33054
DOI: 10.1126/sciadv.abh0050
Resource: RRID:BDSC_33054
Curator: @bdscstockkeepers
SciCrunch record: RRID:BDSC_33054
33054
DOI: 10.1126/sciadv.abh0050
Resource: RRID:BDSC_33054
Curator: @bdscstockkeepers
SciCrunch record: RRID:BDSC_33054
Bloomington Stock #24761
DOI: 10.1111/acel.13493
Resource: RRID:BDSC_24761
Curator: @bdscstockkeepers
SciCrunch record: RRID:BDSC_24761
Bloomington Stock #24787
DOI: 10.1111/acel.13493
Resource: RRID:BDSC_24787
Curator: @bdscstockkeepers
SciCrunch record: RRID:BDSC_24787
Bloomington Stock #7010
DOI: 10.1111/acel.13493
Resource: RRID:BDSC_7010
Curator: @bdscstockkeepers
SciCrunch record: RRID:BDSC_7010
HMS01045 (maskRNAi)
DOI: 10.1093/g3journal/jkab388
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
JF02717 (drkRNAi)
DOI: 10.1093/g3journal/jkab388
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
HMS00904 (TorRNAi#2)
DOI: 10.1093/g3journal/jkab388
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
GL00156 (TorRNAi#1)
DOI: 10.1093/g3journal/jkab388
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
HMS00007 (AktRNAi)
DOI: 10.1093/g3journal/jkab388
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
JF02770 (PI3K92ERNAi)
DOI: 10.1093/g3journal/jkab388
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
HMS05002 (MKK3RNAi#3)
DOI: 10.1093/g3journal/jkab388
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
HMS00173 (ErkRNAi)
DOI: 10.1093/g3journal/jkab388
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
HMS01979 (VavRNAi)
DOI: 10.1093/g3journal/jkab388
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
HMS01294 (RasRNAi#3)
DOI: 10.1093/g3journal/jkab388
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
JF02478 (RasRNAi#2)
DOI: 10.1093/g3journal/jkab388
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
JF01355 (LuciferaseRNAi)
DOI: 10.1093/g3journal/jkab388
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
#64196
DOI: 10.1093/g3journal/jkab388
Resource: RRID:BDSC_64196
Curator: @bdscstockkeepers
SciCrunch record: RRID:BDSC_64196
#8529
DOI: 10.1093/g3journal/jkab388
Resource: RRID:BDSC_8529
Curator: @bdscstockkeepers
SciCrunch record: RRID:BDSC_8529
#9490
DOI: 10.1093/g3journal/jkab388
Resource: RRID:BDSC_9490
Curator: @bdscstockkeepers
SciCrunch record: RRID:BDSC_9490
GL00250 (GckIIIRNAi)
DOI: 10.1093/g3journal/jkab388
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
GL0021 (Ck1αRNAi#2)
DOI: 10.1093/g3journal/jkab388
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
JF01792 (Ck1αRNAi#1)
DOI: 10.1093/g3journal/jkab388
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
UAS-Gfp RNAi
DOI: 10.5607/en21021
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
BL27390
DOI: 10.1093/g3journal/jkab171
Resource: RRID:BDSC_27390
Curator: @bdscstockkeepers
SciCrunch record: RRID:BDSC_27390
BL32551
DOI: 10.1093/g3journal/jkab171
Resource: RRID:BDSC_32551
Curator: @scibot
SciCrunch record: RRID:BDSC_32551
31777
DOI: 10.1073/pnas.2107699118
Resource: RRID:BDSC_31777
Curator: @bdscstockkeepers
SciCrunch record: RRID:BDSC_31777
(BL30727
DOI: 10.1038/s42003-021-02757-z
Resource: RRID:BDSC_30727
Curator: @bdscstockkeepers
SciCrunch record: RRID:BDSC_30727
BL11749
DOI: 10.1038/s42003-021-02757-z
Resource: RRID:BDSC_11749
Curator: @bdscstockkeepers
SciCrunch record: RRID:BDSC_11749
BL42749
DOI: 10.1038/s42003-021-02757-z
Resource: RRID:BDSC_42749
Curator: @bdscstockkeepers
SciCrunch record: RRID:BDSC_42749
BL56555
DOI: 10.1038/s42003-021-02757-z
Resource: RRID:BDSC_56555
Curator: @bdscstockkeepers
SciCrunch record: RRID:BDSC_56555
BL62541
DOI: 10.1038/s42003-021-02757-z
Resource: RRID:BDSC_62541
Curator: @bdscstockkeepers
SciCrunch record: RRID:BDSC_62541
BL84276
DOI: 10.1038/s42003-021-02757-z
Resource: RRID:BDSC_84276
Curator: @bdscstockkeepers
SciCrunch record: RRID:BDSC_84276
BL52268
DOI: 10.1038/s42003-021-02757-z
Resource: RRID:BDSC_52268
Curator: @bdscstockkeepers
SciCrunch record: RRID:BDSC_52268
BL#66499
DOI: 10.1038/s42003-021-02757-z
Resource: RRID:BDSC_66499
Curator: @scibot
SciCrunch record: RRID:BDSC_66499
BL52268
DOI: 10.1038/s42003-021-02757-z
Resource: RRID:BDSC_52268
Curator: @scibot
SciCrunch record: RRID:BDSC_52268
BL11749
DOI: 10.1038/s42003-021-02757-z
Resource: RRID:BDSC_11749
Curator: @scibot
SciCrunch record: RRID:BDSC_11749
BL84276
DOI: 10.1038/s42003-021-02757-z
Resource: RRID:BDSC_84276
Curator: @scibot
SciCrunch record: RRID:BDSC_84276
BL30727
DOI: 10.1038/s42003-021-02757-z
Resource: RRID:BDSC_30727
Curator: @scibot
SciCrunch record: RRID:BDSC_30727
AB_528440
DOI: 10.1038/s42003-021-02757-z
