the future of Southernliberalism might become great.
But in reality not so much
the future of Southernliberalism might become great.
But in reality not so much
Negroes are votingin some places in the South, and white people are tolerating it. In the newannual A.A.A. elections for the crop restriction program they are evenvoting right in the cotton counties of the Black Belt in perhaps even greaterproportion than whites.
The south is not static, but most southerns ideological beliefs are...
The South is also strongly religious.
Always was always will be in fact
woman suffrage andeconomic equality, collective bargaining, labor legislation, progressiveeducation, child welfare, civil service reform, police and court reform,prison refor
But not in the south, explains why they are so far behind today
mainly to the efforts of the federal government.The Southern masses do not generally organize either for advancing theirideals or for protecting their group interests.
What's the matter with kansas
which compels thewhite man to disenfranchise himself in order to take the vote away from the Negro
Lose lose
he fear is that this vast swarm of ignorant, purchasableand credulous voters will be compacted and controlled by desperate and unscrupulouswhite men, and made to hold the balance of power wherever the whites are divided.This fear has kept, and will keep, the whites "solid." It would keep the intelligenceand responsibility of any community, North or South, solid.
I mean racist and they even thought this was a justified reasoning. How about public education pal
Jonathan Furner furtherclarifies that records are not evidence in and ofthemselves, but are defined by their potentiality; they arecapable of serving as evidence in support of claims aboutthe past by a wide range of users
I thought this was a really interesting point, because as we discovered last week, meaning comes from the whole. The record on its own may have some personal value, but the records place in contributing to the whole picture is what makes it important to the archive. This is also where the question of where archives belong in terms of study; humanities? Social Science? It depends on how the record is serving as evidence, and to what purpose. Like Furner claims, they can be used for a wide range of users, and records may serve as different types of evidence to different people.
Be specific. The main points you make about your thesis and the examples you use to expand on those points need to be specific. Use specific examples to provide the evidence and to build upon your general ideas. These types of examples give your reader something narrow to focus on, and if used properly, they leave little doubt about your claim.
Being specific is important to provide evidence for your general ideas. This narrows down the main focus for the reader.
Once you have the structure of your paper figured out, and the main idea you will support, you can start with the introduction and conclusion.
This means you should plan your structure and main idea first, then write your introduction and conclusion.
What incentives do social media companies have to be careless with privacy?
Incentives could be them selling our information to other parties to continue selling more advertisements, or selling it just for money. This is non-consensual most of the time. I also believe that companies might overlook fortifying their security protocols because it might take too much time and money.
their regression coefficients
In the equation below, why isn't the superscript 1 used for the coefficients? It jumps from 0 to 2.
ackers can target individuals with attacks like: Password reuse attacks, where if they find out your password from one site, they try that password on many other sites Hackers tricking a computer into thinking they are another site, for example: the US NSA impersonated Google
Hacking can be very dangerous because many people reuse passwords or create predictable patterns that attackers can exploit. When hackers discover one password, they can use it across multiple sites, increasing the risk of data loss and unauthorized access.
While we have our concerns about the privacy of our information, we often share it with social media platforms under the understanding that they will hold that information securely. But social media companies often fail at keeping our information secure.
This has been a massive problem since the creation and widespread use of social media, and there has never been a definitive resolution to it, as hackers will always find ways to access people's personal information. Additionally, as mentioned, employees will always misuse their access and would also likely sell important personal information, especially if it comes to celebrities.
Hackers finding a vulnerability and inserting, modifying, or downloading information. For example:
When reading this section I'm reminded of a pretty infamous case that happened not long ago. The "Tea App" was used among young girls all across North America, and it was known for being an app where young women would go online and post about men they've been with and their bad, weird, or good attributes- essentially "spilling the tea". It was marketed as a "safe space" for women, where they could post anonomously and communicate potential catfishes, offenders, or overall bad men. However in 2023 a hacker had managed to leak all of the users' information, including but not limited to- credit card information, addresses, and 13,000 government IDs. This happened because the Tea app hadn't properly encrypted or protected the data, allowing the hacker to virtually access alll the users' information.**
While we have our concerns about the privacy of our information, we often share it with social media platforms under the understanding that they will hold that information securely. But social media companies often fail at keeping our information secure.
I find the concept of securty and privacy on social media incredibly intriguing, as users are almost always promised that just by having a username and password, your data is completely protected, or at least we are given that assumption. However, it's relatively easy to hack into anyone's account if you have the right knowledge and know where to look. Especially if you have the capabality to manage an app or website from the back end, having the ability to go through data within an application can lead to data leaks, and private information you thought no one would have can suddenly be given to the world.
