In fact, the UN’s digital cooperation blueprint emphasizes a multi-stakeholder approach, calling for “open, free and secure” digital futures anchored in universal human rights. This implies that improv
I'm just concerned that all that the UN does is makes its "opinion" public knowing fair well that whatever they advocate and is not in line with the power players will be ignored
Over-reliance on AI could even undermine the very trust we seek to build
trust has been eroded to such an extent that some people might be looking for a "new type of trust" precisely using the Ai
But diplomacy is not an engineering problem, and treating it as such can be dangerous.
Certain people see everything as an analytical problem that can be reduced and solved
AI on the Global Stage: Powerful Support, Not a Peacemaker
the source of competitive advantage - clear advantage for the ones that can/ are willing to use it and for the ones who have access to more advanced Ai
The Lure and Limits of Tech-Driven Solutions
An important reason is maybe the fact that most people in high positions are of the "analytical" type and with considerable technological background. That pre-disposes them to look fro solutions within their area of expertise and in line with their mental wiring.
eLife Assessment
Following retinal injury, zebrafish Müller glia reenter the cell cycle and generate replacement cells; this potentially valuable study proposes that injury induces a cxcl18b+ transitional state in Müller cells, which then express nitric oxide, inhibiting Notch signaling and allowing Müller glial cells to reenter the cell cycle. However, the evidence supporting the claims is incomplete, and the authors have made interpretations and conclusions that are not supported by the data. Questions of the temporal expression and function of cxcl18b, as well as the source of potential inflammatory cues before cxcl18b expression, remain unanswered and technical limitations and data inconsistencies raise concerns. Using larval animals complicates the analysis since the retina is still forming, and distinguishing between injury-induced regeneration and ongoing development is complex. With more rigorous testing of the signaling pathways proposed and a clear demonstration of their interdependence, the link between nitric oxide signaling and Notch activity, particularly, would interest those investigating retinal regeneration.
Reviewer #1 (Public review):
Summary:
This study presents a valuable contribution of NO signaling in zebrafish retinal regeneration in larval animals. The data on NO signaling are solid. There are multiple limitations to the study, but these are largely acknowledged by the authors in the revised text.
Strengths:
New data on NO signaling is valuable to the field but may be limited to larval "regeneration".
Weaknesses:
A weakness of the approach is testing cone ablation and regeneration in early larval animals. A near identical study was already done by Hoang et al 2020 in the adult zebrafish, a more relevant biological timepoint.
Reviewer #2 (Public review):
Summary:
In this manuscript Ye at al. examine the sequence of events that occur in the damaged zebrafish Muller glia (MG) in states between quiescence and the onset of proliferation. Using an inducible metronidazole (MTZ) and nitroreductase system to ablate red/green cones in larval zebrafish, they identify a novel transitional MG state that is characterized by the expression of cxcl18b. Using trajectory analysis from single-cell RNA-seq datasets, they find that cxcl18b is expressed before MG expression PCNA and become proliferative. They find that cxcl18b expression peaks in MG at approximately 24 hours post injury (hpi) and rapidly declines as MG proliferate following injury. In a most interesting finding, the authors find a link between nos2b-dependent nitric oxide signaling and cxcl18b-mediated proliferation. Mutagenesis of nos2b decreases MG proliferation. The mechanism linking NO signaling to proliferation was suggested to function via notch signaling as pharmacological inhibition of nitric oxide signaling resulted in elevated Notch activity, thus preventing MG proliferation. The authors suggest a model whereby cxcl18b induces autocrine NO signaling in MG to reduce activity of Notch3, thereby promoting MG proliferation.
Strengths:
The authors utilize a number of sophisticated transgenic approaches and generate novel lines that will have value to the field. The identification of a novel cxcl18b transition state is exciting and the putative link between NO signaling and Notch activity would provide new insight into the drivers of Muller glia proliferation.
Weaknesses:
While the overall model is appealing and may serve as a foundation for future studies, some information gaps remain and certain conclusions rely on correlational data. The cellular expression of nos2b remains unclear as the single-cell RNA-seq data cannot provide expression data that matches RT-PCR results. The temporal sequence of events are based on transgene expression in the Tg(cxcl18b:GFP) lines, where persistence of the GFP fluorescence may not reflect endogenous cxcl18b. The identity of putative cxcl18b receptors on MG to support an autocrine signaling pathway remains unclear. Nevertheless, this is an interesting study that should open new avenues of exploration.
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1:
(1) The authors state that more is known about glial reactivation than cell-cycle re-entry. They are confusing many points here. More gene networks that require cell-cycle re-entry are known. Some of the genes listed for "reactivation" are, in fact, required for cell cycle re-entry/proliferation. And the authors confuse gliosis vs glial reactivation.
We thank the reviewer for this important and constructive comment. We fully agree that clearly distinguishing between the concepts of glial reactivation, glial proliferation, gliosis, and neurogenesis is essential to avoid conceptual confusion in our study.
Injury-induced retinal regeneration in zebrafish:
Glial reactivation refers to the initial response of quiescent Müller glia (MG) to injury, characterized by morphological changes and upregulation of reactive markers (e.g., gfap, ascl1a, lin28a) and activation of signaling pathways such as Notch, Jak/Stat, and Wnt (Lahne et al., 2020; Pollak et al., 2013; Sifuentes et al., 2016; Yao et al., 2016).
Glial proliferation refers to the clonal expansion of these MG-derived progenitor cells, which undergo rapid cell-cycle re-entry and amplify to generate sufficient progenitors for regeneration (Iribarne and Hyde, 2022; Lee et al., 2024; Wan and Goldman, 2016)
Gliosis vs neurogenesis represents a divergent fate decision following proliferation. In zebrafish, MG-derived progenitor cells differentiate into retinal neurons that can replace those damaged or lost due to retinal injury. In contrast, mammalian MG tend to undergo an initial gliotic surge and rapidly revert to a quiescent state, exhibiting gliosis and glial scarring (Thomas et al., 2016; Yin et al., 2024). Thus, we totally agreed that gliosis cannot be confused with glial reactivation because glial reactivation is the very first step of glial injury responses, whereas gliogensis is the very last glial response to the injury.
We agree with the reviewer that many genes typically described as “reactivation markers” (e.g., ascl1a, lin28a, sox2, mycb, mych) are also essential regulators of cell-cycle re-entry (Gorsuch et al., 2017; Hamon et al., 2019; Lee et al., 2024; Lourenço et al., 2021; Pollak et al., 2013; Thomas et al., 2016). Because the glial reactivation is a leading event for glial proliferation, the regulators of glial reactivation are expected to be responsible for glial proliferation as well.
In our study, we focused on the states preceding glial proliferation to understand the mechanism underlying injury-induced glial cell-cycle re-entry. We defined these transitional states and the subsequent proliferative MG states based on single-cell RNA-seq trajectory analysis. (revised lines: 41-58)
(2) A major weakness of the approach is testing cone ablation and regeneration in early larval animals. For example, cones are ablated starting the day that they are born. MG that are responding are also very young, less than 48 hrs old. It is also unclear whether the immune response of microglia is a mature response. All of these assays would be of higher significance if they were performed in the context of a mature, fully differentiated, adult retina. All analysis in the paper is negatively affected by this biological variable.
We thank the reviewer for raising this important point regarding the developmental stage of the retina in our model system. We have carefully considered this concern and now provide additional clarification and justification, as follows:
(1) The glial responses in larval and adult retina:
Previous studies have demonstrated that injury-induced glial responses are largely conserved in larval and adult zebrafish retina, including reactive gliosis marked by gfap upregulation and proliferation(Meyers et al., 2012; Sarich et al., 2025). In our study, G/R cones were ablated beginning at 5 dpf using metronidazole (MTZ), and we observed robust induction of PCNA⁺ MG in the inner nuclear layer, consistent with injury-induced proliferation (Figure 1E). These findings align with previous studies showing that key features of MG regenerative responses are conserved across larval and adult stages.
(2) The microglial responses in larval and adult retina:
Retinal microglia functionally mature at 5 dpf in the zebrafish retina (Mazzolini et al., 2020; Svahn et al., 2013), and prior studies have demonstrated that microglia in larval and adult zebrafish exhibit similar responses to injury, including migration, morphological activation, and phagocytosis(Nagashima and Hitchcock, 2021; White et al., 2017). In our experiments using Tg(mpeg1: GFP) larvae, we observed clear microglial recruitment to the outer nuclear layer (ONL) following cone ablation (Figure 1E and Figure 1-figure supplement 1A), supporting the functional competence of larval microglia in injury-induced immune responses
(3) The contribution using larval animals to study the regeneration program:
We agree that regeneration studies in the adult retina can provide important biological insights, particularly in a fully differentiated tissue environment. Accordingly, we have acknowledged this limitation in our revised manuscript “limitations of this study” section (revised lines 534-540: “1. Our study focuses on larval zebrafish, in which the core features of MG and immune responses are conserved compared to the adult. However, we acknowledge that the adult retina—with its fully matured differentiated retina and immune response—provides irreplaceable biological insight. Nevertheless, larval models offer a powerful platform to uncover conserved regenerative mechanisms and serve as a valuable complement for identifying age-dependent differences in MG-mediated regeneration.”) and have stated our intention to extend future analyses to adult zebrafish, especially to explore age-dependent differences in redox signaling and MG proliferation. At the same time, we believe that the larval model offers unique advantages for uncovering fundamental, conserved mechanisms of regeneration and enables characterization of age-dependent regulatory differences. Thus, our study in larval animals serves as a complementary and informative platform for understanding both the conserved and developmental stage-specific features of injury-induced regeneration.
(4) Related to the above point, the clonal analysis of cxcl18b+ MG is complicated by the fact that new MG are still being born in the CMZ (as are new cones for that matter).
We thank the reviewer for raising this important point regarding potential contributions from CMZ-derived progenitors to the lineage-traced cxcl18b⁺ MG clones. To address this concern, we have implemented evidence to rule out a CMZ origin for the clones analyzed:
Spatial restriction of clones: All clones included in our analysis were located exclusively within the central and dorsal retina, as shown in Figure 2H. From the spatial distribution of reactive MG populations across the retina, we observed a patterned organization in which the vast majority of proliferating MG arose from local mature MG–derived progenitors, rather than from peripheral CMZ-derived progenitors. However, we acknowledge that we cannot entirely exclude the possibility that CMZ-derived progenitors contribute to injury-induced MG proliferation, particularly in the peripheral retina.
We have clarified this point in the revised Methods section (revised lines 756–762: “Clone analysis of cxcl18b<sup>+</sup> lineage-traced MG was restricted to cells located in the central and dorsal region of the zebrafish retina after G/R cone ablation in Figure 2, Figure 6, and their figure supplement. This spatial restriction strongly suggests that the proliferative MG originate from local mature MG, although we cannot completely rule out the possibility that CMZ-derived progenitors contribute to the generation of proliferative MG in the peripheral retina.”) and updated the corresponding figure legends.
(4) A near identical study was already done by Hoang et al., 2020, in adult zebrafish, a more relevant biological timepoint. Did the authors check this published RNA-seq database for their gene(s) of interest?
We thank the reviewer for pointing out the relevance of the study by Hoang et al., 2020, which characterized the transcriptional dynamics of MG reactivation in the adult zebrafish retina. We agree that comparisons with their single-cell RNA-seq dataset are important to confirm the conservation of our findings in larval vs adult zebrafish.
To this end, we examined the adult zebrafish MG dataset reported by Hoang et al., and confirmed that cxcl18b is also present and enriched in their analysis, particularly in activated MG populations under various injury paradigms:
(1) cxcl18b is listed as a differentially expressed gene (DEG) in Supplementary Table ST2, enriched in GFP⁺ MG following injury. It is also significantly upregulated in both NMDA-induced and light damage conditions, as shown in Supplementary Table ST3.
(2) In Supplementary Table ST5, cxcl18b is identified as a classifier of activated MG, with classification power scores of 0.552 (NMDA), 0.632 (light damage), and 0.574 (TNFα + γ-secretase inhibitor treatment), indicating its consistent expression across multiple injury models.
(3). In their pseudotime analysis (Figure 4C and Supplementary Table ST8), cxcl18b is specifically expressed in Module 5, which is expressed earlier along the trajectory than ascl1a. This temporal pattern of cxcl18b preceding ascl1a expression is consistent with our trajectory analysis in larval MG (Figure 1H), further supporting its role as an early marker of the transitional state before proliferation.
These findings underscore the robustness and biological relevance of cxcl18b as a conserved marker of injury-responsive MG in both larval and adult zebrafish. Our data expand upon the prior work by specifically characterizing a cxcl18b-defined transitional MG state preceding cell-cycle re-entry, thereby offering additional insights into the temporal staging of MG activation during regeneration.
(5) KD of cxcl18b did not affect MG proliferation or any other defined outcome. And yet the authors continually state such phrases as "microglia-mediated inflammation is critical for activating the cxcl18b-defined transitional states that drive MG proliferation." This is false. Cxcl18b does not drive MG proliferation at all.