Resource: (DSHB Cat# Prospero (MR1A), RRID:AB_528440)
Curator: @scibot
SciCrunch record: RRID:AB_528440
BL42749
DOI: 10.1038/s42003-021-02757-z
Resource: RRID:BDSC_42749
Curator: @scibot
SciCrunch record: RRID:BDSC_42749
Sgs3-GAL4
DOI: 10.1016/j.ydbio.2021.10.003
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
tub-GAL4
DOI: 10.1016/j.ydbio.2021.10.003
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
93128
DOI: 10.1016/j.ydbio.2021.10.001
Resource: RRID:BDSC_93128
Curator: @bdscstockkeepers
SciCrunch record: RRID:BDSC_93128
93130
DOI: 10.1016/j.ydbio.2021.10.001
Resource: RRID:BDSC_93130
Curator: @bdscstockkeepers
SciCrunch record: RRID:BDSC_93130
93129
DOI: 10.1016/j.ydbio.2021.10.001
Resource: RRID:BDSC_93129
Curator: @bdscstockkeepers
SciCrunch record: RRID:BDSC_93129
Bloomington stock center, #8760
DOI: 10.1016/j.celrep.2021.109769
Resource: RRID:BDSC_8760
Curator: @bdscstockkeepers
SciCrunch record: RRID:BDSC_8760
(arm)-Gal4
DOI: 10.1002/mds.28821
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
elav-Gal4
DOI: 10.1002/mds.28821
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
24B2-Gal4
DOI: 10.1002/mds.28821
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
tub-Gal4
DOI: 10.1002/mds.28821
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
RRID:SCR_001120
DOI: 10.7150/ijms.26329
Resource: Kyoto Encyclopedia of Genes and Genomes Expression Database (RRID:SCR_001120)
Curator: @scibot
SciCrunch record: RRID:SCR_001120
RRID:SCR_008864
DOI: 10.7150/ijms.26329
Resource: GOMO - Gene Ontology for Motifs (RRID:SCR_008864)
Curator: @scibot
SciCrunch record: RRID:SCR_008864
RRID:SCR_010969
DOI: 10.7150/ijms.26329
Resource: GenePix Pro (RRID:SCR_010969)
Curator: @scibot
SciCrunch record: RRID:SCR_010969
RRID:SCR_010848
DOI: 10.7150/ijms.26329
Resource: miRDB (RRID:SCR_010848)
Curator: @scibot
SciCrunch record: RRID:SCR_010848
RRID:SCR_008571
DOI: 10.7150/ijms.26329
Resource: Roche NimbleGen (RRID:SCR_008571)
Curator: @scibot
SciCrunch record: RRID:SCR_008571
RRID:SCR_010845
DOI: 10.7150/ijms.26329
Resource: TargetScan (RRID:SCR_010845)
Curator: @scibot
SciCrunch record: RRID:SCR_010845
RRID:Addgene_12259
DOI: 10.64898/2026.01.06.697868
Resource: RRID:Addgene_12259
Curator: @scibot
SciCrunch record: RRID:Addgene_12259
RRID:Addgene_12260
DOI: 10.64898/2026.01.06.697868
Resource: RRID:Addgene_12260
Curator: @scibot
SciCrunch record: RRID:Addgene_12260
RRID:Addgene_45952
DOI: 10.64898/2026.01.06.697868
Resource: RRID:Addgene_45952
Curator: @scibot
SciCrunch record: RRID:Addgene_45952
RRID:SCR_022613
DOI: 10.64898/2026.01.05.697808
Resource: Fred Hutchinson Cancer Center Flow Cytometry Core Facility (RRID:SCR_022613)
Curator: @scibot
SciCrunch record: RRID:SCR_022613
RRID:SCR_022606
DOI: 10.64898/2026.01.05.697808
Resource: Fred Hutchinson Cancer Center Genomics and Bioinformatics Core Facility (RRID:SCR_022606)
Curator: @scibot
SciCrunch record: RRID:SCR_022606
AB_2563846
DOI: 10.6084/m9.figshare.30598013
Resource: (BioLegend Cat# 139311, RRID:AB_2563846)
Curator: @scibot
SciCrunch record: RRID:AB_2563846
AB_312685
DOI: 10.6084/m9.figshare.30598013
Resource: (BioLegend Cat# 100320, RRID:AB_312685)
Curator: @scibot
SciCrunch record: RRID:AB_312685
AB_493646
DOI: 10.6084/m9.figshare.30598013
Resource: (BioLegend Cat# 100427, RRID:AB_493646)
Curator: @scibot
SciCrunch record: RRID:AB_493646
AB_891395
DOI: 10.6084/m9.figshare.30598013
Resource: (Thermo Fisher Scientific Cat# 35-0193-82, RRID:AB_891395)
Curator: @scibot
SciCrunch record: RRID:AB_891395
AB_312753
DOI: 10.6084/m9.figshare.30598013
Resource: (BioLegend Cat# 100714, RRID:AB_312753)
Curator: @scibot
SciCrunch record: RRID:AB_312753
AB_312791
DOI: 10.6084/m9.figshare.30598013
Resource: (BioLegend Cat# 101208, RRID:AB_312791)
Curator: @scibot
SciCrunch record: RRID:AB_312791
AB_313778
DOI: 10.6084/m9.figshare.30598013
Resource: (BioLegend Cat# 117309, RRID:AB_313778)
Curator: @scibot
SciCrunch record: RRID:AB_313778
AB_2565975
DOI: 10.6084/m9.figshare.30598013
Resource: (BioLegend Cat# 107641, RRID:AB_2565975)
Curator: @scibot
SciCrunch record: RRID:AB_2565975
AB_2562342
DOI: 10.6084/m9.figshare.30598013
Resource: (BioLegend Cat# 103140, RRID:AB_2562342)
Curator: @scibot
SciCrunch record: RRID:AB_2562342
RRID:AB_1642205
DOI: 10.6084/m9.figshare.30598013
Resource: (Cell Signaling Technology Cat# 3683, RRID:AB_1642205)
Curator: @scibot
SciCrunch record: RRID:AB_1642205
RRID:AB_2799843
DOI: 10.6084/m9.figshare.30598013
Resource: (Cell Signaling Technology Cat# 73671, RRID:AB_2799843)
Curator: @scibot
SciCrunch record: RRID:AB_2799843