While we have our concerns about the privacy of our information, we often share it with social media platforms under the understanding that they will hold that information securely. But social media companies often fail at keeping our information secure. For example, the proper security practice for storing user passwords is to use a special individual encryption process for each individual password. This way the database can only confirm that a password was the right one, but it can’t independently look up what the password is or even tell if two people used the same password. Therefore if someone had access to the database, the only way to figure out the right password is to use “brute force,” that is, keep guessing passwords until they guess the right one (and each guess takes a lot of time). But while that is the proper security for storing passwords. So for example, Facebook stored millions of Instagram passwords in plain text, meaning the passwords weren’t encrypted and anyone with access to the database could simply read everyone’s passwords. And Adobe encrypted their passwords improperly and then hackers leaked their password database of 153 million users.
This example illustrates a gap between users’ expectations of privacy and the actual practices of social media companies. Although users consent to sharing data, that consent assumes responsible stewardship, which is violated when companies fail to implement basic security measures.
Phishing attacks, where they make a fake version of a website or app and try to get you to enter your information or password into it. Some people have made malicious QR codes to take you to a phishing site.
I haven't heard the term "social engineering" used for this kind of scam. I have mostly heard it for in person actions, like running into a building and insisting to the security guard that you're late for your interview upstairs and he has to let you in as soon as possible.
Phishing attacks, where they make a fake version of a website or app and try to get you to enter your information or password into it
I remember a story in 2014, where many celebrities' personal photos were leaked because of a simple phishing scam involving an employee. It reminded me that network protection is not the only that needs to be protected but employees need training as well.
We might want to prevent people from stealing our identities or accounts, so we keep information (like passwords) private
This part confirms my thinking on password sharing is not good because the privacy that goes into it is not good. We keep information like phone passwords private to prevent others from stealing our identities or gaining unauthorized access to our accounts or other information. Sharing passwords removes that privacy and increases the risk of theft, financial loss, and misuse of personal information.
For example, a social media application might offer us a way of “Private Messaging” (also called Direct Messaging) with another user. But in most cases those “private” messages are stored in the computers at those companies, and the company might have computer programs that automatically search through the messages, and people with the right permissions might be able to view them directly.
I was aware that direct messaging can store data, not so "private" after all
I don't believe that privacy exists online, because even if there are laws that do not allow these companies to violate our privacy. The governments that make these laws are not as technologically advanced to actually enforce them.
For example, a social media application might offer us a way of “Private Messaging” (also called Direct Messaging) with another user. But in most cases those “private” messages are stored in the computers at those companies, and the company might have computer programs that automatically search through the messages, and people with the right permissions might be able to view them directly.
This is a good reminder that “private” on social media usually just means private from other users, not from the platform itself. It feels like the label creates a sense of safety that isn’t really accurate, especially when companies can still scan or access messages behind the scenes.
For example, a social media application might offer us a way of “Private Messaging” (also called Direct Messaging) with another user. But in most cases those “private” messages are stored in the computers at those companies, and the company might have computer programs that automatically search through the messages, and people with the right permissions might be able to view them directly.
This is a good reminder that “private” on social media usually just means private from other users, not from the platform itself. It feels like the label creates a sense of safety that isn’t really accurate, especially when companies can still scan or access messages behind the scenes.
Some governments and laws protect the privacy of individuals (using a Natural Rights ethical framing). These include the European Union’s General Data Protection Regulation (GDPR), which includes a “right to be forgotten”, and the United State’s Supreme Court has at times inferred a constitutional right to privacy.
This example effectively shows how a Natural Rights ethical framework is reflected in real-world laws, linking abstract ethical principles to concrete legal protections of privacy.
When we use social media platforms though, we at least partially give up some of our privacy.
Eli Pariser's quote in The Filter Bubble, “You’re getting a free service, and the cost is information about you,” is a powerful reminder of the hidden trade-offs in digital life. We often enjoy the convenience of online platforms without realizing we are paying with our personal data. This invisible exchange feels especially concerning because most users never fully understand what they are giving up.
the simple, hearty, sincere style of his exhortations might have edified even better educated persons. But it was in prayer that he especially excelled.
This shows that, although Tom doesn't have formal education or social power, people respond to him because he is sincere. This challenges hierarchies based on race and class as Stowe shows that moral authority does not belong solely to white or educated people.
It seemed to her as if strength poured into her in electric streams, from every gentle touch and movement of the sleeping, confiding child.
Yet another instance of Eliza drawing her endurance from motherhood. The imagery of "strength pour[ing] into her" is almost divine or spiritual. This portrays motherhood as a source of resilience.