We thank the reviewer for raising this concern. We agree with the reviewer and have revised this statement as "These findings suggest that microglia-mediated inflammation may contribute to the activation of cxcl18b-defined transitional states that precede MG proliferation, although a causal relationship remains to be established." (revised lines 251-253).
(6) A technical concern is that intravitreal injections are not routinely performed in larval fish.
We appreciate the reviewer’s technical concern regarding the use of intravitreal injections in larval zebrafish. In our study, we performed intraocular injection according to previously established methods (Alvarez et al., 2009; Giannaccini et al., 2018; Rosa et al., 2023). This approach involves carefully delivering a small volume of viral suspension into the intraocular space by a glass micropipette. To address this concern, we will revise the Materials and Methods section to clearly describe the injection procedure and will cite the relevant references accordingly.
Reviewer #2:
(1) The authors note a peak of PCNA+ Muller glia at 72 hours post injury. This is somewhat surprising as the MG would be expected to generate progenitor cells that would continue proliferating and stain with PCNA. Indeed, only a handful of PCNA+ cells are seen in the INL/ONL layer in Figure 1E2 with few clusters of progenitors present. It would be helpful to stain with a Muller glia marker to confirm these PCNA+ cells are Muller glia. It's also curious that almost all the PCNA+ cells are in the dorsal retina, even though G/R cone loss extends across both dorsal and ventral retina. Is this typical for cone ablation models in larval zebrafish?
We thank the reviewer for their insightful comment regarding the spatial distribution and identity of PCNA⁺ cells following injury.
In our study, we observed that the injury-induced proliferating cells (PCNA⁺) were predominantly located in the central and dorsal regions of the retina at 72 hours post-injury (hpi) (Figure 1E). To verify the identity of these proliferating cells, we performed additional immunostaining using BLBP, and confirmed that the majority of PCNA⁺ cells also express BLBP (Figure 1–figure supplement 1B in our revised Data), these results supporting their MG origin.
The regional bias of MG proliferation towards the central and dorsal retina is consistent with previous findings. Notably, (Krylov et al., 2023) demonstrated that MG exhibit region-specific heterogeneity in their regenerative responses to photoreceptor ablation. Their study identified proliferative MG subpopulations predominantly in the central (fgf24-expressing) and dorsal (efnb2a-expressing) domains, whereas ventral MG showed limited proliferative capacity (Krylov et al., 2023). These observations provide a plausible explanation for the spatially restricted PCNA⁺ MG population observed in our model following cone ablation.
(2) In Line 148: What is meant by "most original MG states" in this context? Original meaning novel? Or original meaning the earliest state MG adopted following injury? The language here is confusing.
We thank the reviewer for pointing out the ambiguous phrasing in our original manuscript. The term “most original MG states” was imprecise and misleading, as it could be interpreted as referring to the quiescent state of MG. In our context, we intended to describe the earliest transitional states in MG respond to injury, as they begin to exit quiescence and enter reactive characteristics. These early transitional MG populations co-express quiescent markers such as cx43 and early reactive markers gfap, as shown in Figure 1H.
To avoid confusion and improve conceptual clarity, we have revised the manuscript by replacing “most original MG states” with “early transitional MG state” (revised line 154) and have added a clearer explanation in the corresponding Results section to define this population more accurately.
(3) Perhaps provide a better image in Figure 2A of the cxcl18b at 48 hpi and 72 hpi. The current images appear virtually identical, with very little cxcl18b expression observed, especially compared to the 24 hpi. This is in contrast to the Tg(cxcl18b:GFP) transgenic line shown in Figure 2D, which indicates either much higher expression in proliferating cells at 48 hpi or the stability of GFP protein. Can the authors provide guidance on the accurate temporal expression of cxcl18b? Does expression peak rapidly at 24 hpi and then rapidly decline or is there persistence of expression to 48-72 hpi?
We appreciate the reviewer’s careful observation regarding the apparent similarity of cxcl18b expression at 48 hpi and 72 hpi in the in situ hybridization (ISH) images (Figure 2A), and the differences compared to the Tg(cxcl18b: GFP) reporter line shown in Figure 2D.
(1) The similarity of ISH images at the 48 hpi and 72 hpi (Figure 2A):
The cxcl18b mRNA signal peaked at 24 hpi, suggesting a rapid transcriptional response after retina injury. By 48 hpi, cxcl18b expression had already declined substantially, and by 72 hpi, the signal was further reduced to near-background levels. This temporal expression pattern explains why the ISH images at 48 hpi and 72 hpi appear nearly identical and much weaker compared to 24 hpi.
(2) The discrepancy between ISH and GFP reporter signal (Figure 2D):
The Tg(cxcl18b: GFP) reporter line shows persistent GFP expression beyond the transcriptional window of cxcl18b mRNA. This may be due to the prolonged stay of GFP protein, which remains detectable even after the endogenous transcription of cxcl18b has diminished. This explanation is also noted in the manuscript (revised lines 198–200). As a result, GFP⁺ MG cells are still visible at 48–72 hpi, and some of them co-label with PCNA.
These findings are consistent with our Pseudotime analysis based on scRNA-seq data (Figure 1H), which shows that cxcl18b expression precedes the induction of proliferative markers such as pcna and ascl1a.
(4) Line 198: The establishment of the Tg(cxcl18b:Cre-vhmc:mcherry::ef1a:loxP-dsRed-loxP-EGFP::lws2:nfsb-mCherry) is considerable but the nomenclature doesn't properly fit. Is the mCherry fused with Cre and driven by the cxcl18b promoter? What is the vhmc component? Finally, while this may provide the ability to clonally track cxcl18b-expressing MG, it does not address the prior question of what is the actual temporal expression of cxcl18b? If anything, this only addresses whether proliferating MG expressed cxcl18b at some point in their history, but does not indicate that cxcl18b expression co-exists in proliferating cells. The most convincing evidence is in Supplemental Figure 2B.
The "vmhc" component refers to the ventricular myosin heavy chain promoter, commonly used to label atrial cardiomyocytes (Jin et al., 2009). We cloned the vmhc upstream region containing its promoter and fusing with mCherry for selection during transgenic fish line construction.
Clone analysis using the Tg(cxcl18b: Cre-vmhc: mCherry::ef1a: loxP-DsRed-loxP-EGFP::lws2: nfsb-mCherry) further indicates that cxcl18b-defined the transitional state is the essential routing for MG proliferation. We have clarified in the revised text that this lineage tracing indicates a “history of injury-induced cxcl18b expression” rather than its ongoing expression during proliferation (revised line 205).
(5) Line 203: The data shown in Figure 2F do not indicate that these MG are cxcl18b+. Rather, the data are consistent with the interpretation that these MG expressed Cre at some prior stage and now express GFP from the ef1a promoter rather than DsRed. Whether these MG continue to express cxcl18b at the time these fish were collected is not addressed by these data. It is not accurate to conclude that these cells are cxcl18b+.
We thank the reviewer for pointing out this important issue. We agreed that the GFP<sup>+</sup> MG shown in Figure 2F represents cells that have previously expressed cxcl18b and thus belong to the cxcl18b-expressing cell lineage, but this does not indicate that they continue to express cxcl18b at the time of sample collection. Performing clonal analysis using the Cre-loxp system, the GFP signal reflects historical cxcl18b promoter activity rather than ongoing transcription. We have revised the relevant sentence in our manuscript to clarify this point and now refer to these GFP<sup>+</sup> cells as "cxcl18b lineage-traced MG" rather than "cxcl18b<sup>+</sup> MG" to avoid any misinterpretation (revised line 207).
(6) Line 213: The statement that proliferative MG mostly originated from cxcl18b+ MG transitional states is a conclusion that does appear fully supported by the data. Whether those MG continue to express cxcl18b remains unanswered by the data in Figure 2 and would likely be inconsistent with the single-cell data in Figure 1.
We thank the reviewer for this valuable comment. We agree that the original statement on Line 213 regarding the lineage relationship between cxcl18b⁺ transitional MG and proliferative MG required clarification.
(1) The cxcl18b expression dynamics:
Our single-cell RNA-seq and ISH analyses consistently show that cxcl18b expression peaks as early as 24 hpi and declines rapidly, with significantly reduced expression by 48 and 72 hpi. These findings suggest that cxcl18b marks an early transitional MG state, rather than being maintained in proliferative MG. Indeed, in our scRNA-seq pseudotime trajectory analysis (Figure 1H), cxcl18b expression is highest in early transitional MG clusters (Clusters 1) and downregulated as cells progress toward proliferative states (Clusters 3/6), supporting a model in which cxcl18b is downregulated before cell-cycle re-entry.
(2) Prolonged stability of GFP protein:
The GFP signal observed in Tg(cxcl18b: GFP) retinas at 72 hpi may be because of the prolonged stability of GFP protein, rather than sustained cxcl18b transcription. The actual expression dynamics of cxcl18b are more directly reflected by our in situ hybridization and single-cell RNA-seq data, both showing a rapid decline after its early peak at 24 hpi. This explanation is also noted in the manuscript (revised lines 196–197).
(7) Line 246: The use of Dexamethasone to block inflammation is a widely used approach. However, dexamethasone is a broad-spectrum anti-inflammatory molecule that works through glucocorticoid signaling that may involve more than microglia. The observation that microglia recruitment and cxcl18a expression are both reduced is correlative but does not prove causation. Thus, the data are not sufficient to conclude that microglia-mediated inflammation is critical for activating cxcl18b expression. Indeed, data in Figure 1 indicate that cxcl18b expression occurs prior to microglia migration to the ONL.
We thank the reviewer for this thoughtful and important comment. We fully acknowledge that dexamethasone is a broad-spectrum anti-inflammatory agent that acts via glucocorticoid receptor signaling and may influence multiple immune and non-immune pathways beyond microglia.
In our study, dexamethasone treatment led to a reduction in both microglial recruitment and the number of cxcl18b<sup>+</sup> MG at 72 hpi, suggesting a potential association between inflammation and cxcl18b activation. However, we agree that this observation remains correlative and is not sufficient to establish a direct link between microglia activity and cxcl18b induction. Our time-course analysis indicates that cxcl18b expression peaks at 24 hpi, preceding robust microglial accumulation in the ONL, further highlighting the need to clarify the temporal dynamics and cellular sources of inflammatory cues.
To address this question more conclusively, selective ablation of microglia during cone injury would be necessary. However, implementing such an approach would require a complex intersection of three transgenic lines—Tg(mpeg1: nfsB-mCherry) for microglia ablation, Tg(lws2: nfsB-mCherry) for cone ablation, and Tg(cxcl18b: GFP) for reporting—posing substantial genetic and experimental challenges.
We have revised the Results section accordingly to state: “These findings suggest that microglia-mediated inflammation may contribute to the activation of cxcl18b-defined transitional states that precede MG proliferation, although a causal relationship remains to be established.” (revised lines 251–253). We also added a new paragraph in the “Result: Clonal analysis reveals injury-induced MG proliferation via cxcl18b-defined transitional states associated with inflammation” as “While dexamethasone suppressed both microglial recruitment and cxcl18b<sup>+</sup> MG generation, its broad anti-inflammatory action precludes definitive conclusions about microglial causality. Dissecting this relationship would require concurrent ablation of microglia and cone photoreceptors using a triple-transgenic strategy, which is beyond the scope of the current study. Targeted approaches will be necessary to resolve the specific role of microglia in initiating cxcl18b expression.” (revised lines 251–258) to explicitly acknowledge this limitation and the need for future studies using microglia-specific ablation models to resolve the mechanism.
(8) Could the authors clarify the basis of investigating NO signaling, given the relative expression of the genes by either cxcl18b+ MG or uninjured MG? Based on the expression illustrated in Supplemental Figure 4A, there is almost no expression of nos1 or nos2b in any MG. The authors are encouraged to revisit the earlier single-cell data sets to identify those cells that express components of NO signaling to determine the source(s) of NO that could be impacting the Muller glia.
We thank the reviewer for raising these important points.
Nitric oxide (NO) signaling has been implicated in the regeneration of multiple zebrafish tissues, including the heart (Rochon et al., 2020; Yu et al., 2024), spinal cord (Bradley et al., 2010), and fin (Matrone et al., 2021). Based on these findings, we hypothesized that NO signaling might also contribute to retinal regeneration.
As described in the manuscript, we compiled a redox-related gene list and systematically screened their roles in injury-induced MG proliferation using CRISPR-Cas9-mediated gene disruption. Among the candidates, disruption of nos genes significantly reduced the number of PCNA<sup>+</sup> MG cells following G/R cone ablation (Figure 4), prompting us to further investigate the role of NO signaling.
(9) Line 319-320: this sentence appears to be missing text as "while no influenced across the nos mutants and gsnor mutants" does not make sense.
We appreciate the reviewer’s observation and agree that the original sentence was unclear. We have revised the sentence in the manuscript as follows:
“In contrast, no significant change in MG proliferation was observed in nos1, nos2a, or gsnor mutants compared to wild type (Figures 4F–4I)” (revised lines 326-328).