RRID:SCR_002798
DOI: 10.6084/m9.figshare.30598013
Resource: GraphPad Prism (RRID:SCR_002798)
Curator: @scibot
SciCrunch record: RRID:SCR_002798
RRID:AB_2809227
DOI: 10.6084/m9.figshare.30598013
Resource: None
Curator: @scibot
SciCrunch record: RRID:AB_2809227
RRID:AB_772206
DOI: 10.6084/m9.figshare.30598013
Resource: (GE Healthcare Cat# NA934, RRID:AB_772206)
Curator: @scibot
SciCrunch record: RRID:AB_772206
AB_467251
DOI: 10.6084/m9.figshare.30598013
Resource: (Thermo Fisher Scientific Cat# 14-0451-82, RRID:AB_467251)
Curator: @scibot
SciCrunch record: RRID:AB_467251
AB_1236494
DOI: 10.6084/m9.figshare.30598013
Resource: (BioLegend Cat# 127606, RRID:AB_1236494)
Curator: @scibot
SciCrunch record: RRID:AB_1236494
RRID:AB_2799831
DOI: 10.6084/m9.figshare.30598013
Resource: (Cell Signaling Technology Cat# 72971, RRID:AB_2799831)
Curator: @scibot
SciCrunch record: RRID:AB_2799831
RRID:AB_312801
DOI: 10.6084/m9.figshare.30598013
Resource: (BioLegend Cat# 101302, RRID:AB_312801)
Curator: @scibot
SciCrunch record: RRID:AB_312801
RRID:SCR_003070
DOI: 10.6084/m9.figshare.30598013
Resource: ImageJ (RRID:SCR_003070)
Curator: @scibot
SciCrunch record: RRID:SCR_003070
RRID:AB_2773013
DOI: 10.6084/m9.figshare.30598013
Resource: (Cell Signaling Technology Cat# 29047, RRID:AB_2773013)
Curator: @scibot
SciCrunch record: RRID:AB_2773013
RRID:AB_10693472
DOI: 10.6084/m9.figshare.30598013
Resource: (Cell Signaling Technology Cat# 5483, RRID:AB_10693472)
Curator: @scibot
SciCrunch record: RRID:AB_10693472
BL26504
DOI: 10.3390/ijms222010997
Resource: RRID:BDSC_26504
Curator: @scibot
SciCrunch record: RRID:BDSC_26504
BL55110
DOI: 10.3389/fcell.2021.739357
Resource: RRID:BDSC_55110
Curator: @scibot
SciCrunch record: RRID:BDSC_55110
BL28281
DOI: 10.3389/fcell.2021.739357
Resource: RRID:BDSC_28281
Curator: @scibot
SciCrunch record: RRID:BDSC_28281
BL7018
DOI: 10.3389/fcell.2021.739357
Resource: RRID:BDSC_7018
Curator: @scibot
SciCrunch record: RRID:BDSC_7018
BL6902
DOI: 10.3389/fcell.2021.739357
Resource: RRID:BDSC_6902
Curator: @scibot
SciCrunch record: RRID:BDSC_6902
BL7011
DOI: 10.3389/fcell.2021.739357
Resource: RRID:BDSC_7011
Curator: @scibot
SciCrunch record: RRID:BDSC_7011
BL6313
DOI: 10.3389/fcell.2021.739357
Resource: RRID:BDSC_6313
Curator: @scibot
SciCrunch record: RRID:BDSC_6313
BL49248
DOI: 10.3389/fcell.2021.739357
Resource: RRID:BDSC_49248
Curator: @scibot
SciCrunch record: RRID:BDSC_49248
BL25041
DOI: 10.3389/fcell.2021.739357
Resource: RRID:BDSC_25041
Curator: @scibot
SciCrunch record: RRID:BDSC_25041
BL59829
DOI: 10.3389/fcell.2021.739357
Resource: RRID:BDSC_59829
Curator: @scibot
SciCrunch record: RRID:BDSC_59829
BL8546
DOI: 10.3389/fcell.2021.739357
Resource: RRID:BDSC_8546
Curator: @scibot
SciCrunch record: RRID:BDSC_8546
BL8492
DOI: 10.3389/fcell.2021.739357
Resource: RRID:BDSC_8492
Curator: @scibot
SciCrunch record: RRID:BDSC_8492
BL78361
DOI: 10.3389/fcell.2021.739357
Resource: RRID:BDSC_78361
Curator: @scibot
SciCrunch record: RRID:BDSC_78361
BL3605
DOI: 10.3389/fcell.2021.739357
Resource: RRID:BDSC_3605
Curator: @scibot
SciCrunch record: RRID:BDSC_3605
BL30140
DOI: 10.3389/fcell.2021.739357
Resource: RRID:BDSC_30140
Curator: @scibot
SciCrunch record: RRID:BDSC_30140
BL# 24511
DOI: 10.1242/jcs.258891
Resource: RRID:BDSC_24511
Curator: @scibot
SciCrunch record: RRID:BDSC_24511
BL# 5133
DOI: 10.1242/jcs.258891
Resource: RRID:BDSC_5133
Curator: @scibot
SciCrunch record: RRID:BDSC_5133
BL# 63802
DOI: 10.1242/jcs.258891
Resource: RRID:BDSC_63802
Curator: @scibot
SciCrunch record: RRID:BDSC_63802
BL 25786
DOI: 10.1242/jcs.258891
Resource: RRID:BDSC_25786
Curator: @scibot
SciCrunch record: RRID:BDSC_25786
BL#57084
DOI: 10.1242/jcs.258891
Resource: RRID:BDSC_57084
Curator: @scibot
SciCrunch record: RRID:BDSC_57084
BL# 23862
DOI: 10.1242/jcs.258891
Resource: RRID:BDSC_23862
Curator: @scibot
SciCrunch record: RRID:BDSC_23862
BL# 64459
DOI: 10.1242/jcs.258891
Resource: RRID:BDSC_64459
Curator: @scibot
SciCrunch record: RRID:BDSC_64459
BL9898
DOI: 10.1242/bio.058910
Resource: RRID:BDSC_9898
Curator: @scibot
SciCrunch record: RRID:BDSC_9898
BL33611
DOI: 10.1242/bio.058910
Resource: RRID:BDSC_33611
Curator: @scibot
SciCrunch record: RRID:BDSC_33611
BL9376
DOI: 10.1242/bio.058910
Resource: RRID:BDSC_9376
Curator: @scibot
SciCrunch record: RRID:BDSC_9376
BL23736
DOI: 10.1242/bio.058910
Resource: RRID:BDSC_23736
Curator: @scibot
SciCrunch record: RRID:BDSC_23736
BL26820
DOI: 10.1242/bio.058910
Resource: RRID:BDSC_26820
Curator: @scibot
SciCrunch record: RRID:BDSC_26820
BL58778
DOI: 10.1242/bio.058910
Resource: RRID:BDSC_58778