I thought I could gild it over,—I thought, by kindness, and care, and instruction
The phrase "gild it over" suggests covering something ugly with a layer of gold, implying that kindness only disguises injustice and does not fix it. This is an example of Stowe's critique of so-called "benevolent" slaveholding.
of the real limits and fragility of the wo
As another article in this week’s reading suggests, the nature and condition of things, such as their fragility and state of breakdown, are often only noticed once everyday life and object has already been damaged. Only when people recognize this fragility and the state of breakdown can reflection and repair take place.
reading materials ahead of time in order to make the lecture and any complex concepts easier to follow. If
Some professors encourage reading the materials ahead of time, so that if questions do arise, which they will, they can be answered or better understood during the teaching. Even if the reading material is completely foreign, reading ahead of time gives the opportunity to be prepared and better understanding of the material.
stressful, but it is important to not feel overwhelmed by the anxiety of the situation so that you can make a carefully calculated decision based on the value and impact of your choice.
There should be coping techniques when anxiety arises, as it can be debilitating.
Figure 3.9 The Eisenhower Matrix can help organize priorities and ensure that you focus on the correct tasks.
Is this a daily chart ?
this, it is critically important to understand exactly what needs to be done to complete a task before you determine its priority.
Precisely! Or even what the task entails, what is it asking you to do.
at least making decisions based on strictly emotional reactions. It
Its always best to think before reacting, it lessens the chances of mistakes.
Why is prioritization important? What are the steps involved in prioritization? How do I deal with situation where others’ priorities are not the same as my own? What do I do when priorities conflict? What are the best ways to make sure I complete tasks?
These are all great questions to ask yourself and even asking your professor or another aid?
I think this is important because this would be the only tool that even users would have to stand up for their rights. to have a public specialist that to regulate the privacy rules and makes sure it makes sense to the average person.
9.3. Additional Privacy Violations# Besides hacking, there are other forms of privacy violations, such as: Unclear Privacy Rules: Sometimes privacy rules aren’t made clear to the people using a system. For example: If you send “private” messages on a work system, your boss might be able to read them. When Elon Musk purchased Twitter, he also was purchasing access to all Twitter Direct Messages Others Posting Without Permission: Someone may post something about another person without their permission. See in particular: The perils of ‘sharenting’: The parents who share too much Metadata: Sometimes the metadata that comes with content might violate someone’s privacy. For example, in 2012, former tech CEO John McAfee was a suspect in a murder in Belize, John McAfee hid out in secret. But when Vice magazine wrote an article about him, the photos in the story contained metadata with the exact location in Guatemala. Deanonymizing Data: Sometimes companies or researchers release datasets that have been “anonymized,” meaning that things like names have been removed, so you can’t directly see who the data is about. But sometimes people can still deduce who the anonymized data is about. This happened when Netflix released anonymized movie ratings data sets, but at least some users’ data could be traced back to them. Inferred Data: Sometimes information that doesn’t directly exist can be inferred through data mining (as we saw last chapter), and the creation of that new information could be a privacy violation. This includes the creation of Shadow Profiles, which are information about the user that the user didn’t provide or consent to Non-User Information: Social Media sites might collect information about people who don’t have accounts, like how Facebook does
I was surprised that privacy violations don’t only come from hacking, but also from normal everyday systems and data practices. Things like unclear rules, metadata, or “anonymous” datasets can still expose people without them realizing it. The example of photo metadata revealing someone’s hidden location shows how small technical details can create serious risks. This reminds me that data mining and sharing data always have hidden consequences, so companies should be much more responsible and transparent.
Deanonymizing Data: Sometimes companies or researchers release datasets that have been “anonymized,” meaning that things like names have been removed, so you can’t directly see who the data is about. But sometimes people can still deduce who the anonymized data is about. This happened when Netflix released anonymized movie ratings data sets, but at least some users’ data could be traced back to them.
This part stood out to me because it shows how fragile “anonymization” actually is. Even when obvious identifiers are removed, patterns in the data can still point back to real people. It makes anonymized data feel a lot less safe than it’s usually presented to be.
Unclear Privacy Rules: Sometimes privacy rules aren’t made clear to the people using a system. For example: If you send “private” messages on a work system, your boss might be able to read them. When Elon Musk purchased Twitter, he also was purchasing access to all Twitter Direct Messages Others Posting Without Permission: Someone may post something about another person without their permission. See in particular: The perils of ‘sharenting’: The parents who share too much Metadata: Sometimes the metadata that comes with content might violate someone’s privacy. For example, in 2012, former tech CEO John McAfee was a suspect in a murder in Belize, John McAfee hid out in secret. But when Vice magazine wrote an article about him, the photos in the story contained metadata with the exact location in Guatemala. Deanonymizing Data: Sometimes companies or researchers release datasets that have been “anonymized,” meaning that things like names have been removed, so you can’t directly see who the data is about. But sometimes people can still deduce who the anonymized data is about. This happened when Netflix released anonymized movie ratings data sets, but at least some users’ data could be traced back to them. Inferred Data: Sometimes information that doesn’t directly exist can be inferred through data mining (as we saw last chapter), and the creation of that new information could be a privacy violation. This includes the creation of Shadow Profiles, which are information about the user that the user didn’t provide or consent to Non-User Information: Social Media sites might collect information about people who don’t have accounts, like how Facebook does
I would like to say that I have a suspicion that our privacy was never secured or private, especially to the companies that are running the social media platform. The reason is that I feel like our activities are always under surveillance to prevent anything bad from happening or is going to happen.