(10) Line 326-328: The text should be rewritten as the current meaning would suggest there was no significant loss of photoreceptors in the nos2b mutants. That is incorrect. Rather, there was no significant difference between WT and the nos2b mutants in the number of photoreceptors lost at 72 hpi following MTZ treatment. Both groups lost photoreceptors, but the number lost in nos2b hets and homozygotes was the same as WT.
We agree with the suggestion and have revised our manuscript. We have revised the sentence in the manuscript as follows:
“We observed no significant difference in the loss of cone photoreceptor at 72 hpi between nos2b mutants and WT, indicating that the reduced MG proliferation observed in nos2b mutants is independent of the injury (WT: 45 ± 8 remaining cones, n = 24; nos2b⁺/⁻: 49 ± 12, n = 20; nos2b⁻/⁻: 46 ± 9, n = 20; mean ± SEM) (Figure 4K).” (revised lines 331-335).
(11) There is concern over the inconsistencies with some of the data. In Figure 4, Supplement 1A, the single-cell data found virtually no expression of nos2b in either uninjured MG or cxcl18b+ MG. In contrast, the authors find nos2b expression by RT-PCR in the cxcl18b:GFP+ MG. The in situ expression of nos2b in Figure 5 - Supplement 1 is not persuasive. The red puncta are seen in a single cxcl18b:GFP+ cell but also in the plexiform layer and is other non cxcl18b:GFP+ cells.
We appreciate the concern regarding the apparent inconsistencies in nos2b expression across different datasets. We provide the following explanations:
(1) Low expression of nos2b in scRNA-seq data:
We propose a potential explanation: Nitric oxide (NO) signaling is known to exert its biological functions in a dose-dependent manner and is tightly regulated post-transcriptionally, especially in inducible nitric oxide synthase (iNOS) (Bogdan, 2001; Nathan and Xie, 1994; Thomas et al., 2008). Thus, even modest changes in nos2b expression may exert meaningful biological effects without producing strong transcriptional signals detectable by scRNA-seq, which could fall below the detection threshold of scRNA-seq methods. Supporting this idea, our functional assay (Figure 4J) reveals a clear concentration-dependent effect of NO on MG proliferation, consistent with the biological relevance of Nos2b activity despite its low transcript abundance.
(2) Regarding the in situ hybridization data:
We used both commercially available in situ hybridization probes from (HCR<sup>TM</sup>) and RNAscope<sup>TM</sup> (data not shown) to detect nos2b transcripts. While the nos2b signal was observed in other retinal cell types, including cells in the plexiform layer, our primary study was focused on examining its expression within the cxcl18b<sup>+</sup> MG lineage.
(3) Regarding RT-PCR detection of nos2b in cxcl18b: GFP<sup>+</sup> MG:
To enhance detection sensitivity, we enriched cxcl18b: GFP<sup>+</sup> MG by FACS at 72 hpi and performed cDNA amplification before RT-PCR. This approach allowed the detection of low-abundance transcripts such as nos2b. It is also important to note that RT-PCR reflects fold changes in expression compared to MG to other retina cell type. The subtle but biologically upregulated of nos2b expression may not be readily captured by in situ hybridization or scRNA-seq.
(12) Line 356 - there is a disagreement over the interpretation of the current data. The statement that nos2b was specifically expressed in cxcl18b+ transitional MG states is not entirely accurate. This conclusion is based on expression of GFP from a cxcl18b promoter, which may reflect persistence of the GFP protein and not evidence of cxcl18b expression. Even assuming that the nos2b in situ hybridization and RT-PCR data are correct, the data would indicate that nos2b is expressed in proliferating MG that are derived from the cxcl18b+ transitional states. The single-cell trajectory analysis in Figure 2 indicates that cxcl18b is not co-expressed with PCNA. Furthermore, the single-cell data in Figure 4, Supplement 1, indicates no expression of nos2b in cxcl18b+ MG. The authors need to reconcile these seemingly contradictory pieces of data.
We thank the reviewer for this thoughtful and important comment. We agree that clarification is needed to accurately interpret the relationship between cxcl18b, nos2b, and MG proliferation, particularly considering the different temporal and technical contexts of our datasets.
(1) Lineage labeling and interpretation of GFP expression:
We acknowledge that in the Tg(cxcl18b: Cre-vhmc: mcherry::ef1a: loxP-dsRed-loxP-EGFP::lws2: nfsb-mCherry) line, GFP expression reflects historical activity of the cxcl18b promoter, rather than ongoing transcription. This GFP signal, due to its prolonged stay, may persist beyond the time window of endogenous cxcl18b expression. Accordingly, we have revised the manuscript to replace “cxcl18b⁺ MG” with “cxcl18b⁺ lineage-traced MG” throughout the relevant sections to prevent potential misinterpretation.
(2) Functional experiments support a lineage relationship between cxcl18b⁺ states and nos2b activity:
To further investigate the regulatory relationship between cxcl18b and nos2b, we conducted NO scavenging experiments using C-PTIO in the Tg(cxcl18b: GFP) background. We observed that the generation of cxcl18b: GFP⁺ MG after injury was not affected by NO depletion, indicating that cxcl18b activation precedes NO signaling (data not shown). However, PCNA⁺ MG was significantly reduced under the same treatment, suggesting that NO signaling is not required for cxcl18b⁺ transitional state formation, but is necessary for proliferation. Together with our MG-specific nos2b knockout data, these results support a model in which nos2b-derived NO acts downstream of the cxcl18b⁺ transitional state to promote MG cell-cycle re-entry.
(3) The scRNA-seq data with nos2b expression:
We agree with the reviewer that our scRNA-seq dataset shows minimal overlap between cxcl18b and pcna expression, which is consistent with our interpretation that cxcl18b expression marks a transitional phase before cell-cycle entry. Furthermore, nos2b transcripts were not robustly detected in cxcl18b⁺ MG clusters in our scRNA dataset. This discrepancy may be caused by technical limitations of scRNA-seq in capturing low-abundance or transient transcripts such as nos2b, as discussed in response to comment #11.
(13) The data in Figure 7 are interesting and suggest a link between NO signaling and notch activity. The use of the C-PTIO NO scavenger is not specific to MG, which limits the conclusions related to autocrine NO signaling in cxcl18b+ MG.
We acknowledge that the use of C-PTIO cannot distinguish between NO signaling within MG and paracrine effects from other retinal cells. Currently, technical limitations prevent MG-specific NO depletion. We have discussed this limitation accordingly in our revised “Limitations of this study” section (revised lines 540-545: “2. While our data suggest that injury-induced NO suppresses Notch signaling activation and promotes MG proliferation, the use of a general NO scavenger (C-PTIO) does not allow us to determine whether this regulation occurs in an autocrine or paracrine manner. The specific role of NO signaling within cxcl18b⁺ MG requires further validation using MG-specific NO depletion.”)
(14) Line 446-448. As mentioned before, the data do not support a causative link between microglia recruitment and cxcl18b induction. More specifically, dexamethasone is a broad-spectrum anti-inflammatory drug that blocks microglia activation and recruitment. Critically, the authors demonstrate that expression of cxcl18b occurs prior to microglia recruitment (see Figure 1, Supplement 1). Thus, the statement that cxcl18b induction depends on microglia recruitment is not accurate.
We thank the reviewer for reiterating this important point. We fully agree that the current data do not support a direct causal relationship between microglia recruitment and cxcl18b induction. As also addressed in our response to Comment 7, dexamethasone, as a broad-spectrum anti-inflammatory agent, cannot distinguish microglia-specific effects from those of other immune components. We have revised the text in revised lines 251–258 to clarify that microglia-mediated inflammation is associated with—but not required for—activation of cxcl18b-defined transitional MG states.
Reference:
Bogdan, C. (2001). Nitric oxide and the immune response. Nature immunology 2, 907-916.
Bradley, S., Tossell, K., Lockley, R., and McDearmid, J.R. (2010). Nitric oxide synthase regulates morphogenesis of zebrafish spinal cord motoneurons. The Journal of neuroscience : the official journal of the Society for Neuroscience 30, 16818-16831.
Gorsuch, R.A., Lahne, M., Yarka, C.E., Petravick, M.E., Li, J., and Hyde, D.R. (2017). Sox2 regulates Müller glia reprogramming and proliferation in the regenerating zebrafish retina via Lin28 and Ascl1a. Experimental eye research 161, 174-192.
Hamon, A., García-García, D., Ail, D., Bitard, J., Chesneau, A., Dalkara, D., Locker, M., Roger, J.E., and Perron, M. (2019). Linking YAP to Müller Glia Quiescence Exit in the Degenerative Retina. Cell reports 27, 1712-1725.e1716.
Iribarne, M., and Hyde, D.R. (2022). Different inflammation responses modulate Müller glia proliferation in the acute or chronically damaged zebrafish retina. Frontiers in cell and developmental biology 10, 892271.
Jin, D., Ni, T.T., Hou, J., Rellinger, E., and Zhong, T.P. (2009). Promoter analysis of ventricular myosin heavy chain (vmhc) in zebrafish embryos. Developmental dynamics : an official publication of the American Association of Anatomists 238, 1760-1767.
Krylov, A., Yu, S., Veen, K., Newton, A., Ye, A., Qin, H., He, J., and Jusuf, P.R. (2023). Heterogeneity in quiescent Müller glia in the uninjured zebrafish retina drive differential responses following photoreceptor ablation. Frontiers in molecular neuroscience 16, 1087136.
Lahne, M., Nagashima, M., Hyde, D.R., and Hitchcock, P.F. (2020). Reprogramming Müller Glia to Regenerate Retinal Neurons. Annual review of vision science 6, 171-193.
Lee, M.S., Jui, J., Sahu, A., and Goldman, D. (2024). Mycb and Mych stimulate Müller glial cell reprogramming and proliferation in the uninjured and injured zebrafish retina. Development (Cambridge, England) 151.
Lourenço, R., Brandão, A.S., Borbinha, J., Gorgulho, R., and Jacinto, A. (2021). Yap Regulates Müller Glia Reprogramming in Damaged Zebrafish Retinas. Frontiers in cell and developmental biology 9, 667796.
Matrone, G., Jung, S.Y., Choi, J.M., Jain, A., Leung, H.E., Rajapakshe, K., Coarfa, C., Rodor, J., Denvir, M.A., Baker, A.H., et al. (2021). Nuclear S-nitrosylation impacts tissue regeneration in zebrafish. Nat Commun 12, 6282.
Mazzolini, J., Le Clerc, S., Morisse, G., Coulonges, C., Kuil, L.E., van Ham, T.J., Zagury, J.F., and Sieger, D. (2020). Gene expression profiling reveals a conserved microglia signature in larval zebrafish. Glia 68, 298-315.
Meyers, J.R., Hu, L., Moses, A., Kaboli, K., Papandrea, A., and Raymond, P.A. (2012). β-catenin/Wnt signaling controls progenitor fate in the developing and regenerating zebrafish retina. Neural development 7, 30.
Nagashima, M., and Hitchcock, P.F. (2021). Inflammation Regulates the Multi-Step Process of Retinal Regeneration in Zebrafish. Cells 10.
Nathan, C., and Xie, Q.W. (1994). Nitric oxide synthases: roles, tolls, and controls. Cell 78, 915-918.
Pollak, J., Wilken, M.S., Ueki, Y., Cox, K.E., Sullivan, J.M., Taylor, R.J., Levine, E.M., and Reh, T.A. (2013). ASCL1 reprograms mouse Muller glia into neurogenic retinal progenitors. Development (Cambridge, England) 140, 2619-2631.
Rochon, E.R., Missinato, M.A., Xue, J., Tejero, J., Tsang, M., Gladwin, M.T., and Corti, P. (2020). Nitrite Improves Heart Regeneration in Zebrafish. Antioxidants & redox signaling 32, 363-377.
Sarich, S.C., Sreevidya, V.S., Udvadia, A.J., Svoboda, K.R., and Gutzman, J.H. (2025). The transcription factor Jun is necessary for optic nerve regeneration in larval zebrafish. PloS one 20, e0313534.
Sifuentes, C.J., Kim, J.W., Swaroop, A., and Raymond, P.A. (2016). Rapid, Dynamic Activation of Müller Glial Stem Cell Responses in Zebrafish. Investigative ophthalmology & visual science 57, 5148-5160.
Svahn, A.J., Graeber, M.B., Ellett, F., Lieschke, G.J., Rinkwitz, S., Bennett, M.R., and Becker, T.S. (2013). Development of ramified microglia from early macrophages in the zebrafish optic tectum. Developmental neurobiology 73, 60-71.
Thomas, D.D., Ridnour, L.A., Isenberg, J.S., Flores-Santana, W., Switzer, C.H., Donzelli, S., Hussain, P., Vecoli, C., Paolocci, N., Ambs, S., et al. (2008). The chemical biology of nitric oxide: implications in cellular signaling. Free radical biology & medicine 45, 18-31.
Thomas, J.L., Ranski, A.H., Morgan, G.W., and Thummel, R. (2016). Reactive gliosis in the adult zebrafish retina. Experimental eye research 143, 98-109.
Wan, J., and Goldman, D. (2016). Retina regeneration in zebrafish. Current opinion in genetics & development 40, 41-47.