Curator: @scibot
SciCrunch record: RRID:BDSC_58778
BL58776
DOI: 10.1242/bio.058910
Resource: RRID:BDSC_58776
Curator: @scibot
SciCrunch record: RRID:BDSC_58776
BL51666
DOI: 10.1242/bio.058910
Resource: RRID:BDSC_51666
Curator: @scibot
SciCrunch record: RRID:BDSC_51666
RRID:SCR_024440
DOI: 10.1186/s13024-025-00919-9
Resource: Allen Brain Cell Atlas (RRID:SCR_024440)
Curator: @scibot
SciCrunch record: RRID:SCR_024440
Bloomington #5138
DOI: 10.1186/s12915-021-01154-6
Resource: RRID:BDSC_5138
Curator: @scibot
SciCrunch record: RRID:BDSC_5138
RRID:AB_262044
DOI: 10.1126/scisignal.adw1483
Resource: (Sigma-Aldrich Cat# F1804, RRID:AB_262044)
Curator: @scibot
SciCrunch record: RRID:AB_262044
BL3605
DOI: 10.1080/21505594.2021.1980989
Resource: RRID:BDSC_3605
Curator: @scibot
SciCrunch record: RRID:BDSC_3605
RRID:SCR_007345
DOI: 10.1038/s41598-025-34820-8
Resource: PhysioNet (RRID:SCR_007345)
Curator: @scibot
SciCrunch record: RRID:SCR_007345
RRID:AB_3095039
DOI: 10.1038/s41598-025-29548-4
Resource: (DRG International Cat# EIA5069R, RRID:AB_3095039)
Curator: @scibot
SciCrunch record: RRID:AB_3095039
RRID:SCR_016379
DOI: 10.1038/s41598-025-29548-4
Resource: Illumina MiSeq System (RRID:SCR_016379)
Curator: @scibot
SciCrunch record: RRID:SCR_016379
RRID:SCR_022982
DOI: 10.1038/s41598-025-29548-4
Resource: HTGQC (RRID:SCR_022982)
Curator: @scibot
SciCrunch record: RRID:SCR_022982
RRID:SCR_006431
DOI: 10.1038/s41531-025-01248-w
Resource: Parkinson's Progression Markers Initiative (RRID:SCR_006431)
Curator: @scibot
SciCrunch record: RRID:SCR_006431
RRID:AB_2889867
DOI: 10.1007/s40618-025-02710-5
Resource: (DiaSorin Cat# 310450, RRID:AB_2889867)
Curator: @scibot
SciCrunch record: RRID:AB_2889867
RRID:SCR_014242
DOI: 10.1007/s11894-023-00883-8
Resource: JMP (RRID:SCR_014242)
Curator: @scibot
SciCrunch record: RRID:SCR_014242
RRID:SCR_022647
DOI: 10.1007/s11894-023-00883-8
Resource: PICRUSt2 (RRID:SCR_022647)
Curator: @scibot
SciCrunch record: RRID:SCR_022647
RRID:SCR_001905
DOI: 10.1007/s11894-023-00883-8
Resource: R Project for Statistical Computing (RRID:SCR_001905)
Curator: @scibot
SciCrunch record: RRID:SCR_001905
RRID:SCR_006423
DOI: 10.1007/s11894-023-00883-8
Resource: SILVA (RRID:SCR_006423)
Curator: @scibot
SciCrunch record: RRID:SCR_006423
RRID:SCR_021258
DOI: 10.1007/s11894-023-00883-8
Resource: QIIME2 (RRID:SCR_021258)
Curator: @scibot
SciCrunch record: RRID:SCR_021258
RRID:SCR_018257
DOI: 10.1007/s11894-023-00883-8
Resource: QuPath (RRID:SCR_018257)
Curator: @scibot
SciCrunch record: RRID:SCR_018257
RRID:SCR_023241
DOI: 10.1007/s11894-023-00883-8
Resource: MaAsLin2 (RRID:SCR_023241)
Curator: @scibot
SciCrunch record: RRID:SCR_023241
RRID:SCR_003070
DOI: 10.1007/s11064-025-04647-w
Resource: ImageJ (RRID:SCR_003070)
Curator: @scibot
SciCrunch record: RRID:SCR_003070
RRID:AB_2892682
Traceback (most recent call last): File "/home/ubuntu/dashboard/py/create_release_tables.py", line 54, in format_anno_for_release parsedanno = HypothesisAnnotation(anno) File "/home/ubuntu/dashboard/py/hypothesis.py", line 192, in init t = row['document']['title'] TypeError: string indices must be integers
RRID:AB_3718609
Traceback (most recent call last): File "/home/ubuntu/dashboard/py/create_release_tables.py", line 54, in format_anno_for_release parsedanno = HypothesisAnnotation(anno) File "/home/ubuntu/dashboard/py/hypothesis.py", line 192, in init t = row['document']['title'] TypeError: string indices must be integers
RRID:AB_955447
Traceback (most recent call last): File "/home/ubuntu/dashboard/py/create_release_tables.py", line 54, in format_anno_for_release parsedanno = HypothesisAnnotation(anno) File "/home/ubuntu/dashboard/py/hypothesis.py", line 192, in init t = row['document']['title'] TypeError: string indices must be integers
RRID:AB_2753196
Traceback (most recent call last): File "/home/ubuntu/dashboard/py/create_release_tables.py", line 54, in format_anno_for_release parsedanno = HypothesisAnnotation(anno) File "/home/ubuntu/dashboard/py/hypothesis.py", line 192, in init t = row['document']['title'] TypeError: string indices must be integers
RRID:AB_2107448
Traceback (most recent call last): File "/home/ubuntu/dashboard/py/create_release_tables.py", line 54, in format_anno_for_release parsedanno = HypothesisAnnotation(anno) File "/home/ubuntu/dashboard/py/hypothesis.py", line 192, in init t = row['document']['title'] TypeError: string indices must be integers
RRID:AB_302459