Others Posting Without Permission: Someone may post something about another person without their permission. See in particular: The perils of ‘sharenting’: The parents who share too much
I am very interested in how the law evolves around sharing content of children online. The area is ripe for exploitation, child abuse, and human rights violations, we're already seeing horrifying stories unfold. How many tots today are going to sue their parents tomorrow for publicly posting videos of them they find embarrassing, traumatizing, or invasive?
Unclear Privacy Rules: Sometimes privacy rules aren’t made clear to the people using a system. For example: If you send “private” messages on a work system, your boss might be able to read them. When Elon Musk purchased Twitter, he also was purchasing access to all Twitter Direct Messages
This quote made me think about the long and complex consent agreements that users must accept before using many platforms. These documents often hide important privacy rules in dense language, making it difficult for users to fully understand what they are agreeing to. It raises the concern that companies might intentionally design these agreements to obscure their true data practices, taking advantage of users' lack of time or legal knowledge.
Others Posting Without Permission: Someone may post something about another person without their permission. See in particular: The perils of ‘sharenting’: The parents who share too much
This reminds me of how teenagers at times would screenshot dms. A lot of times it was evidence for an argument, but other times it would be malicious. This isn't wholly a bad thing however, this could be used to expose creeps.
Dan et al. (2021) highlights how deepfakes pose a direct threat toelectoral integrity—if voters cannot trust the authenticity of political content, democraticparticipation itself is jeopardized
Dan et al (2024)- if voters can't trust the authenticity of political content, democratic participation itself is jeopardised
growing cultureof distrust
growing culture of distrust is seeping into political institutions, distorting foundations of knowledge (Coeckelbergh (2024)) - loss of epistemic agency and one's confidence in independently assessing the truth which is essential to democracy
directly weakens societal trust by fueling disinformation
AI technology directly weakens societal trust by fueling disinformation- what is real? who can be a trusted source? is the person i'm seeing actually the person who's relaying the information?
AI enables the subtle fabrication of misleading content
ISSUE: AI enables fabrication and misleading content which undermines our collective belief in governmental transparency; erodes trust in democratic institutions
Hetherington’s (2005)
Hetherington (2005)- political trust is rooted in the perception that the government will act within the public's interests - ensure transparency, authenticity and accountability are more than ideals; but principles that are actively practiced
Hawley (2014)
Hawley (2014)- institutional trust exceeds belief in government goodwill- also includes the expectation that they are abled to complete democratic duties in good faith
BOSTON
The Civil War was between the North and the South. Boston is in Union territory; in the North
eLife Assessment
This study provides valuable findings on how the activity of the E3 ubiquitin ligase Highwire (Hiw/Phr1) is regulated and its impact on synaptic growth. The authors propose that impaired endocytosis leads to condensation of Hiw, resulting in increased synaptic growth. They also integrate such a mechanism within the known JNK (c-JUN N-terminal Kinase) and BMP (Bone Morphogenetic Protein) signalling pathways involved in synapse regulation. While the work raises an interesting mechanistic framework, several aspects of the experimental design and methodology are incomplete, and key conclusions, particularly those regarding the liquid-liquid phase separation of the E3 ubiquitin ligase, are not fully supported by the presented data.
Joint Public Review:
Pippadpally et al. investigate how the conserved E3 ubiquitin ligase Highwire (Hiw/Phr1), a well-established negative regulator of synaptic growth, is functionally and spatially regulated. Using a GFP-tagged Hiw transgene in Drosophila, the authors report that disruption of endocytosis via loss of AP-2, synaptojanin, or Rab11-mediated recycling endosome function leads to accumulation of Hiw in neuronal cell bodies as enlarged foci, altogether accompanied by synaptic overgrowth. Provided that the Hiw foci are sensitive to aliphatic alcohol treatment, the authors propose that impaired endocytosis promotes liquid-liquid phase separation of the E3 ubiquitin ligase, reducing its ability to degrade the MAPKKK Wallenda and thereby activating JNK signalling. Crosstalk with BMP signalling and roles for autophagy are also explored within this framework.
Strengths
The work provides a novel tool, the GFP-tagged Hiw transgene, to study the spatio-temporal regulation of the E3 ubiquitin ligase Highwire (Hiw/Phr1) in Drosophila, and its impact on synaptic growth. The results presented point to a potentially thought-provoking connection between endocytic defects, Hiw condensation, Hiw down-regulation and synaptic overgrowth. The specific effects of the endocytic mutants on the redistribution of the Hiw to the neuronal cell body and the genetic interactions between the endocytosis and JNK pathway mutants are convincing.