White, D.T., Sengupta, S., Saxena, M.T., Xu, Q., Hanes, J., Ding, D., Ji, H., and Mumm, J.S. (2017). Immunomodulation-accelerated neuronal regeneration following selective rod photoreceptor cell ablation in the zebrafish retina. Proceedings of the National Academy of Sciences of the United States of America 114, E3719-e3728.
Yao, K., Qiu, S., Tian, L., Snider, W.D., Flannery, J.G., Schaffer, D.V., and Chen, B. (2016). Wnt Regulates Proliferation and Neurogenic Potential of Müller Glial Cells via a Lin28/let-7 miRNA-Dependent Pathway in Adult Mammalian Retinas. Cell reports 17, 165-178.
Yin, Z., Kang, J., Xu, H., Huo, S., and Xu, H. (2024). Recent progress of principal techniques used in the study of Müller glia reprogramming in mice. Cell regeneration (London, England) 13, 30.
Yu, C., Li, X., Ma, J., Liang, S., Zhao, Y., Li, Q., and Zhang, R. (2024). Spatiotemporal modulation of nitric oxide and Notch signaling by hemodynamic-responsive Trpv4 is essential for ventricle regeneration. Cellular and molecular life sciences : CMLS 81, 60.
eLife Assessment
This important study focuses on the molecular mechanisms underlying the generation of neuronal diversity. Taking advantage of a well-defined neuroblast lineage in Drosophila, the authors provide convincing evidence that two transcription factors of the conserved forkhead box (FOX) family provide a mechanistic link between transient spatial cues that initially specify neuroblast identity and terminal selector genes that define post-mitotic neuron identity. The findings will be of interest to developmental neurobiologists.
Reviewer #1 (Public review):
Summary:
Lai and Doe address the integration of spatial information with temporal patterning and genes that specify cell fate. They identify the Forkhead transcription factor Fd4 as a lineage-restricted cell fate regulator that bridges transient spatial transcription factors to terminal selector genes in the developing Drosophila ventral nerve cord. The experimental evidence convincingly demonstrates that Fd4 is both necessary for late-born NB7-1 neurons, but also sufficient to transform other neural stem cell lineages toward the NB7-1 identity. This work addresses an important question that will be of interest to developmental neurobiologists: How can cell identities defined by initial transient developmental cues be maintained in the progeny cells, even if the molecular mechanism remains to be investigated? In addition, the study proposes a broader concept of lineage identity genes that could be utilized in other lineages and regions in the Drosophila nervous system and in other species.
Strengths:
While the spatial factors patterning the neuroepithelium to define the neuroblast lineages in the Drosophila ventral nerve cord are known, these factors are sometimes absent or not required during neurogenesis. In the current work, Lai and Doe identified Fd4 in the NB7-1 lineage that bridges this gap and explains how NB7-1 neurons are specified after Engrailed (En) and Vnd cease their expression. They show that Fd4 is transiently co-expressed with En and Vnd and is present in all nascent NB7-1 progenies. They further demonstrate that Fd4 is required for later-born NB7-1 progenies and sufficient for the induction of NB7-1 markers (Eve and Dbx) while repressing markers of other lineages when force-expressed in neural progenitors, e.g., in the NB5-6 lineage and in the NB7-3 lineage. They also demonstrate that, when Fd4 is ectopically expressed in NB7-3 and NB5-6 lineages, this leads to the ectopic generation of dorsal muscle-innervating neurons. The inclusion of functional validation using axon projections demonstrates that the transformed neurons acquire appropriate NB7-1 characteristics beyond just molecular markers. Quantitative analyses are thorough and well-presented for all experiments.
Weaknesses:
(1) While Fd4 is required and sufficient for several later-born NB7-1 progeny features, a comparison between early-born (Hb/Eve) and later-born (Run/Eve) appears missing for pan-progenitor gain of Fd4 (with sca-Gal4; Figure 4) and for the NB7-3 lineage (Figure 6). Having a quantification for both could make it clearer whether Fd4 preferentially induces later-born neurons or is sufficient for NB7-1 features without temporal restriction.
(2) Fd4 and Fd5 are shown to be partially redundant, as Fd4 loss of function alone does not alter the number of Eve+ and Dbx+ neurons. This information is critical and should be included in Figure 3.
(3) Several observations suggest that lineage identity maintenance involves both Fd4-dependent and Fd4-independent mechanisms. In particular, the fact that fd4-Gal4 reporter remains active in fd4/fd5 mutants even after Vnd and En disappear indicates that Fd4's own expression, a key feature of NB7-1 identity, is maintained independently of Fd4 protein. This raises questions about what proportion of lineage identity features require Fd4 versus other maintenance mechanisms, which deserves discussion.
(4) Similarly, while gain of Fd4 induces NB7-1 lineage markers and dorsal muscle innervation in NB5-6 and NB7-3 lineages, drivers for the two lineages remain active despite the loss of molecular markers, indicating some regulatory elements retain activity consistent with their original lineage identity. It is therefore important to understand the degree of functional conversion in the gain-of-function experiments. Sparse labeling of Fd4 overexpressing NB5-6 and NB7-3 progenies, as was done in Seroka and Doe (2019), would be an option.
(5) The less-penetrant induction of Dbx+ neurons in NB5-6 with Fd4-overexpression is interesting. It might be worth the authors discussing whether it is an Fd4 feature or an NB5-6 feature by examining Dbx+ neuron number in NB7-3 with Fd4-overexpression.
(6) It is logical to hypothesize that spatial factors specify early-born neurons directly, so only late-born neurons require Fd4, but it was not tested. The model would be strengthened by examining whether Fd4-Gal4-driven Vnd rescues the generation of later-born neurons in fd4/fd5 mutants.
(7) It is mentioned that Fd5 is not sufficient for the NB7-1 lineage identity. The observation is intriguing in how similar regulators serve distinct roles, but the data are not shown. The analysis in Figure 4 should be performed for Fd5 as supplemental information.
Reviewer #2 (Public review):
Summary:
Via a detailed expression analysis, they find that Fd4 is selectively expressed in embryonic NB7-1 and newly born neurons within this lineage. They also undertake a comprehensive genetic analysis to provide evidence that fd4 is necessary and sufficient for the identity of NB7-1 progeny.
Strengths:
The analysis is both careful and rigorous, and the findings are of interest to developmental neurobiologists interested in molecular mechanisms underlying the generation of neuronal diversity. Great care was taken to make the figures clear and accessible. This work takes great advantage of years of painstaking descriptive work that has mapped embryonic neuroblast lineages in Drosophila.
Weaknesses:
The argument that Fd4 is necessary for NB7-1 lineage identity is based on a Fd4/Fd5 double mutant. Loss of fd4 alone did not alter the number of NB7-1-derived Eve+ or Dbx+ neurons. The authors clearly demonstrate redundancy between fd4 and fd5, and the fact that the LOF analysis is based on a double mutant should be better woven through the text. The authors generated an Fd5 mutant. I assume that Fd5 single mutants do not display NB7-1 lineage defects, but this is not stated. The focus on Fd4 over Fd5 is based on its highly specific expression profile and the dramatic misexpression phenotypes. But the LOF analysis demonstrates redundancy, and the conclusions in the abstract and through the results should reflect the existence of Fd5 in the conclusions of this manuscript.
It is notable that Fd4 overexpression can rewire motor circuits. This analysis adds another dimension to the changes in transcription factor expression and, importantly, demonstrates functional consequences. Could the authors test whether U4 and U5 motor axon targeting changes in the fd4/fd5 double mutant? To strengthen claims regarding the importance of fd4/fd5 for lineage identity, it would help to address terminal features of U motorneuron identity in the LOF condition.
Reviewer #3 (Public review):
The goal of the work is to establish the linkage between the spatial transcription factors (STFs) that function transiently to establish the identities of the individual NBs and the terminal selector genes (typically homeodomain genes) that appear in the newborn post-mitotic neurons. How is the identity of the NB maintained and carried forward after the spatial genes have faded away? Focusing on a single neuroblast (NB 7-1), the authors present evidence that the fork-head transcription factor, fd4, provides a bridge linking the transient spatial cues that initially specified neuroblast identity with the terminal selector genes that establish and maintain the identity of the stem cell's progeny.
The study is systematic, concise, and takes full advantage of 40+ years of work on the molecular players that establish neuronal identities in the Drosophila CNS. In the embryonic VNC, fd4 is expressed only in the NB 7-1 and its lineage. They show that Fd4 appears in the NB while the latter is still expressing the Spatial Transcription Factors and continues after the expression of the latter fades out. Fd4 is maintained through the early life of the neuronal progeny but then declines as the neurons turn on their terminal selector genes. Hence, fd4 expression is compatible with it being a bridging factor between the two sets of genes.
Experimental support for the "bridging" role of Fd4 comes from a set of loss-of-function and gain-of-function manipulations. The loss of function of Fd4, and the partially redundant gene Fd5, from lineage 7-1 does not affect the size of the lineage, but terminal markers of late-born neuronal phenotypes, like Eve and Dbx, are reduced or missing. By contrast, ectopic expression of fd4, but not fd5, results in ectopic expression of the terminal markers eve and Dbx throughout diverse VNC lineages.
A detailed test of fd4's expression was then carried out using lineages 7-3 and 5-6, two well-characterized lineages in Drosophila. Lineage 7-3 is much smaller than 7-1 and continues to be so when subjected to fd4 misexpression. However, under the influence of ectopic Fd4 expression, the lineage 7-3 neurons lost their expected serotonin and corazonin expression and showed Eve expression as well as motoneuron phenotypes that partially mimic the U motoneurons of lineage 7-1.
Ectopic expression of Fd4 also produced changes in the 5-6 lineage. Expression of apterous, a feature of lineage 5-6, was suppressed, and expression of the 7-1 marker, Eve, was evident. Dbx expression was also evident in the transformed 5-6 lineages, but extremely restricted as compared to a normal 7-1 lineage. Considering the partial redundancy of fd4 and fd5, it would have been interesting to express both genes in the 5-6 lineage. The anatomical changes that are exhibited by motoneurons in response to Fd4 expression confirm that these cells do, indeed, show a shift in their cellular identity.
Author response:
Reviewer #1 (Public Review):
Lai and Doe address the integration of spatial information with temporal patterning and genes that specify cell fate. They identify the Forkhead transcription factor Fd4 as a lineage-restricted cell fate regulator that bridges transient spatial transcription factors to terminal selector genes in the developing Drosophila ventral nerve cord. The experimental evidence convincingly demonstrates that Fd4 is both necessary for lateborn NB7-1 neurons, but also sufficient to transform other neural stem cell lineages toward the NB7-1 identity. This work addresses an important question that will be of interest to developmental neurobiologists: How can cell identities defined by initial transient developmental cues be maintained in the progeny cells, even if the molecular mechanism remains to be investigated? In addition, the study proposes a broader concept of lineage identity genes that could be utilized in other lineages and regions in the Drosophila nervous system and in other species.
Thanks for the accurate summary and positive comments!
While the spatial factors patterning the neuroepithelium to define the neuroblast lineages in the Drosophila ventral nerve cord are known, these factors are sometimes absent or not required during neurogenesis. In the current work, Lai and Doe identified Fd4 in the NB7-1 lineage that bridges this gap and explains how NB7-1 neurons are specified after Engrailed (En) and Vnd cease their expression. They show that Fd4 is transiently co-expressed with En and Vnd and is present in all nascent NB7-1 progenies. They further demonstrate that Fd4 is required for later-born NB7-1 progenies and sufficient for the induction of NB7-1 markers (Eve and Dbx) while repressing markers of other lineages when force-expressed in neural progenitors, e.g., in the NB56 lineage and in the NB7-3 lineage. They also demonstrate that, when Fd4 is ectopically expressed in NB7-3 and NB5-6 lineages, this leads to the ectopic generation of dorsal muscle-innervating neurons. The inclusion of functional validation using axon projections demonstrates that the transformed neurons acquire appropriate NB7-1 characteristics beyond just molecular markers. Quantitative analyses are thorough and well-presented for all experiments.
Thanks for the positive comments!
(1) While Fd4 is required and sufficient for several later-born NB7-1 progeny features, a comparison between early-born (Hb/Eve) and later-born (Run/Eve) appears missing for pan-progenitor gain of Fd4 (with sca-Gal4; Figure 4) and for the NB7-3 lineage (Figure 6). Having a quantification for both could make it clearer whether Fd4 preferentially induces later-born neurons or is sufficient for NB7-1 features without temporal restriction.
We quantified the percentage of Hb+ and Runt+ cells among Eve+ cells with sca-gal4, and the results are shown in Figure 4-figure supplement 1. We found that the proportion of early-born cells is slightly reduced but the proportion of later-born cells remain similar. Interestingly, we also found a subset of Eve+ cells with a mixed fate (Hb+Runt+) but the reason remains unclear.
(2) Fd4 and Fd5 are shown to be partially redundant, as Fd4 loss of function alone does not alter the number of Eve+ and Dbx+ neurons. This information is critical and should be included in Figure 3.