Traceback (most recent call last): File "/home/ubuntu/dashboard/py/create_release_tables.py", line 54, in format_anno_for_release parsedanno = HypothesisAnnotation(anno) File "/home/ubuntu/dashboard/py/hypothesis.py", line 192, in init t = row['document']['title'] TypeError: string indices must be integers
RRID:AB_2756528
Traceback (most recent call last): File "/home/ubuntu/dashboard/py/create_release_tables.py", line 54, in format_anno_for_release parsedanno = HypothesisAnnotation(anno) File "/home/ubuntu/dashboard/py/hypothesis.py", line 192, in init t = row['document']['title'] TypeError: string indices must be integers
RRID:AB_2894870
Traceback (most recent call last): File "/home/ubuntu/dashboard/py/create_release_tables.py", line 54, in format_anno_for_release parsedanno = HypothesisAnnotation(anno) File "/home/ubuntu/dashboard/py/hypothesis.py", line 192, in init t = row['document']['title'] TypeError: string indices must be integers
RRID:AB_10597232
Traceback (most recent call last): File "/home/ubuntu/dashboard/py/create_release_tables.py", line 54, in format_anno_for_release parsedanno = HypothesisAnnotation(anno) File "/home/ubuntu/dashboard/py/hypothesis.py", line 192, in init t = row['document']['title'] TypeError: string indices must be integers
RRID:AB_2289842
Traceback (most recent call last): File "/home/ubuntu/dashboard/py/create_release_tables.py", line 54, in format_anno_for_release parsedanno = HypothesisAnnotation(anno) File "/home/ubuntu/dashboard/py/hypothesis.py", line 192, in init t = row['document']['title'] TypeError: string indices must be integers
RRID:AB_3083804
Traceback (most recent call last): File "/home/ubuntu/dashboard/py/create_release_tables.py", line 54, in format_anno_for_release parsedanno = HypothesisAnnotation(anno) File "/home/ubuntu/dashboard/py/hypothesis.py", line 192, in init t = row['document']['title'] TypeError: string indices must be integers
RRID:AB_2210545
Traceback (most recent call last): File "/home/ubuntu/dashboard/py/create_release_tables.py", line 54, in format_anno_for_release parsedanno = HypothesisAnnotation(anno) File "/home/ubuntu/dashboard/py/hypothesis.py", line 192, in init t = row['document']['title'] TypeError: string indices must be integers
RRID:AB_2801561
Traceback (most recent call last): File "/home/ubuntu/dashboard/py/create_release_tables.py", line 54, in format_anno_for_release parsedanno = HypothesisAnnotation(anno) File "/home/ubuntu/dashboard/py/hypothesis.py", line 192, in init t = row['document']['title'] TypeError: string indices must be integers
RRID:AB_955417
Traceback (most recent call last): File "/home/ubuntu/dashboard/py/create_release_tables.py", line 54, in format_anno_for_release parsedanno = HypothesisAnnotation(anno) File "/home/ubuntu/dashboard/py/hypothesis.py", line 192, in init t = row['document']['title'] TypeError: string indices must be integers
RRID:AB_2714032
Traceback (most recent call last): File "/home/ubuntu/dashboard/py/create_release_tables.py", line 54, in format_anno_for_release parsedanno = HypothesisAnnotation(anno) File "/home/ubuntu/dashboard/py/hypothesis.py", line 192, in init t = row['document']['title'] TypeError: string indices must be integers
RRID:AB_2076150
Traceback (most recent call last): File "/home/ubuntu/dashboard/py/create_release_tables.py", line 54, in format_anno_for_release parsedanno = HypothesisAnnotation(anno) File "/home/ubuntu/dashboard/py/hypothesis.py", line 192, in init t = row['document']['title'] TypeError: string indices must be integers
RRID:AB_10644283
Traceback (most recent call last): File "/home/ubuntu/dashboard/py/create_release_tables.py", line 54, in format_anno_for_release parsedanno = HypothesisAnnotation(anno) File "/home/ubuntu/dashboard/py/hypothesis.py", line 192, in init t = row['document']['title'] TypeError: string indices must be integers
RRID:AB_3678465
Traceback (most recent call last): File "/home/ubuntu/dashboard/py/create_release_tables.py", line 54, in format_anno_for_release parsedanno = HypothesisAnnotation(anno) File "/home/ubuntu/dashboard/py/hypothesis.py", line 192, in init t = row['document']['title'] TypeError: string indices must be integers
RRID:AB_3532206
DOI: 10.1007/s00737-025-01657-z
Resource: (Diagnostics Biochem Canada Cat# CAN-L-4260, RRID:AB_3532206)
Curator: @scibot
SciCrunch record: RRID:AB_3532206
the
Artistic and Iconographic Motifs: olmecs
p.s. Want the visual map? You'll find a full FieldNote Sketch Summary of this inside the 'hidden' layer. Click this highlight to see the synthesis, share it with someone you know needs it and save a copy for yourself.
You made it to the end. Here is the map of the territory we just covered.