Weaknesses
Several conclusions are insufficiently supported at this point. For example, evidence that the Hiw foci represent bona fide liquid-liquid phase (LLP) separated condensates is limited. Sensitivity to 1,6-hexanediol is not definitive proof of their liquid condensate nature, and their recovery kinetics after 1,6-hexanediol wash-out and their morphology are inconsistent with a pure liquid behaviour. Furthermore, the claim that the Hiw foci are non-vesicular is not strongly supported, as it is only based on the lack of colocalization with a handful of endosomal proteins.
Importantly, the appearance of the putative condensates is correlative rather than causative for synaptic overgrowth, and in the absence of a mechanistic link between endocytosis and Hiw condensation, the causality is difficult to address. Of note is that the putative condensates are already present (albeit to a lesser extent) in the absence of endocytic defects and that the conclusions rely heavily on overexpressed GFP-Hiw, which may perturb normal protein behaviour and artificially induce condensation or aggregation.
The use of hypomorphic mutants in genetic experiments also introduces some ambiguity in their interpretation, as the results may reflect dosage effects from multiple pathways rather than pathway order. Finally, the manuscript would benefit from a more comprehensive reference to relevant literature on JNKKKs and BMP signalling, as well as on the recycling endosome function in synaptic growth and the regulation of the aforementioned pathways.
Overall, while the work presents thought-provoking observations and a potentially interesting regulatory model, additional experimental rigor and broader contextualization are needed to substantiate the proposed mechanism and its biological relevance.
Author response:
Weaknesses:
(1) Several conclusions are insufficiently supported at this point. For example, evidence that the Hiw foci represent bona fide liquid-liquid phase (LLP) separated condensates is limited. Sensitivity to 1,6-hexanediol is not definitive proof of their liquid condensate nature, and their recovery kinetics after 1,6-hexanediol wash-out and their morphology are inconsistent with a pure liquid behaviour. Furthermore, the claim that the Hiw foci are non-vesicular is not strongly supported, as it is only based on the lack of colocalization with a handful of endosomal proteins.
We agree that, at the current stage of the manuscript, we have presented data only on Hiw foci in the VNC and shown that they are sensitive to 1,6-HD but not to 2,5-HD. To further provide definitive proof that these are bona fide condensates, we will now perform in vitro analysis of different domains of Hiw and the Hiw IDR region. In addition, we will also investigate the Hiw-GFP behavior in non-neuronal and transiently transfected cell lines using FRAP and other protocols previously applied to condensate-forming proteins.
Finally, we will perform an in-depth analysis of the Hiw condensates for their colocalization with endocytic proteins and cellular compartments and determine whether they are part of any known vesicular structures.
(2) Importantly, the appearance of the putative condensates is correlative rather than causative for synaptic overgrowth, and in the absence of a mechanistic link between endocytosis and Hiw condensation, the causality is difficult to address. Of note is that the putative condensates are already present (albeit to a lesser extent) in the absence of endocytic defects and that the conclusions rely heavily on overexpressed GFP-Hiw, which may perturb normal protein behaviour and artificially induce condensation or aggregation.
To investigate the formation of condensates and their relation to synaptic growth, we will perform a time-course analysis of changes at the NMJ and correlate with the Hiw condensate appearance in the VNC of shi<sup>ts</sup> expressing GFP-Hiw, along with appropriate controls. The GFP transgene used is a functional transgene and well established for studying Hiw behaviour. The Hiw condensates do not form when expressed on an otherwise wild-type background. We will further assess the formation of Hiw condensates in other endocytic mutants with appropriate controls.
(3) The use of hypomorphic mutants in genetic experiments also introduces some ambiguity in their interpretation, as the results may reflect dosage effects from multiple pathways rather than pathway order. Finally, the manuscript would benefit from a more comprehensive reference to relevant literature on JNKKKs and BMP signalling, as well as on the recycling endosome function in synaptic growth and the regulation of the aforementioned pathways.
We will perform genetic analysis using homozygous mutants of the wit and saxophone genes to further support epistatic interactions between the BMP signaling pathway and synaptic growth. We will strengthen the discussion part.
eLife Assessment
The authors use sequencing of nascent DNA (DNA linked to an RNA primer, “SNS-Seq”) to localise DNA replication origins in Trypanosoma brucei, so this work will be of interest to those studying either Kinetoplastids or DNA replication. The paper presents the SNS-seq results for only part of the genome, and there are significant discrepancies between the SNS-Seq results and those from other, previously-published results obtained using other origin mapping methods. The reasons for the differences are unknown and from the data available, it is not possible to assess which origin-mapping method is most suitable for origin mapping in T. brucei. Thus at present, the evidence that origins are distributed as the authors claim - and not where previously mapped - is inadequate.