Because every hemisegment in an fd4 single mutant is normal, we just added it as the following text: “In fd4 mutants, we observe no change in the number of Eve+ neurons or Dbx+ neurons (n=40 hemisegments).”
(3) Several observations suggest that lineage identity maintenance involves both Fd4dependent and Fd4-independent mechanisms. In particular, the fact that fd4-Gal4 reporter remains active in fd4/fd5 mutants even after Vnd and En disappear indicates that Fd4's own expression, a key feature of NB7-1 identity, is maintained independently of Fd4 protein. This raises questions about what proportion of lineage identity features require Fd4 versus other maintenance mechanisms, which deserves discussion.
We agree, thanks for raising this point. We add the following text to the Discussion. “Interestingly, the fd4 fd5 mutant maintains expression of fd4:gal4, suggesting that the fd4/fd5 locus may have established a chromatin state that allows “permanent” expression in the absence of Vnd, En, and Fd4/Fd5 proteins.”
(4) Similarly, while gain of Fd4 induces NB7-1 lineage markers and dorsal muscle innervation in NB5-6 and NB7-3 lineages, drivers for the two lineages remain active despite the loss of molecular markers, indicating some regulatory elements retain activity consistent with their original lineage identity. It is therefore important to understand the degree of functional conversion in the gain-of-function experiments. Sparse labeling of Fd4 overexpressing NB5-6 and NB7-3 progenies, as was done in Seroka and Doe (2019), would be an option.
We agree it is interesting that the NB7-3 and NB5-6 drivers remain on following Fd4 misexpression. To explore this, we used sca-gal4 to overexpress Fd4 and observed that Lbe expression persisted while Eg was largely repressed (see Author response image 1 below). The results show that Lbe and Eg respond differently to Fd4. A non-mutually exclusive possibility is that the continued expression of lbe-Gal4 UAS-GFP or eg-Gal4 UAS-GFP may be due to the lengthy perdurance of both Gal4 and GFP.
Author response image 1.
(5) The less-penetrant induction of Dbx+ neurons in NB5-6 with Fd4-overexpression is interesting. It might be worth the authors discussing whether it is an Fd4 feature or an NB56 feature by examining Dbx+ neuron number in NB7-3 with Fd4-overexpression.
In the NB7-3 lineages misexpressing Fd4, only 5 lineages generated Dbx+ cells (0.1±0.4, n=64 hemisegments), suggesting that the low penetrance of Dbx+ induction is an intrinsic feature of Fd4 rather than lineage context. We have added this information in the results section.
(6) It is logical to hypothesize that spatial factors specify early-born neurons directly, so only late-born neurons require Fd4, but it was not tested. The model would be strengthened by examining whether Fd4-Gal4-driven Vnd rescues the generation of laterborn neurons in fd4/fd5 mutants.
When we used en-gal4 driver to express UAS-vnd in the fd4/fd5 mutant background, we found an average 7.4±2.2 Eve+ cells per hemisegment (n=36), significantly higher than fd4/fd5 mutant alone (3.9±0.8 cells, n=52, p=2.6x10<sup.-11</sup>) (Figure 3J). In addition, 0.2±0.5 Eve+ cells were ectopic Hb+ (excluding U1/U2), indicating that Vnd-En integration is sufficient to generate both early-born and late-born Eve+ cells in the fd4/fd5 mutants. We have added the results to the text.
(7) It is mentioned that Fd5 is not sufficient for the NB7-1 lineage identity. The observation is intriguing in how similar regulators serve distinct roles, but the data are not shown. The analysis in Figure 4 should be performed for Fd5 as supplemental information.
Thanks for the suggestion. Because the results are exactly the same as the wild type, we don’t think it is necessary to provide an additional images or analysis as supplemental information.
Reviewer #2 (Public review):
Via a detailed expression analysis, they find that Fd4 is selectively expressed in embryonic NB7-1 and newly born neurons within this lineage. They also undertake a comprehensive genetic analysis to provide evidence that fd4 is necessary and sufficient for the identity of NB7-1 progeny.
Thanks for the accurate summary!
The analysis is both careful and rigorous, and the findings are of interest to developmental neurobiologists interested in molecular mechanisms underlying the generation of neuronal diversity. Great care was taken to make the figures clear and accessible. This work takes great advantage of years of painstaking descriptive work that has mapped embryonic neuroblast lineages in Drosophila.
Thanks for the positive comments!
The argument that Fd4 is necessary for NB7-1 lineage identity is based on a Fd4/Fd5 double mutant. Loss of fd4 alone did not alter the number of NB7-1-derived Eve+ or Dbx+ neurons. The authors clearly demonstrate redundancy between fd4 and fd5, and the fact that the LOF analysis is based on a double mutant should be better woven through the text.
The authors generated an Fd5 mutant. I assume that Fd5 single mutants do not display NB7-1 lineage defects, but this is not stated. The focus on Fd4 over Fd5 is based on its highly specific expression profile and the dramatic misexpression phenotypes. But the LOF analysis demonstrates redundancy, and the conclusions in the abstract and through the results should reflect the existence of Fd5 in the conclusions of this manuscript.
We agree, and have added new text to clarify the single mutant phenotypes (there are none) and the double mutant phenotype (loss of NB7-1 molecular and morphological features. The following text is added to the manuscript: “Not surprisingly, we found that fd4 single mutants or fd5 single mutants had no phenotype (Eve+ neurons were all normal). Thus, to assess their roles, we generated a fd4 and fd5 double mutant. Because many Eve+ and Dbx+ cells are generated outside of NB7-1 lineage, it was also essential to identify the Eve+ or Dbx+ cells within NB7-1 lineage in wild type and fd4 mutant embryos. To achieve this, we replaced the open reading frame of fd4 with gal4 (called fd4-gal4) (see Methods); this stock simultaneously knocked out both fd4 and fd5 (called fd4/fd5 mutant hereafter) while specifically labeling the NB7-1 lineage. For the remainder of this paper we use the fd4/fd5 double mutant to assay for loss of function phenotypes.”
It is notable that Fd4 overexpression can rewire motor circuits. This analysis adds another dimension to the changes in transcription factor expression and, importantly, demonstrates functional consequences. Could the authors test whether U4 and U5 motor axon targeting changes in the fd4/fd5 double mutant? To strengthen claims regarding the importance of fd4/fd5 for lineage identity, it would help to address terminal features of U motorneuron identity in the LOF condition.
Thanks for raising this important point. We examined the axon targeting on body wall muscles in both wild type and in fd4/fd5 mutant background and added the results in Figure 3-figure supplement 2. We found that the axon targeting in the late-born neuron region (LL1) is significantly reduced, suggesting that the loss of late-born neurons in fd4/fd5 mutant leads to the absence of innervation of corresponding muscle targets.
Reviewer #3 (Public review):
The goal of the work is to establish the linkage between the spatial transcription factors (STFs) that function transiently to establish the identities of the individual NBs and the terminal selector genes (typically homeodomain genes) that appear in the newborn postmitotic neurons. How is the identity of the NB maintained and carried forward after the spatial genes have faded away? Focusing on a single neuroblast (NB 7-1), the authors present evidence that the fork-head transcription factor, fd4, provides a bridge linking the transient spatial cues that initially specified neuroblast identity with the terminal selector genes that establish and maintain the identity of the stem cell's progeny.
Thanks for the positive comments!
The study is systematic, concise, and takes full advantage of 40+ years of work on the molecular players that establish neuronal identities in the Drosophila CNS. In the embryonic VNC, fd4 is expressed only in the NB 7-1 and its lineage. They show that Fd4 appears in the NB while the latter is still expressing the Spatial Transcription Factors and continues after the expression of the latter fades out. Fd4 is maintained through the early life of the neuronal progeny but then declines as the neurons turn on their terminal selector genes. Hence, fd4 expression is compatible with it being a bridging factor between the two sets of genes.
Thanks for the accurate summary!
Experimental support for the "bridging" role of Fd4 comes from a set of loss-of-function and gain-of-function manipulations. The loss of function of Fd4, and the partially redundant gene Fd5, from lineage 7-1 does not aoect the size of the lineage, but terminal markers of late-born neuronal phenotypes, like Eve and Dbx, are reduced or missing. By contrast, ectopic expression of fd4, but not fd5, results in ectopic expression of the terminal markers eve and Dbx throughout diverse VNC lineages.
Thanks for the accurate summary!
A detailed test of fd4's expression was then carried out using lineages 7-3 and 5-6, two well-characterized lineages in Drosophila. Lineage 7-3 is much smaller than 7-1 and continues to be so when subjected to fd4 misexpression. However, under the influence of ectopic Fd4 expression, the lineage 7-3 neurons lost their expected serotonin and corazonin expression and showed Eve expression as well as motoneuron phenotypes that partially mimic the U motoneurons of lineage 7-1.
Thanks for the positive comments!
Ectopic expression of Fd4 also produced changes in the 5-6 lineage. Expression of apterous, a feature of lineage 5-6, was suppressed, and expression of the 7-1 marker, Eve, was evident. Dbx expression was also evident in the transformed 5-6 lineages, but extremely restricted as compared to a normal 7-1 lineage. Considering the partial redundancy of fd4 and fd5, it would have been interesting to express both genes in the 5-6 lineage. The anatomical changes that are exhibited by motoneurons in response to Fd4 expression confirm that these cells do, indeed, show a shift in their cellular identity.
We appreciate the positive comments. We agree double misexpression of Fd4 and Fd5 might give a stronger phenotype (as the reviewer says) but the lack of this experiment does not change the conclusions that Fd4 can promote NB7-1 molecular and morphological aspects at the expense of NB5-6 molecular markers.
eLife Assessment
This study presents a valuable open-source and cost-effective method for automating the quantification of male aggression and courtship in Drosophila melanogaster. The work as presented provides solid evidence that the use of the behavioral setup that the authors designed - using readily available laboratory equipment and standardised high-performing classifiers they developed using existing software packages - accurately and reliably characterises social behavior in Drosophila. The work will be of interest to Drosophila neurobiologists and particularly to those working on male social behaviors.
Reviewer #1 (Public review):
The study introduces an open-source, cost-effective method for automating the quantification of male social behaviors in Drosophila melanogaster. It combines machine-learning based behavioral classifiers developed using JAABA (Janelia Automatic Animal Behavior Annotator) with inexpensive hardware constructed from off-the-shelf components. This approach addresses the limitations of existing methods, which often require expensive hardware and specialized setups. The authors demonstrate that their new "DANCE" classifiers accurately identify aggression (lunges) and courtship behaviors (wing extension, following, circling, attempted copulation, and copulation), closely matching manually annotated ground-truth data. Furthermore, DANCE classifiers outperform existing rule-based methods in accuracy. Finally, the study shows that DANCE classifiers perform as well when used with low-cost experimental hardware as with standard experimental setups across multiple paradigms, including RNAi knockdown of the neuropeptide Dsk and optogenetic silencing of dopaminergic neurons.
The authors make creative use of existing resources and technology to develop an inexpensive, flexible, and robust experimental tool for the quantitative analysis of Drosophila behavior. A key strength of this work is the thorough benchmarking of both the behavioral classifiers and the experimental hardware against existing methods. In particular, the direct comparison of their low-cost experimental system with established systems across different experimental paradigms is compelling. A weakness of the study is that the use of JAABA-based classifiers to analyze aggression and courtship is not novel (Tao et al., J. Neurosci., 2024; Sten et al., Cell, 2023; Chiu et al., Cell, 2021; Isshi et al., eLife, 2020; Duistermars et al., Neuron, 2018). However, the demonstration the JAABA classifiers they developed work as well without expensive experimental hardware opens the door to more low-cost systems for quantitative behavior analysis.
In summary, this work provides a practical and accessible approach to quantifying Drosophila behavior, reducing the economic barriers to the study of the neural and molecular mechanisms underlying social behavior.
Reviewer #2 (Public review):
Summary:
This manuscript addresses the development of a low-cost behavioural setup and standardised open-source high performing classifiers for aggression and courtship behaviour. It does so by using readily available laboratory equipment and previously developed software packages. By comparing the performance of the setup and the classifiers to previously developed ones, this study shows the classifier's overperformance and the reliability of the low-cost setup in recapitulating previously described effects of different manipulations on aggression and courtship.
Strengths:
The newly developed classifiers for lunges, wing extension, attempted copulation, copulation, following, circling, perform better than previously available developed ones. The behavioural setup developed is low cost and reliably allows analysis of both aggression and courtship behaviour, validated through social experience manipulation (social isolation), gene knock (Dsk in Dilp2 neurons) and neuronal inactivation (dopaminergic neurons) know to affect courtship and aggression.
Weaknesses:
This framework only encompasses analysis of lunges, while aggression encompasses multiple behaviours. Even though DANCE can serve as a template allowing future development of additional classifiers, the current study compares performance to CADABRA which analyses further aggression behaviours, making the comparisons incomplete.