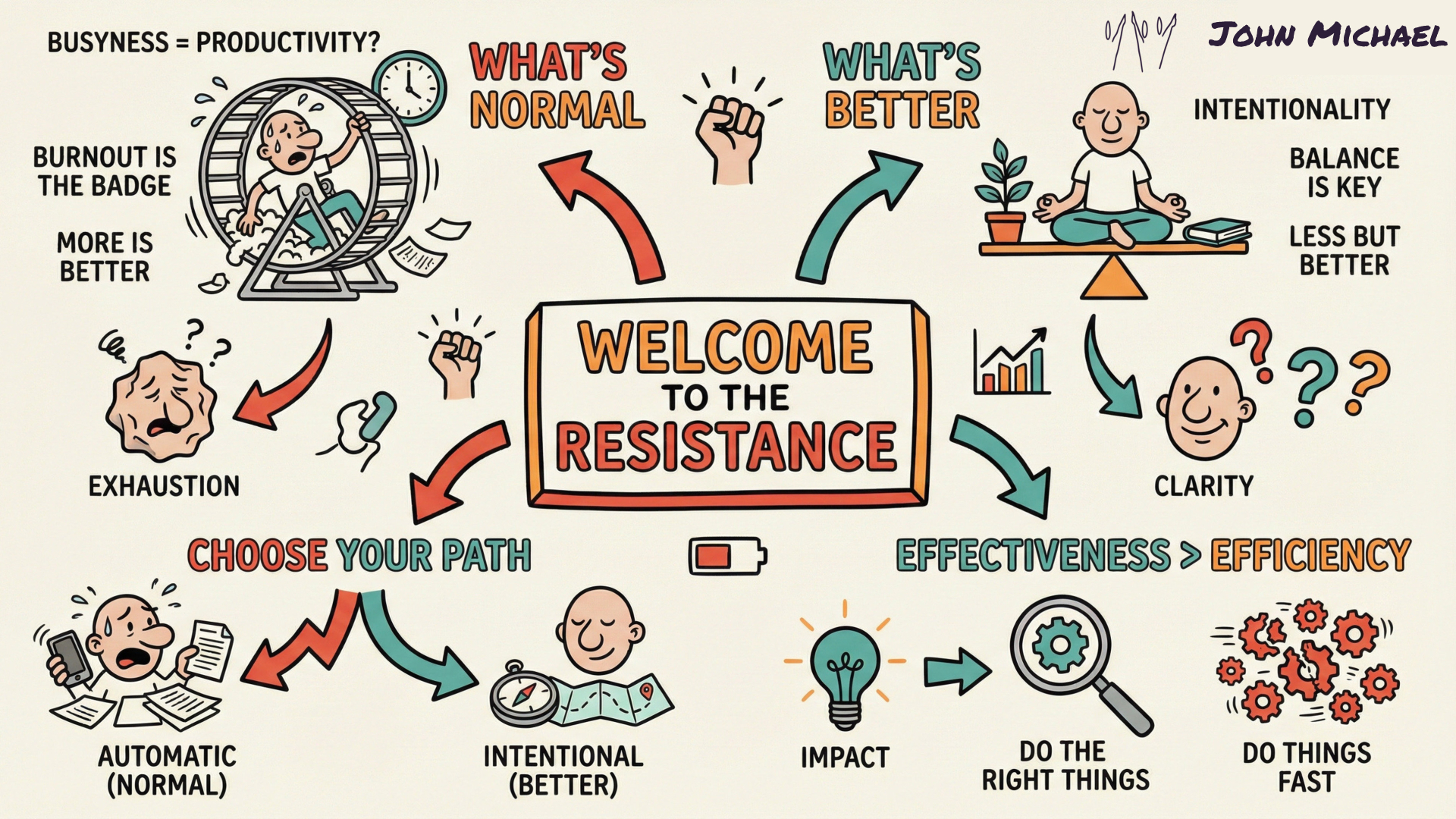 Save it, share it, or keep it as a reminder that the Resistance is just proof that you’re onto something big.
Save it, share it, or keep it as a reminder that the Resistance is just proof that you’re onto something big.
The Override Protocol You cannot out-argue the loop; you must overwrite it with Source Code.
This is the pivot. Everything changes when we spot the lie. If you're stuck here, use the counter-measure - it seems to easy to fix it, but it works!
We tend to think our demons are complex
I almost didn't hit publish on this. The Resistance told me it was too 'woo-woo' or too niche. If you're feeling that 'not-enoughness' right now, you’re in the right place.
THE RESISTANCE
For me, Resistance usually looks like a notification ping that distracts my attention right in the middle of my "flow" when I'm writing or sketching. What does your 'gatekeeper' look like today?"
7. ORDER BY If an order is specified by the ORDER BY clause, the rows are then sorted by the specified data in either ascending or descending order. Since all the expressions in the SELECT part of the query have been computed, you can reference aliases in this clause. 8. LIMIT / OFFSET Finally, the rows that fall outside the range specified by the LIMIT and OFFSET are discarded, leaving the final set of rows to be returned from the query.
最后先 order by 再 limit / offset
5. SELECT Any expressions in the SELECT part of the query are finally computed. 6. DISTINCT Of the remaining rows, rows with duplicate values in the column marked as DISTINCT will be discarded.
接下来先 select 再 distinct
If the query has a GROUP BY clause, then the constraints in the HAVING clause are then applied to the grouped rows, discard the grouped rows that don't satisfy the constraint. Like the WHERE clause, aliases are also not accessible from this step in most databases.
接下来是 having,进一步过滤 group by 之后的结果。
having 可以拿到聚合函数的值,但是不能使用 select 之后的别名。
As a result of the grouping, there will only be as many rows as there are unique values in that column. Implicitly, this means that you should only need to use this when you have aggregate functions in your query.
接下来是 group by,只有用聚合函数的时候才需要使用 group by
Each of the constraints can only access columns directly from the tables requested in the FROM clause. Aliases in the SELECT part of the query are not accessible in most databases since they may include expressions dependent on parts of the query that have not yet executed.