Reviewer #1 (Public review):
In this paper, Stanojcic and colleagues attempt to map sites of DNA replication initiation in the genome of the African trypanosome, Trypanosoma brucei. Their approach to this mapping is to isolate 'short-nascent strands' (SNSs), a strategy adopted previously in other eukaryotes (including in the related parasite Leishmania major), which involves isolation of DNA molecules whose termini contain replication-priming RNA. By mapping the isolated and sequenced SNSs to the genome (SNS-seq), the authors suggest that they have identified origins, which they localise to intergenic (strictly, inter-CDS) regions within polycistronic transcription units and suggest display very extensive overlap with previously mapped R-loops in the same loci. Finally, having defined locations of SNS-seq mapping, they suggest they have identified G4 and nucleosome features of origins, again using previously generated data. Though there is merit in applying a new approach to understand DNA replication initiation in T. brucei, where previous work has used MFA-seq and ChIP of a subunit of the Origin Replication Complex (ORC), there are two significant deficiencies in the study that must be addressed to ensure rigour and accuracy.
(1) The suggestion that the SNS-seq data is mapping DNA replication origins that are present in inter-CDS regions of the polycistronic transcription units of T. brucei is novel and does not agree with existing data on the localisation of ORC1/CDC6, and it is very unclear if it agrees with previous mapping of DNA replication by MFA-seq due to the way the authors have presented this correlation. For these reasons, the findings essentially rely on a single experimental approach, which must be further tested to ensure SNS-seq is truly detecting origins. Indeed, in this regard, the very extensive overlap of SNS-seq signal with RNA-DNA hybrids should be tested further to rule out the possibility that the approach is mapping these structures and not origins.
(2) The authors' presentation of their SNS-seq data is too limited and therefore potentially provides a misleading view of DNA replication in the genome of T. brucei. The work is presented through a narrow focus on SNS-seq signal in the inter-CDS regions within polycistronic transcription units, which constitute only part of the genome, ignoring both the transcription start and stop sites at the ends of the units and the large subtelomeres, which are mainly transcriptionally silent. The authors must present a fuller and more balanced view of SNS-seq mapping across the whole genome to ensure full understanding and clarity.
Reviewer #2 (Public review):
Summary:
Stanojcic et al. investigate the origins of DNA replication in the unicellular parasite Trypanosoma brucei. They perform two experiments, stranded SNS-seq and DNA molecular combing. Further, they integrate various publicly available datasets, such as G4-seq and DRIP-seq, into their extensive analysis. Using this data, they elucidate the structure of the origins of replication. In particular, they find various properties located at or around origins, such as polynucleotide stretches, G-quadruplex structures, regions of low and high nucleosome occupancy, R-loops, and that origins are mostly present in intergenic regions. Combining their population-level SNS-seq and their single-molecule DNA molecular combing data, they elucidate the total number of origins as well as the number of origins active in a single cell.
Strengths:
(1) A very strong part of this manuscript is that the authors integrate several other datasets and investigate a large number of properties around origins of replication. Data analysis clearly shows the enrichment of various properties at the origins, and the manuscript concludes with a very well-presented model that clearly explains the authors' understanding and interpretation of the data.
(2) The DNA combing experiment is an excellent orthogonal approach to the SNS-seq data. The authors used the different properties of the two experiments (one giving location information, one giving single-molecule information) well to extract information and contrast the experiments.
(3) The discussion is exemplary, as the authors openly discuss the strengths and weaknesses of the approaches used. Further, the discussion serves its purpose of putting the results in both an evolutionary and a trypanosome-focused context.
Weaknesses:
I have major concerns about the origin of replication sites determined from the SNS-seq data. As a caveat, I want to state that, before reading this manuscript, SNS-seq was unknown to me; hence, some of my concerns might be misplaced.
(1) I do not understand why SNS-seq would create peaks. Replication should originate in one locus, then move outward in both directions until the replication fork moving outward from another origin is encountered. Hence, in an asynchronous population average measurement, I would expect SNS data to be broad regions of + and -, which, taken together, cover the whole genome. Why are there so many regions not covered at all by reads, and why are there such narrow peaks?
(2) I am concerned that up to 96% percent of all peaks are filtered away. If there is so much noise in the data, how can one be sure that the peaks that remain are real? Specifically, if the authors placed the same number of peaks as was measured randomly in intergenic regions, would 4% of these peaks pass the filtering process by chance?