Reviewer #3 (Public review):
The study by Yadav et al. describes a new setup to quantify a number of aggression and mating behaviors in Drosophila melanogaster. The investigation of these behaviors requires the analysis of large number of videos to identify each kind of behavior displayed by a fly. Several approaches to automatize this process have been published before, but each of them has their limitations. The authors set out to develop a new setup that includes a very low-cost, easy to acquire hardware and open-source machine-learning classifiers to identify and quantify the behavior.
Strengths:
(1) The study demonstrates that their cheap, simple, and easy to obtain hardware works just as well as custom-made, specialized hardware for analyzing aggression and mating behavior. This enables the setup to be used in a wide range of settings, from research with limited resources to classroom teaching.
(2) The authors used previously published software to train new classifiers for detecting a range of behaviors related to aggression and mating and make them freely available. The classifiers are very positively benchmarked against a manually acquired ground-truth as well as existing algorithms.
(3) The study demonstrates the applicability of the setup (hardware and classifiers) to common methods in the field by confirming a number of expected phenotypes with their setup.
Taken together, this work can greatly facilitate research of aggression and mating in Drosophila. The combination of low-cost, off-the-shelf hardware and open-source, robust software enables researchers with very little funding or technical expertise to contribute to the scientific process, and also allows large-scale experiments, for example, in classroom teaching with many students, or for systematic screenings.
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public review):
The study introduces an open-source, cost-effective method for automating the quantification of male social behaviors in Drosophila melanogaster. It combines machine-learning-based behavioral classifiers developed using JAABA (Janelia Automatic Animal Behavior Annotator) with inexpensive hardware constructed from off-the-shelf components. This approach addresses the limitations of existing methods, which often require expensive hardware and specialized setups. The authors demonstrate that their new "DANCE" classifiers accurately identify aggression (lunges) and courtship behaviors (wing extension, following, circling, attempted copulation, and copulation), closely matching manually annotated groundtruth data. Furthermore, DANCE classifiers outperform existing rule-based methods in accuracy. Finally, the study shows that DANCE classifiers perform as well when used with low-cost experimental hardware as with standard experimental setups across multiple paradigms, including RNAi knockdown of the neuropeptide Dsk and optogenetic silencing of dopaminergic neurons.
The authors make creative use of existing resources and technology to develop an inexpensive, flexible, and robust experimental tool for the quantitative analysis of Drosophila behavior. A key strength of this work is the thorough benchmarking of both the behavioral classifiers and the experimental hardware against existing methods. In particular, the direct comparison of their low-cost experimental system with established systems across different experimental paradigms is compelling.
While JAABA-based classifiers have been previously used to analyze aggression and courtship (Tao et al., J. Neurosci., 2024; Sten et al., Cell, 2023; Chiu et al., Cell, 2021; Isshi et al., eLife, 2020; Duistermars et al., Neuron, 2018), the demonstration that they work as well without expensive experimental hardware opens the door to more low-cost systems for quantitative behavior analysis.
We thank the reviewer for their positive assessment and constructive suggestions. We have cited these additional JAABA studies in the Introduction. We clarified that several prior JAABA-based classifiers were developed using specialized machinevision cameras or custom setups, and that in some cases the original code and classifiers were not made publicly available, which limits reproducibility and wider adoption. To address this, we explicitly note in the revised manuscript that DANCE was developed with accessibility in mind.
Although the study provides a detailed evaluation of DANCE classifier performance, its conclusions would be strengthened by a more comprehensive analysis. The authors assess classifier accuracy using a bout-level comparison rather than a frame-level analysis, as employed in previous studies (Kabra et al., Nat Methods, 2013). They define a true positive as any instance where a DANCE-detected bout overlaps with a manually annotated ground-truth bout by at least one frame. This criterion may inflate true positive rates and underestimate false positives, particularly for longer-duration courtship behaviors. For example, a 15-second DANCE-classified wing extension bout that overlaps with ground truth for only one frame would still be considered a true positive. A frame-level analysis performance would help address this possibility.
We thank the reviewer for raising this important point. Our original use of bout-level analysis followed existing literature (Duistermars et al., 2018; Ishii et al., 2020; Chiu et al., 2021; Tao et al., 2024; Hindmarsh Sten et al., 2025). While our lunge classifier already operates at the frame level, we have now performed additional frame-level evaluations for the duration based courtship classifiers. These analyses revealed only minor differences in precision, recall, and F1 scores compared with the original bout-level approach (see new Figure 5—Figure Supplement 3). Details of this analysis are now included in the Materials and Methods.
In summary, this work provides a practical and accessible approach to quantifying Drosophila behavior, reducing the economic barriers to the study of the neural and molecular mechanisms underlying social behavior.
We thank the reviewer for their encouraging comments and for recognizing the accessibility and practical value of our approach. We appreciate the constructive suggestions, which have helped strengthen the manuscript.
Reviewer #2 (Public review):
Summary:
This manuscript addresses the development of a low-cost behavioural setup and standardised open-source high-performing classifiers for aggression and courtship behaviour. It does so by using readily available laboratory equipment and previously developed software packages. By comparing the performance of the setup and the classifiers to previously developed ones, this study shows the classifier's overperformance and the reliability of the low-cost setup in recapitulating previously described effects of different manipulations on aggression and courtship.
Strengths:
The newly developed classifiers for lunges, wing extension, attempted copulation, copulation, following, and circling, perform better than available previously developed ones. The behavioural setup developed is low cost and reliably allows analysis of both aggression and courtship behaviour, validated through social experience manipulation (social isolation), gene knock (Dsk in Dilp2 neurons) and neuronal inactivation (dopaminergic neurons) known to affect courtship and aggression.
We thank the reviewer for the clear summary of our work and for highlighting its strengths. We appreciate these positive comments and suggestions, which have helped improve the clarity of the manuscript.
Weaknesses:
Aggression encompasses multiple defined behaviours, yet only lunges were analysed. Moreover, the CADABRA software to which DANCE was compared analyses further aggression behaviours, making their comparisons incomplete. In addition, though DANCE performs better than CADABRA and Divider in classifying lunges in the behavioural setup tested, it did not yield very high recall and F1 scores.
We thank the reviewer for raising this important point. We focused on lunges because they are widely used as a standard proxy for male aggression across multiple laboratories (Agrawal et al., 2020; Asahina et al., 2014; Chiu et al., 2021; Chowdhury et al., 2021; Dierick et al., 2007; Hoyer et al., 2008; Jung et al., 2020; Nilsen et al., 2004; Watanabe et al., 2017). As noted in the Discussion, our study also provides a template for the future development of additional aggression classifiers (fencing, wing flick, tussle, chase, female headbutt) and courtship classifiers (tapping, licking, rejection), which can be trained and shared through the same DANCE framework. Developing and validating these was beyond the scope of the present work.
To address the concern regarding precision, recall, and F1 scores, we performed additional analyses across all training videos and compiled these results in the new Figure 2—Figure Supplement 2. Our earlier lunge classifier had performance metrics obtained after training on a total of 11 videos. Our analysis shows performance metrics for classifiers trained on four independent datasets (Videos 8– 11). We found that the classifier trained on nine videos provided the best balance of precision, recall, and F1 (78.73%, 73.07%, and 75.79%, respectively), which was slightly better than the earlier classifier. We therefore updated the main figure, text, and Materials and Methods to use this version and uploaded the corresponding classifier and training details to the GitHub repository.
DANCE is of limited use for neuronal circuit-level enquiries, since mechanisms for intensity and temporally controlled optogenetic manipulations, which are nowadays possible with open-source software and low-cost hardware, were not embedded in its development.
We thank the reviewer for this valuable point. The primary aim of DANCE is to provide an accessible, modular, and low-cost behavioural recording and analysis platform. It was designed so that users can readily integrate additional components such as optogenetic control when needed. As a proof of concept, we implemented optogenetic silencing of dopaminergic neurons using the DANCE hardware and confirmed that this manipulation increased aggression (Figure 7R).
To facilitate adoption, we now provide schematic diagrams, LED control code, and instructions on our GitHub page and setup photographs in the manuscript (see new Figure 7—Figure Supplement 1). The released code allows programmable timing and intensity control, enabling users to reproduce temporally precise optogenetic protocols or extend the system for other stimulation paradigms.
Reviewer #3 (Public review):
The preprint by Yadav et al. describes a new setup to quantify a number of aggression and mating behaviors in Drosophila melanogaster. The investigation of these behaviors requires the analysis of a large number of videos to identify each kind of behavior displayed by a fly. Several approaches to automatize this process have been published before, but each of them has its limitations. The authors set out to develop a new setup that includes very low-cost, easy-to-acquire hardware and open-source machine-learning classifiers to identify and quantify the behavior.
Strengths:
(1) The study demonstrates that their cheap, simple, and easy-to-obtain hardware works just as well as custom-made, specialized hardware for analyzing aggression and mating behavior. This enables the setup to be used in a wide range of settings, from research with limited resources to classroom teaching.
(2) The authors used previously published software to train new classifiers for detecting a range of behaviors related to aggression and mating and to make them freely available. The classifiers are very positively benchmarked against a manually acquired ground truth as well as existing algorithms.
(3) The study demonstrates the applicability of the setup (hardware and classifiers) to common methods in the field by confirming a number of expected phenotypes with their setup.
We thank the reviewer for the positive assessment of our work and for highlighting its strengths. We appreciate these encouraging comments and suggestions, which have helped improve the clarity and presentation of the manuscript.
Weaknesses:
(1) When measuring the performance of the duration-based classifiers, the authors count any bout of behavior as true positive if it overlaps with a ground-truth positive for only 1 frame - despite the minimal duration of a bout is 10 frames, and most bouts are much longer. That way, true positives could contain cases that are almost totally wrong as long there was an overlap of a single frame. For the mating behaviors that are classified in ongoing bouts, I think performance should be evaluated based on the % of correctly classified frames, not bouts.
We thank the reviewer for raising this concern. In response to this point, and to Reviewer #1’s similar comment, we performed a frame-level evaluation of all duration-based courtship classifiers. The analysis revealed only minor differences compared with the original bout-level metrics (see new Figure 5—Figure Supplement 3), confirming the robustness of our classifiers. We have also added a description of this analysis in the Materials and Methods section.
(2) In the methods part, only one of the pre-existing algorithms (MateBook), is described. Given that the comparison with those algorithms is a so central part of the manuscript, each of them should be briefly explained and the settings used in this study should be described.
We thank the reviewer for this helpful suggestion. In the revised manuscript, we expanded the Materials and Methods to include concise descriptions and parameter settings for all pre-existing algorithms used for comparison. This includes dedicated subsections for CADABRA and the Divider assay, with explicit reference to their rulebased or geometric features. For MateBook, we specified the persistence filters used and the adjustments made for fair benchmarking. These changes ensure transparency and reproducibility.
Taken together, this work can greatly facilitate research on aggression and mating in Drosophila. The combination of low-cost, off-the-shelf hardware and open-source, robust software enables researchers with very little funding or technical expertise to contribute to the scientific process and also allows large-scale experiments, for example in classroom teaching with many students, or for systematic screenings.
We thank the reviewer for the encouraging comments and for recognizing the accessibility and broad applicability of DANCE. We believe these revisions have further strengthened the manuscript.
Reviewer #1 (Recommendations for the authors):
The following comments highlight areas where additional context, clarification, or further analysis could strengthen the manuscript. I hope these suggestions will be useful in refining your work.
(1) Lines 71-73: The authors state that Ctrax "leads to frequent identity switches among tracked flies, which is not the case while using FlyTracker." However, Ctrax was specifically designed to minimize identity errors, and Kabra et al. (2013) reported a low frequency of such errors-approximately one per five fly-hours in 10-fly videos. In contrast, Caltech FlyTracker does not correct identity errors automatically, requiring manual corrections, as noted in the Methods section of this study. If this is not an oversight, please provide further context to clarify this distinction.
We thank the reviewer for raising this clarification. As reported by Bentzur et al. (2021), when groups of flies were tracked simultaneously, Ctrax often generated multiple identities for the same individual, sometimes producing more trajectories than the actual number of flies. To prevent ambiguity, we revised the text to read: “While both Ctrax and FlyTracker (Eyjolfsdottir et al., 2014) may produce identity switches, when groups of flies were tracked simultaneously, Ctrax led to inaccuracies that required manual correction using specialized algorithms such as FixTrax (Bentzur et al., 2021).” We also quantified FlyTracker identity-switch rates in our datasets and report them in new Supplementary File 5, confirming that such events were rare (< 2% of tracked intervals). We believe, this updated version provides the necessary context and ensures accuracy in describing each tracker’s limitations.
(2) Line 85: Providing additional context on how this study builds on previous work using JAABA-based classifiers for fly social behavior and comparing these classifiers to rule-based methods would more accurately situate it within the field. The authors state that "recently, a few JAABA-based classifiers have been developed for measuring aggression and courtship" and cite four related studies. However, this statement seems to underrepresent the use of JAABA-based classifiers for quantifying fly social behavior, which has become common in the field. Several additional studies (as noted in the public review) have developed JAABA-based classifiers for scoring aggression or courtship. Furthermore, other studies have compared the performance of JAABA-based classifiers with rule-based classifiers like CADABRA (e.g., Chowdhury et al., Comm Biology 2021; Leng et al., PlosOne 2020; Kabra et al., Nat Methods 2013). Mentioning the similar findings in those studies and your own helps strengthen the conclusion that machine-learning-based classifiers outperform rule-based classifiers in several experimental contexts.