第一道执行的是 where 语句,只能获取到 from 语句里得到的列,select 里的别名获取不到
1. FROM and JOINs The FROM clause, and subsequent JOINs are first executed to determine the total working set of data that is being queried. This includes subqueries in this clause, and can cause temporary tables to be created under the hood containing all the columns and rows of the tables being joined.
from 和 join 第一个被执行,用来确定 working data set
如果有子查询的话会执行
Luckily, SQL allows us to do this by adding an additional HAVING clause which is used specifically with the GROUP BY clause to allow us to filter grouped rows from the result set.
HAVING 语句是用来 filter group by 之后的结果
COUNT(*), COUNT(column) A common function used to counts the number of rows in the group if no column name is specified. Otherwise, count the number of rows in the group with non-NULL values in the specified column.
count 组内的数量,* 是所有,指定了列就是 non-NULL 的行
Without a specified grouping, each aggregate function is going to run on the whole set of result rows and return a single value.
如果不带 grouping 的话,就是在整个表上计算,返回一个结果
In addition to the simple expressions that we introduced last lesson, SQL also supports the use of aggregate expressions (or functions) that allow you to summarize information about a group of rows of data.
aggregation 是聚合一系列的行
Sometimes, it's also not possible to avoid NULL values, as we saw in the last lesson when outer-joining two tables with asymmetric data.
有时候 null 是不可避免的,比如 join 的时候两张表的 asymmetric
In such cases, SQL provides a convenient way to discard rows that have a duplicate column value by using the DISTINCT keyword.
distinct the combination or the first following column?
answer: the combination
Another clause which is commonly used with the ORDER BY clause are the LIMIT and OFFSET clauses, which are a useful optimization to indicate to the database the subset of the results you care about. The LIMIT will reduce the number of rows to return, and the optional OFFSET will specify where to begin counting the number rows from.
limit 限制 return 的行数 offset 制定从哪开始 count row
offset 从 1 开始:limit 1 offset 1 返回第二个
When an ORDER BY clause is specified, each row is sorted alpha-numerically based on the specified column's value. In some databases, you can also specify a collation to better sort data containing international text.
by default it's sorted alpha-numerically
some db provide collation for better sorting for i18n text, but what?
Some of the buildings are new, so they don't have any employees in them yet, but we need to find some information about them regardless.
this is an example of growing independently:
new building can be without employees
You might see queries with these joins written as LEFT OUTER JOIN, RIGHT OUTER JOIN, or FULL OUTER JOIN, but the OUTER keyword is really kept for SQL-92 compatibility and these queries are simply equivalent to LEFT JOIN, RIGHT JOIN, and FULL JOIN respectively.
outer 关键字是和 SQL-92 兼容,等同于不写
When joining table A to table B, a LEFT JOIN simply includes rows from A regardless of whether a matching row is found in B. The RIGHT JOIN is the same, but reversed, keeping rows in B regardless of whether a match is found in A. Finally, a FULL JOIN simply means that rows from both tables are kept, regardless of whether a matching row exists in the other table.
left join right join full join
For example, the arts can connect to school concerns such as character education/bullying, collaboration, habits of mind, or multiple intelligences.
I love that the arts can be used to describe various topics!!
By their nature, the arts engage students in learning through observing, listening, and moving and offer learners various ways to acquire information and act on it to build understanding
This is an amazing idea that arts can provide to us in the classroom. It is so awesome that these skills are all used to help students gain a better understanding.
You might see queries where the INNER JOIN is written simply as a JOIN. These two are equivalent, but we will continue to refer to these joins as inner-joins because they make the query easier to read once you start using other types of joins,
inner join is join
After the tables are joined, the other clauses we learned previously are then applied.
first join
and allows for data in the database to grow independently of each other (ie. Types of car engines can grow independent of each type of car). As a trade-off, queries get slightly more complex since they have to be able to find data from different parts of the database, and performance issues can arise when working with many large tables.
normalization pros and cons
normalization
don't really understand normalization & orthogonal theory
June 13, 2019
and reflects the views of the author
After the primordium forms, SRY—a gene on the Y chromosome discovered in 1990, thanks to the participation of intersex XX males and XY females—might be activated.*
The popular belief that your sex arises only from your chromosomal makeup is wrong.The truth is, your biological sex isn’t carved in stone, but a living system with the potential for change.
Nearly everyone in middle school biology learned that if you’ve got XX chromosomes, you’re a female; if you’ve got XY, you’re a male. T
Contrary to popular belief, scientific research helps us better understand the unique and real transgender experience. Specifically, through three subjects: (1) genetics, (2) neurobiology and (3) endocrinology. So, hold onto your parts, whatever they may be. It’s time for “the talk.”
Antiscientific sentiment bombards our politics, or so says the Intellectual Dark Web (IDW)
3.This first line shows the exigence of the article, or what motivated the author to write this article. It essentially stemmed from groups on the internet spreading hateful, false rhetoric in order to influence the opinions of their readers. The author of this article, however, recognized the potential dangers of this, and therefore, wrote this article.
The real world consequences are stacking up: the trans military ban, bathroom bills, and removal of workplace and medical discrimination protections, a 41-51 percent suicide attempt rate and targeted fatal violence .
the Parisian soul
"The Parisian soul" .... Jayzus. "Wine-stained" ... please, no.
We should note that while most database implementations are quite efficient when using these operators, full-text search is best left to dedicated libraries like Apache Lucene or Sphinx.
数据库避免全文搜索对吗?
Apache Lucene 和 Sphinx 是啥?
IN (…) String exists in a list col_name IN ("A", "B", "C") NOT IN (…) String does not exist in a list col_name NOT IN ("D", "E", "F")
in and not in also for text data
_ Used anywhere in a string to match a single character (only with LIKE or NOT LIKE) col_name LIKE "AN_" (matches "AND", but not "AN")
_ is for one character used with like, not like
% Used anywhere in a string to match a sequence of zero or more characters (only with LIKE or NOT LIKE) col_name LIKE "%AT%" (matches "AT", "ATTIC", "CAT" or even "BATS")
used with like, not like 0 or more characters
LIKE Case insensitive exact string comparison
like is case insensitive comparison
= Case sensitive exact string comparison (notice the single equals) col_name = "abc"
In addition to making the results more manageable to understand, writing clauses to constrain the set of rows returned also allows the query to run faster due to the reduction in unnecessary data being returned.