(3) There are 3 previous studies that map origins of replication in T. brucei. Devlin et al. 2016, Tiengwe et al. 2012, and Krasiļņikova et al. 2025 (https://doi.org/10.1038/s41467-025-56087-3), all with a different technique: MFA-seq. All three previous studies mostly agree on the locations and number of origins. The authors compared their results to the first two, but not the last study; they found that their results are vastly different from the previous studies (see Supplementary Figure 8A). In their discussion, the authors defend this discrepancy mostly by stating that the discrepancy between these methods has been observed in other organisms. I believe that, given the situation that the other studies precede this manuscript, it is the authors' duty to investigate the differences more than by merely pointing to other organisms. A conclusion should be reached on why the results are different, e.g., by orthogonally validating origins absent in the previous studies.
(4) Some patterns that were identified to be associated with origins of replication, such as G-quadruplexes and nucleosomes phasing, are known to be biases of SNS-seq (see Foulk et al. Characterizing and controlling intrinsic biases of lambda exonuclease in nascent strand sequencing reveals phasing between nucleosomes and G-quadruplex motifs around a subset of human replication origins. Genome Res. 2015;25(5):725-735. doi:10.1101/gr.183848.114).
Are the claims well substantiated?:
My opinion on whether the authors' results support their conclusions depends on whether my concerns about the sites determined from the SNS-seq data can be dismissed. In the case that these concerns can be dismissed, I do think that the claims are compelling.
Impact:
If the origins of replication prove to be distributed as claimed, this study has the potential to be important for two fields. Firstly, in research focused on T. brucei as a disease agent, where essential processes that function differently than in mammals are excellent drug targets. Secondly, this study would impact basic research analyzing DNA replication over the evolutionary tree, where T. brucei can be used as an early-divergent eukaryotic model organism.
Author response:
eLife Assessment
The authors use sequencing of nascent DNA (DNA linked to an RNA primer, "SNS-Seq") to localise DNA replication origins in Trypanosoma brucei, so this work will be of interest to those studying either Kinetoplastids or DNA replication. The paper presents the SNS-seq results for only part of the genome, and there are significant discrepancies between the SNS-Seq results and those from other, previously-published results obtained using other origin mapping methods. The reasons for the differences are unknown and from the data available, it is not possible to assess which origin-mapping method is most suitable for origin mapping in T. brucei. Thus at present, the evidence that origins are distributed as the authors claim - and not where previously mapped - is inadequate.
We would like to clarify a few points regarding our study. Our primary objective was to characterise the topology and genome-wide distribution of short nascent-strand (SNS) enrichments. The stranded SNS-seq approach provides the high strand-specific resolution required to analyse origins. The observation that SNS-seq peaks (potential origins) are most frequently found in intergenic regions is not an artefact of analysing only part of the genome; rather, it is a result of analysing the entire genome.
We agree that orthogonal validation is necessary. However, neither MFA-seq nor TbORC1/CDC6 ChIP-on-chip has yet been experimentally validated as definitive markers of origin activity in T. brucei, nor do they validate each other.
Public Reviews:
Reviewer #1 (Public review):
In this paper, Stanojcic and colleagues attempt to map sites of DNA replication initiation in the genome of the African trypanosome, Trypanosoma brucei. Their approach to this mapping is to isolate 'short-nascent strands' (SNSs), a strategy adopted previously in other eukaryotes (including in the related parasite Leishmania major), which involves isolation of DNA molecules whose termini contain replication-priming RNA. By mapping the isolated and sequenced SNSs to the genome (SNS-seq), the authors suggest that they have identified origins, which they localise to intergenic (strictly, inter-CDS) regions within polycistronic transcription units and suggest display very extensive overlap with previously mapped R-loops in the same loci. Finally, having defined locations of SNS-seq mapping, they suggest they have identified G4 and nucleosome features of origins, again using previously generated data.
Though there is merit in applying a new approach to understand DNA replication initiation in T. brucei, where previous work has used MFA-seq and ChIP of a subunit of the Origin Replication Complex (ORC), there are two significant deficiencies in the study that must be addressed to ensure rigour and accuracy.
(1) The suggestion that the SNS-seq data is mapping DNA replication origins that are present in inter-CDS regions of the polycistronic transcription units of T. brucei is novel and does not agree with existing data on the localisation of ORC1/CDC6, and it is very unclear if it agrees with previous mapping of DNA replication by MFA-seq due to the way the authors have presented this correlation. For these reasons, the findings essentially rely on a single experimental approach, which must be further tested to ensure SNS-seq is truly detecting origins. Indeed, in this regard, the very extensive overlap of SNS-seq signal with RNA-DNA hybrids should be tested further to rule out the possibility that the approach is mapping these structures and not origins.
(2) The authors' presentation of their SNS-seq data is too limited and therefore potentially provides a misleading view of DNA replication in the genome of T. brucei. The work is presented through a narrow focus on SNS-seq signal in the inter-CDS regions within polycistronic transcription units, which constitute only part of the genome, ignoring both the transcription start and stop sites at the ends of the units and the large subtelomeres, which are mainly transcriptionally silent. The authors must present a fuller and more balanced view of SNS-seq mapping across the whole genome to ensure full understanding and clarity.