We thank the reviewer for this helpful suggestion. We have revised the Introduction to include additional references to studies that applied JAABA-based classifiers for aggression and courtship and made textual edits to reflect this. We further noted that, unlike several previous studies, all DANCE classifiers and analysis code are publicly available.
Reviewer #2 (Recommendations for the authors):
(1) Suggestions for improved or additional experiments, data or analyses: As mentioned in the description of the effect of optogenetic inactivation of dopaminergic neurons, in the conclusion and also reported in the literature, there are other important identified aggression behaviours, such as fencing, wing flick, tussle, and chase. Similarly, for courtship, tapping and licking have also been defined. This study, as opposed to proposed future studies, would benefit from creating opensource classifiers for these established behaviours, which are important for the analysis of aggression and courtship.
We thank the reviewer for this valuable suggestion. As clarified in the Discussion, this manuscript intentionally focuses on six core, well-validated aggression and courtship behaviors to demonstrate DANCE’s modularity and reproducibility. Developing additional classifiers such as fencing, wing flick, tussle, chase, tapping, and licking would require extensive annotation and validation beyond the present scope. To address this point, we explicitly note in the revised text that the DANCE pipeline is readily extendable, allowing the community to build new classifiers within the same framework.
In terms of observer bias assessment for ground-truthing in courtship, this was only presented for circling and it would be beneficial to have encompassed all behaviours analysed.
We thank the reviewer for this suggestion. Observer-bias comparisons for all six classifiers are presented in Figure 2—Figure Supplement 1 (panels A–F). We clarified in the Results that annotations from two independent evaluators were compared for all classifiers, with no significant differences observed, confirming their robustness.
Finally, intensity and temporal optogenetic control are important for neuronal circuit analysis of underlying behaviour. The authors could embed this aspect in DANCE by integrating control of the green light LED strip used in this study using, for example, the open-source visual reactive programming software Bonsai (Lopes et al., 2015) and open-source electronics platform Arduino. This is an important and valuable addition in line with maintaining low cost.
We thank the reviewer for this valuable suggestion. DANCE was designed to be modular, allowing integration of temporal optogenetic control. To support immediate adoption, we now provide Arduino LED control code, setup schematics, and photographs (new Figure 7—Figure Supplement 1) along with step-by-step instructions on our GitHub page. We also note that Bonsai and Arduino frameworks are compatible with DANCE, enabling future extensions for closed-loop or behaviortriggered stimulation.
(2) Minor corrections to the text and figures:
Figure Supplement 1 refers only to Figure 2, yet panels D-F refer to the behaviour circling in courtship and therefore should be assigned to the respective figure.
Thanks, we have corrected this.
In lines 315-316, the cumbersome task of fluon coating for aggression assays seems to be ubiquitous across assays which is not the case, and therefore the sentence should include the word 'some'.
Thanks, we have edited this.
The cost of the phone and/or tablet should be included in the DANCE setup costs, as presumably these devices will be dedicated to the behavioural studies, for consistency purposes.
We thank the reviewer for this comment. We intentionally did not include smartphones or tablets in the setup cost because, in our experiments, these devices were not dedicated exclusively to DANCE but were repurposed from routine personal use. Our aim was to leverage readily available consumer electronics so that their cost does not become a barrier to adoption. We confirmed that commonly available Android phones capable of 30 fps at 1080p in H.264 format, as well as tablets or phones running a simple white-screen light app, are sufficient for reliable behavior classification and illumination. Since these devices can be returned to regular use after recordings, including their cost in the setup would not accurately reflect the intended accessibility of DANCE. For consistency, we now clarify in the Materials and Methods that such devices should be placed in airplane mode during recordings.
Reviewer #3 (Recommendations for the authors):
(1) For my taste, the authors put too much emphasis on the point that their method outperforms existing methods. I understand the value in comparing to published methods and it is of course fully justified to state the advantages of the new method. But the whole preprint is set up as a competition with the old algorithms, and the conclusion that the new classifier is better is repeated in each figure caption and after each paragraph of the results. This competitive mindset also extends to the selection of which results are presented as main figures and which as supplements - all cases in which the previous methods actually perform well are only presented in the supplement. I think this is simply unnecessary as the authors' results speak for themselves, and do not need the continuous competitive comparison.
We thank the reviewer for this thoughtful suggestion. Our intention was to benchmark DANCE rigorously against existing methods, not to frame the study competitively. We agree that repeated emphasis on relative performance was unnecessary. In the revised version, we streamlined figure captions and text throughout the manuscript to balance comparisons and removed redundant phrasing. Instances where other methods performed well are now presented with equal clarity to maintain a neutral and informative tone.
(2) When describing the DANCE hardware, as a reader I would find it interesting to also read about potential issues that the authors encountered. For example, how difficult is it to handle the materials without breaking or deforming them, which could affect the behavioral assays? How critical is it to use specific blister packs - the availability of which will likely vary strongly between countries? Did the authors try different sizes, and products? Such information, even as a supplement, could be very helpful for the widespread use of the hardware.
We thank the reviewer for this important point. To address this, we conducted additional tests comparing DANCE arenas of different diameters (new Figure 7— Figure Supplement 3A–C and new Figure 7—Figure Supplement 4A–L). We also consulted colleagues in multiple countries and verified that the blister packs used in our assays are readily available. The Materials and Methods now include practical handling notes: blister foils can be reused ~30–40 times for aggression assays and ~10–15 times for courtship assays before deformation. We also describe how to prevent agar surface damage during assembly and how to wash and dry the arenas for optimal reusability.
(3) I find the arrows pointing to several videos in a number of figures rather distracting and redundant, and suggest omitting them.
Thanks, we have omitted these arrows from all relevant figures and clarified the figure legends to enhance readability.
(4) P8, line 169 ff: this is a very long sentence that should be separated into several sentences.
We have rewritten this as follows: “DANCE scores remained comparable to groundtruth scores across all categories, whereas CADABRA and Divider underestimated the lunge counts (Figure 2B–E). Correlation analysis revealed a strong relationship between DANCE and ground-truth scores (Figure 2F, Supplementary File 2). In comparison, CADABRA and the Divider assay classifier showed a weaker correlation (Figure 2G-H, Supplementary File 2).”
(5) P10, line 216: please explain, here and in the methods, how these behavioral indices are calculated. I did not find this information anywhere in the paper.
We thank the reviewer for pointing this out. We now define the behavioral index explicitly in Materials and Methods: “For each assay, a behavioral index was calculated as the proportion of frames in which the male engaged in the specified behavior. This was obtained by dividing the total number of frames annotated for that behavior by the total number of frames in the recording.”
(6) P11, line 253: I don't understand the modifications to MateBook regarding attempted copulations, neither in the results nor the methods section. I would ask the authors to explain more explicitly what was done.
We thank the reviewer for this helpful suggestion. We have re-written several parts of the Materials and methods to clarify these details and streamline the text. To train the attempted copulation classifier, we combined datasets from assays with mated and decapitated virgin females, using manual annotations as ground truth. We also adapted MateBook’s persistence filters (Ribeiro et al., 2018) and defined thresholds explicitly: mounting lasting >45 s (>1350 frames at 30 fps) was defined as copulation, whereas abdominal curling without mounting, or mounting lasting 0.33– 45 s, was defined as attempted copulation.
(7) Figure 7F: this is the only case with a significant difference between the two setups. What explanations do the authors have for the discrepancy?
We thank the reviewer for raising this point. After repeating the experiments, we no longer found a significant difference between the setups. Figure 7 and its legend have been updated to reflect these results.
(8) Figure 2 - Supplement 1: I do not understand why the boxes for Observer 1 have different colors in different figures. Does this have a meaning?
Thanks for pointing this out. The color differences had no intended meaning, and we have corrected the figure for consistency across panels.
(9) P22, line 517ff: It would be interesting to know how frequently identity switches occurred. For large-scale, automatic behavioral screenings that step could be a crucial bottleneck.
We thank the reviewer for this valuable suggestion. We analyzed identity switches using the FlyTracker “Visualizer” package, which flags frames with possible overlaps or jumps. Flagged intervals were manually verified, and we report these data in new Supplementary File 5. Identity switch rates were very low: 0.66% for high-resolution recordings and 1.9% for smartphone DANCE videos in the most challenging decapitated-virgin dataset. These findings demonstrate robust tracking performance under both setups.
The tag does not have to be a plain identifier. You can use any expression with precedence greater than 16, which includes property access, function call, new expression, or even another tagged template literal.
einde
eLife Assessment
This important study presents a compelling theoretical framework for understanding condensation or phase separation of membrane-bound proteins, with a focus on the organization of tight junction components. By incorporating non-dilute binding effects into thermodynamic models and validating the model's predictions with in vitro experiments on the tight junction protein ZO-1, the authors provide a quantitative tool that combines theory and experiments and will help researchers in the field quantitatively interpret their findings. Given that phase separation of membrane bound molecules is becoming key in signaling, spanning from immune signaling to cell-cell adhesion, this work will be of broad interest for cell biologists and biophysicists.
Reviewer #1 (Public review):
Summary:
Biomolecular condensates are essential part of cellular homeostatic regulation. In this manuscript, authors develop a theoretical framework for phase separation of membrane bound proteins. They show the effect of non-dilute surface binding and phase separation on tight junction protein organization.
Strengths:
It is an important study considering the phase separation of membrane bound molecules are taking the center stage of signaling, spanning from immune signaling to cell-cell adhesion. A theoretical framework will help biologists to quantitatively interpret their findings.
Weaknesses:
Understandably, authors used one system to test their theory (ZO-1). However, to establish a theoretical framework, this is sufficient.
Comments on revisions:
I do not recommend new experiments. The manuscript is clear and establishes a new step in understanding the physical chemistry of biomolecular condensates.
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public review):
Summary:
Biomolecular condensates are an essential part of cellular homeostatic regulation. In this manuscript, the authors develop a theoretical framework for the phase separation of membrane-bound proteins. They show the effect of non-dilute surface binding and phase separation on tight junction protein organization.
Strengths:
It is an important study, considering that the phase separation of membrane-bound molecules is taking the center stage of signaling, spanning from immune signaling to cell-cell adhesion. A theoretical framework will help biologists to quantitatively interpret their findings.
Weaknesses:
Understandably, the authors used one system to test their theory (ZO-1). However, to establish a theoretical framework, this is sufficient.
We acknowledge this limitation. While we agree that additional systems would strengthen the generality of our theory, we note that the focus of this work is to introduce and validate a theoretical framework. As the reviewer notes, this is sufficient for establishing the framework. Nonetheless, we are open to further collaborations or future studies to test the model with other systems.
Reviewer #2 (Public review):
Summary:
The authors present a clear expansion of biophysical (thermodynamic) theory regarding the binding of proteins to membrane-bound receptors, accounting for higher local concentration effects of the protein. To partially test the expanded theory, the authors perform in vitro experiments on the binding of ZO1 proteins to Claudin2 C-terminal receptors anchored to a supported lipid bilayer, and capture the effects that surface phase separation of ZO1 has on its adsorption to the membrane.
Strengths:
(1) The derived theoretical framework is consistent and largely well-explained.
(2) The experimental and numerical methodologies are transparent.
(3) The comparison between the best parameterized non-dilute theory is in reasonable agreement with experiments.
Weaknesses:
(1) In the theoretical section, what has previously been known, compared to which equations are new, should be made more clear.
We have revised the theory section to clearly distinguish previously established formulations from novel contributions following equation (4), which is .
(2) Some assumptions in the model are made purely for convenience and without sufficient accompanying physical justification. E.g., the authors should justify, on physical grounds, why binding rate effects are/could be larger than the other fluxes.
For our problem, binding is relevant together with diffusive transport in each phase. Each process is accompanied by kinetic coefficients that we estimate for the experimental system. For the considered biological systems (and related ones), it is difficult to determine whether other fluxes (see, e.g., Eq. 8(e)) have relaxed or not. We note that their effects are, of course, included in the kinetic model applied to the coarsening of ZO1 surface condensates as boundary conditions. But we cannot exclude that the corresponding kinetic coefficient in the actual biological system is large enough such that, e.g., Eq. (9e) does not vanish to zero “quasi-statically”. We have now added a sentence to the outlook highlighting the relevance of testing those flux-force relationships in biological systems.
(3) I feel that further mechanistic explanation as to why bulk phase separation widens the regime of surface phase separation is warranted.
We have discussed the mechanistic explanation related to bulk protein interaction strength in the manuscript in the section: “Effects of binding affinity and interactions on surface phase separation”. We explained how the bulk interaction parameter affects the binding equilibrium.
(4) The major advantage of the non-dilute theory as compared with a best parameterized dilute (or homogenous) theory requires further clarification/evidence with respect to capturing the experimental data.