返回的行数少了,SQL 就快了是吗?
IN (…) Number exists in a list col_name IN (2, 4, 6) NOT IN (…) Number does not exist in a list col_name NOT IN (1, 3, 5)
in and not in used for numeric data
BETWEEN … AND … Number is within range of two values (inclusive) col_name BETWEEN 1.5 AND 10.5 NOT BETWEEN … AND … Number is not within range of two values (inclusive) col_name NOT BETWEEN 1 AND 10
between ... and ... and not between ... and ...
used for numerical data inclusive
My colleagues and I at Sourcegraph are firmly in the camp that believes that you are the agent who will wrangle the task graph, above the level of the leaf nodes. We believe the wrangler will be human, not a model. We are not sitting around to wait for the research on autonomous agent
He seems to have shifted that position over the last year (Gas Town and other efforts)
And due to its simplicity, SQL databases provide safe and scalable storage for millions of websites and mobile applications.
不太理解为什么 SQL 数据库是 safe 和 scale 的
和存文件来比?
bát cháo hành của thị Nở làm hắn suy nghĩ nhiều
Bát cháo hành là biểu hiện của sự quan tâm giản dị nhưng có sức đánh thức phần người còn sót lại trong Chí Phèo
Tức mình hắn chửi ngay tất cả làng Vũ Ðại. Nhưng cả làng Vũ Ðại ai cũng nhủ, "Chắc nó trừ mình ra!" Không ai lên tiếng cả.
Tiếng chửi của Chí Phèo thể hiện sự cô độc của một con người bị xã hội ruồng bỏ
Good99
Good99 gia nhap san choi ca cuoc uy tin va chuyen nghiep. Noi mang den cho anh em nhung san pham giai tri hap dan cung co hoi rinh thuong lon.
Dia chi: 863 D. Le Duc Tho, Phuong 16, Go Vap, Ho Chi Minh, Viet Nam
Email: wildalutz760@gmail.com
Website: https://good99.top/
Dien thoai: (+84) 856212415
Social Links:
https://good99.top/
https://www.facebook.com/good99top/
https://www.youtube.com/channel/UChIKMLC40tE38i8HlKNqsaA
https://www.reddit.com/user/good99top/
https://www.pinterest.com/good99top/
https://ameblo.jp/good99top/entry-12920180461.html
https://gravatar.com/good99top
https://www.band.us/band/99472083/intro
https://www.blogger.com/profile/04997327806621680460
https://wildalutz760.wixsite.com/good99top
https://www.tumblr.com/good99top
https://good99top-cstxa.wordpress.com/
https://www.twitch.tv/good99top/about
https://sites.google.com/view/good99top/home
https://bookmarksclub.com/backlink/good99top/
https://good99top.mystrikingly.com/
https://good99top.amebaownd.com/
https://telegra.ph/good99top-08-01
https://mk.gta5-mods.com/users/good99top
https://688c628a577eb.site123.me/
https://scholar.google.com/citations?hl=vi&user=Ch1ROxIAAAAJ
https://www.pearltrees.com/good99top/item728363385
https://good99top.localinfo.jp/
https://good99top.shopinfo.jp/
https://good99top.hashnode.dev/good99top
https://good99top.themedia.jp/
https://rapidapi.com/user/good99top
https://fliphtml5.com/homepage/good99top/good99top/
https://good99top.therestaurant.jp/
https://ask.mallaky.com/?qa=user/good99top
https://good99top.website3.me/
https://www.quora.com/profile/Good99top
https://good99top.pixieset.com/
https://good99top.gumroad.com/
https://flipboard.com/@good99top/good99top-221hijnty
https://www.threadless.com/@good99top/activity
https://wakelet.com/@good99top
https://www.magcloud.com/user/good99top
https://hackmd.io/@good99top/good99top
https://good99top.blogspot.com/2025/08/good99top.html
https://good99top.doorkeeper.jp/
https://good99top.storeinfo.jp/
https://velog.io/@good99top/about
https://bato.to/u/2869294-good99top
https://quicknote.io/362f0e60-6ea8-11f0-9a10-ebfd0a0b8f55/
https://community.fabric.microsoft.com/t5/user/viewprofilepage/user-id/1326639
https://bulkwp.com/support-forums/users/good99top/
https://orcid.org/0009-0002-6150-8943
https://community.cisco.com/t5/user/viewprofilepage/user-id/1904032
https://archive.org/details/@good99top/web-archive
https://wpfr.net/support/utilisateurs/good99top
https://plaza.rakuten.co.jp/good99top/diary/202508010000/
https://pad.darmstadt.social/s/yQgbEkQ9c
https://pixabay.com/users/51573085/
https://disqus.com/by/good99top/about/
https://www.reverbnation.com/artist/good99top
https://es.gta5-mods.com/users/good99top
https://www.gamblingtherapy.org/forum/users/good99top/
https://forum.m5stack.com/user/good99top/
https://app.readthedocs.org/profiles/good99top/
https://public.tableau.com/app/profile/good99.top/viz/good99top/Sheet1#1
https://connect.garmin.com/modern/profile/6e1b2d49-4014-4ccf-b012-52144d3534cf
https://www.pixiv.net/en/users/118528628
https://community.amd.com/t5/user/viewprofilepage/user-id/518767
https://readtoto.com/u/2869294-good99top
https://qna.habr.com/user/good99top
https://id.gta5-mods.com/users/good99top
https://www.bark.com/en/gb/company/good99top/j8XK0N/
https://pastebin.com/u/good99top
https://www.storeboard.com/good99top
https://etextpad.com/jdcskgxpjj
https://md.darmstadt.ccc.de/s/ASS5MEKRl
https://comicvine.gamespot.com/profile/good99top/
https://padlet.com/good99top/good99top
https://3dwarehouse.sketchup.com/by/good99top
https://muckrack.com/good99-top/bio
https://hedgedoc.k8s.eonerc.rwth-aachen.de/s/G0976HjhV
https://nl.gta5-mods.com/users/good99top
https://openlibrary.org/people/good99top