We thank reviewer for this helpful question. To address this point, we have added new paragraphs in the conclusion section, which explicitly discuss the necessity of employing the non-dilute theory for interpreting the experimental data.
(5) Discrete (particle-based) molecular modelling could help to delineate the quantitative improvements that the non-dilute theory has over the previous state-of-the-art. Also, this could help test theoretical statements regarding the roles of bulk-phase separation, which were not explored experimentally.
We appreciate the suggestion and agree that such modeling would be valuable. However, this is beyond the scope of the current study.
(6) Discussion of the caveats and limitations of the theory and modelling is missing from the text.
We sincerely appreciate the reviewer’s helpful comment. We have added a discussion in the conclusion section outlining the caveats and limitations of our modeling approach.
Reviewing Editor Comments:
Upon discussing with the reviewers, we feel that this manuscript could significantly be improved if testing the model with a different model system (beyond ZO1/tight junctions), in which case we foresee that we could enhance the strength of evidence from "compelling" to "exceptional". But of course, this is up to the authors to go for it or not, the paper is already very good.
Reviewer #2 (Recommendations for the authors):
(1) Lines 132-134: Re-word, the use of "complex" is confusing.
We have rephrased the sentence for clarity. The revised version reads: ṽ<sub>_𝑃𝑅</sub>_ are the molecular volume and area of the protein-receptor complex ѵ<sub>𝑃𝑅</sub>, respectively”, and the changes have been in the revised manuscript.
(2) Line 154 use of ""\nu"" for volume and area could be avoided for better clarity.
We thank the reviewer for this helpful suggestion. We have removed the statement involving ""\nu"" as these quantities have already been defined in the preceding context.
(3) Line 158 the total "Helmholtz" free energy F...
We have added the word "Helmholtz" to the sentence.
(4) Line 160 typo "In specific,..."
We carefully checked this sentence but could not identify a typo.
(5) For equation 5 explain the physical origins of each term, or provide a reference if this equation is explained elsewhere.
Thank you very much for your valuable suggestions. We have carefully rephrased Equation (5) and added a paragraph immediately afterward to provide a detailed explanation of its physical meaning.
(6) Derivation on lines 163-174 is poorly written. Make the logical flow between the equations clearer.
We greatly appreciate your insightful suggestions. Equation (6) has been carefully revised for clarity, and the explanation has been rewritten to ensure better readability. All modifications are Done.
(7) Define bold "t" in Equation 6.
The variable “t” has been explicitly defined in the context for clarity.
(8) In equations. 7b-7c the nablas (gradients) should be the 2D versions.
We have updated the gradient operators in Equations (7b) and (7c) [Eq. (9) in revised manuscript] to their 2D forms for consistency.
(9) Line 190, avoid referring to the future Equation 14, and state in words what is meant by "thermodynamic equilibrium".
We have added the explanation of “thermodynamic equilibrium” and remove the reference to equation accordingly.
(10) In Equation 11 you don't explain what you are doing ( which is a perturbation around the minimum of the free energy).
We have revised the paragraph before equation (11) [Eq. (13) in revised manuscript] to clarify that the expression represents a perturbation around the minimum of the free energy.
(11) In Equation 12, doesn't this also depend on how you have written equation 6 (not just equation 5).
Eq. (12) [Eq. (14) in revised manuscript] is derived directly from the variation of the total free energy F. In contrast, Eq. (6) contains the time derivative of free energies that were not written in their final form. In the revised version, we have now given the conjugate forces and fluxes in Eqs. (7) and (8) for clarity.
(12) Line 206 specify the threshold of local concentration (or provide a reference).
We have specified the threshold of local concentration in the revised text, and the corresponding statement has been highlighted.
(13) Line 223 is the deviation from ideality captured in a pair-wise fashion? I presume it does not account for N many-body interactions?
Yes, our model is formulated within a mean-field framework that incorporates pairwise (second order) interaction coefficients. For example, 𝜒<sub>𝑃𝑅 -𝑅</sub> characterizes the interaction between the complex 𝑃𝑅 and the free receptor 𝑅, 𝜒<sub>𝑅 -L</sub> the interaction between free receptor 𝑅 and free lipid 𝐿, 𝜒<sub>𝑃𝑅-𝐿</sub> the interaction between complex 𝑃𝑅and free lipid 𝐿. We have stressed this choice of free energy in the revised manuscript.
(14) Line 274, how do the authors know the secondary effects (of which they should mention a few) do not significantly impact the observed behaviour?
We sincerely thank the reviewer for the helpful comment. First, the parameters 𝜒<sub>𝑅 -L</sub> and 𝜒<sub>𝑃𝑅 -𝑅</sub> are not essential based on the experimental observations. For more information, please see our revised paragraph on the choice of the specific parameter values, which has been in the following Eq. (21).
(15) It's not clear how Figures 3 b and c are generated with reference to which parameters are changed to investigate with/without bulk phase separation.
To improve clarity, we have revised Figure 3 to display the corresponding parameter values directly in each panel. Figures 3b and 3c were generated by computing the surface binding curves (as shown in Fig. 2) for each binding affinity 𝜔<sub>𝑃𝑅</sub> and membrane-complex interaction strength 𝜒<sub>𝑃𝑅-𝐿</sub>, under different bulk interaction strengths chi, to compare the cases with and without bulk phase separation.
(16) The jump between theory and the "Mechanism in ..." section is too much. The authors should include the biological context of tight junctions and ZO1 in the main introduction.
We appreciate the reviewer’s suggestion. Following this comment, we have added an extended discussion in the main introduction to provide the necessary biological context of tight junctions and ZO1. In addition, we inserted new bridging paragraphs between the theoretical section and the section “Mechanism in tight junction formation” to create a smoother transition from theory to experiments. These revisions help to better connect the theoretical framework with the biological phenomena discussed in the later section.
eLife Assessment
This useful study raises interesting questions but provides inadequate evidence of an association between atovaquone-proguanil use (as well as toxoplasmosis seropositivity) and reduced Alzheimer's dementia risk. The findings are intriguing but they are correlative and hypothesis-generating with the strong possibility of residual confounding.
Reviewer #1 (Public review):
Summary:
This useful study provides incomplete evidence of an association between atovaquone-proguanil use (as well as toxoplasmosis seropositivity) and reduced Alzheimer's dementia risk. The study reinforces findings that VZ vaccine lowers AD risk and suggests that this vaccine may be an effect modifier of A-P's protective effect. Strengths of the study include two extremely large cohorts, including a massive validation cohort in the US. Statistical analyses are sound, and the effect sizes are significant and meaningful. The CI curves are certainly impressive.
Weaknesses include the inability to control for potentially important confounding variables. In my view, the findings are intriguing but remain correlative / hypothesis generating rather than causative. Significant mechanistic work needs to be done to link interventions which limit the impact of Toxoplasmosis and VZV reactivation on AD.
Weaknesses:
Major:
(1) Most of the individuals in the study received A-P for malaria prophylaxis as it is not first line for Toxo treatment. Many (probably most) of these individuals were likely to be Toxo negative (~15% seropositive in the US), thereby eliminating a potential benefit of the drug in most people in the cohort. Finally, A-P is not a first line treatment for Toxo because of lower efficacy.
(2) A-P exposure may be a marker of subtle demographic features not captured in the dataset such as wealth allowing for global travel and/or genetic predisposition to AD. This raises my suspicion of correlative rather than casual relationships between A-P exposure and AD reduction. The size of the cohort does not eliminate this issue, but rather narrows confidence intervals around potentially misleading odds ratios which have not been adjusted for the multitude of other variables driving incident AD.
(3) The relationship between herpes virus reactivation and Toxo reactivation seems speculative.
(4) A direct effect on A-P on AD lesions independent on infection is not considered as a hypothesis. Given the limitations above and effects on metabolic pathways, it probably should be. The Toxo hypothesis would be more convincing if the authors could demonstrate an enhanced effect of the drug in Toxo positive individuals without no effect in Toxo negative individuals.
Minor:
(5) "Clinically meaningful" should be eliminated from the discussion given that this is correlative evidence.
Reviewer #2 (Public review):
Summary:
This manuscript examines the association between atovaquone/proguanil use, zoster vaccination, toxoplasmosis serostatus and Alzheimer's Disease, using 2 databases of claims data. The manuscript is well written and concise. The major concerns about the manuscript center around the indications of atovaquone/proguanil use, which would not typically be active against toxoplasmosis at doses given, and the lack of control for potential confounders in the analysis.
Strengths:
(1) Use of 2 databases of claims data.
(2) Unbiased review of medications associated with AD, which identified zoster vaccination associated with decreased risk of AD, replicating findings from other studies.
Weaknesses:
(1) Given that atovaquone/proguanil is likely to be given to a healthy population who is able to travel, concern that there are unmeasured confounders driving the association.
(2) The dose of atovaquone in atovaquone/proguanil is unlikely to be adequate suppression of toxo (much less for treatment/elimination of toxo), raising questions about the mechanism.
(3) Unmeasured bias in the small number of people who had toxoplasma serology in the TriNetX cohort.
Passival GerundConstruction only has a subject; that its -ing-form is nominal; and that, when used with want, the matrix verb has a slightlydifferent meaning – ‘require, need’ – instead of its usual volitional semantics.
You (1 soggetto che è anche oggetto di remind) need no reminding (nominale perché dipende da need). The club wants retiling > come in italiano "qualcosa chiama/vuole qualcos'altro" ne ha bisogno disperato > WANT PASSA DA VOLERE A AVERE BISOGNO
the recent ruling by the international court of justice last month that basically said two things. That nations absolutely have an obligation to address climate change
for - climate crisis - International Court of Justice ruling - climate change
is not peer-reviewed
не проходит экспертную оценку; контекст:" the study is not peer-reviewed..."
A recent MIT Media Lab study reported that “excessive reliance on AI-driven solutions” may contribute” to “cognitive atrophy” and shrinking of critical thinking abilities. The study is small and is not peer-reviewed, and yet it delivers a warning that even artificial intelligence assistants are willing to acknowledge. When we asked ChatGPT whether AI can make us dumber or smarter, it answered, “It depends on how we engage with it: as a crutch or a tool for growth.”
Введение, центральная проблема, затрагиваемая в статье и для кого она написана. Тема того, что сейчас с развитием ИИ технологий, наш мозг отвыкнет думать и принимать решения самостоятельно. Уже с начала статьи автор привлекает внимание потенциального читателя "громкими словами", соответствующими публицистике
I am less worried about AI as an aid to guide expert-level reasoning in targeted domains, as when, for example, a doctor might use AI in diagnosis to ensure that she hasn’t overlooked an unusual disease. But the problem is that there is too much hype surrounding LLMs as general reasoners upon which we can (at least partially) offload our thinking about any topic whatsoever. It’s in the interest of people producing the technology to make us think that its possibilities are limitless and that it will usher in a wonderful new future for everyone. We should be cautious before we too enthusiastically pin our hopes on the latest technological trend.
Джефф Бэхрендс подытоживает прошлые высказывания и говорит о своих опасениях, что современные технологии становятся настолько распространенными, что мы полагаемся на его работу без сомнений. Нам рекламируют способности ИИ как то, что не может ошибаться и давать не точною информацию, однако это не так
AI is forcing us to think differently about the various components of critical thinking. For instance, AI can be a helpful partner in analyzing and inferring, as well as with certain types of problem-solving, but it’s not always that successful at evaluating, and reflecting can’t (yet) be outsourced to AI.
Карен Торнбер продолжает мысль предыдущих авторов, ИИ лишен критического мышление, он-машина для сбора данных, анализа по заданным критериям, но он не может оценить масштаб и сложность процессов в глобальном масштабе, у него нет того набора жизненного опята,что и у человека
the key to having the owl be a positive force instead of a negative one is not to let it do your thinking for you. We know that generative AI doesn’t understand the human context, so it’s not going to provide wisdom about social, emotional, and contextual events, because those are not part of its repertoire. However, GenAI is very good at absorbing large amounts of data and making calculative predictions in ways that can augment your thinking.
Кристофер Дед проводит параллель с богиней и совой на её плече, чтобы высказать ту же мысль связанную с ИИ, мы не можем позволить инструменту думать за нас, но пользоваться им, как вспомогательным помощником в нашей работе и становится мудрее
At the end of the day, if you think you’re in school to produce outputs, then you might be OK with AI helping you produce those outputs. But if you’re in school because you want to be learning, remember that the output is just a vehicle through which that learning is going to happen. The output is typically not the ultimate goal. When you confuse those two things, you might use AI in ways that are not conducive to learning.
Дэн Леви высказывает конкретную мысль о том, для чего создан ИИ, если ваша цель в обучении- простое получение результатов, а не получение знаний, то он конечно является подручным средством. Если вы хотите учиться, то ИИ-полезный инструмент, который действительно может помочь повысить эффективность вашего обучения
Summary of key points
This is nice to summarise and review the section. However, it should appear in each section consistently?
Before you watch the video on Data Collection, consider these questions:
Shall we begin this sentence with the below?
'Data collection is the first step in data processing, and the collected data is then prepared for the subsequent processing stages.'