Regarding comparisons with previous work:
Two other attempts to identify origins in T. brucei —ORC1/CDC6 binding sites (ChIP-on-chip, PMID: 22840408) and MFA-seq (PMID: 22840408, 27228154)—were both produced by the McCulloch group. These methods do not validate each other; in fact, MFA-seq origins overlap with only 4.4% of the 953 ORC1/CDC6 sites (PMID: 29491738). Therefore, low overlap between SNS-seq peaks and ORC1/CDC6 sites cannot disqualify our findings. Similar low overlaps are observed in other parasites (PMID: 38441981, PMID: 38038269, PMID: 36808528) and in human cells (PMID: 38567819).
We also would like to emphasize that the ORC1/CDC6 dataset originally published (PMID: 22840408) is no longer available; only a re-analysis by TritrypDB exists, which differs significantly from the published version (personal communication from Richard McCulloch). While the McCulloch group reported a predominant localization of ORC1/CDC6 sites within SSRs at transcription start and termination regions, our re-analysis indicates that only 10.3% of TbORC1/CDC6-12Myc sites overlapped with 41.8% of SSRs.
MFA-seq does not map individual origins, it rather detects replicated genomic regions by comparing DNA copy number between S- and G1-phases of the cell cycle (PMID: 36640769; PMID: 37469113; PMID: 36455525). The broad replicated regions (0.1–0.5 Mbp) identified by MFA-seq in T. brucei are likely to contain multiple origins, rather than just one. In that sense we disagree with the McCulloch's group who claimed that there is a single origin per broad peak. Our analysis shows that up to 50% of the origins detected by stranded SNS-seq locate within broad MFA-seq regions. The methodology used by McCulloch’s group to infer single origins from MFA-seq regions has not been published or made available, as well as the precise position of these regions, making direct comparison difficult.
Finally, the genomic features we describe—poly(dA/dT) stretches, G4 structures and nucleosome occupancy patterns—are consistent with origin topology described in other organisms.
On the concern that SNS-seq may map RNA-DNA hybrids rather than replication origins: Isolation and sequencing of short nascent strands (SNS) is a well-established and widely used technique for high-resolution origin mapping. This technique has been employed for decades in various laboratories, with numerous publications documenting its use. We followed the published protocol for SNS isolation (Cayrou et al., Methods, 2012, PMID: 22796403). RNA-DNA hybrids cannot persist through the multiple denaturation steps in our workflow, as they melt at 95°C (Roberts and Crothers, Science, 1992; PMID: 1279808). Even in the unlikely event that some hybrids remained, they would not be incorporated into libraries prepared using a single-stranded DNA protocol and therefore would not be sequenced (see Figure 1B and Methods).
Furthermore, our analysis shows that only a small proportion (1.7%) of previously reported RNA-DNA hybrids overlap with SNS-seq origins. It is important to note that RNA-primed nascent strands naturally form RNA-DNA hybrids during replication initiation, meaning the enrichment of RNA-DNA hybrids near origins is both expected and biologically relevant.
On the claim that our analysis focuses narrowly on inter-CDS regions and ignores other genomic compartments: this is incorrect. We mapped and analyzed stranded SNS-seq data across the entire genome of T. brucei 427 wild-type strain (Müller et al., Nature, 2018; PMID: 30333624), including both core and subtelomeric regions. Our findings indicate that most origins are located in intergenic regions, but all analyses were performed using the full set of detected origins, regardless of location.
We did not ignore transcription start and stop sites (TSS/TTS). The manuscript already includes origin distribution across genomic compartments as defined by TriTrypDB (Fig. 2C) and addresses overlap with TSS, TTS and HT in the section “Spatial coordination between the activity of the origin and transcription”. While this overlap is minimal, we have included metaplots in the revised manuscript for clarity.
Reviewer #2 (Public review):
Summary:
Stanojcic et al. investigate the origins of DNA replication in the unicellular parasite Trypanosoma brucei. They perform two experiments, stranded SNS-seq and DNA molecular combing. Further, they integrate various publicly available datasets, such as G4-seq and DRIP-seq, into their extensive analysis. Using this data, they elucidate the structure of the origins of replication. In particular, they find various properties located at or around origins, such as polynucleotide stretches, G-quadruplex structures, regions of low and high nucleosome occupancy, R-loops, and that origins are mostly present in intergenic regions. Combining their population-level SNS-seq and their single-molecule DNA molecular combing data, they elucidate the total number of origins as well as the number of origins active in a single cell.
Strengths:
(1) A very strong part of this manuscript is that the authors integrate several other datasets and investigate a large number of properties around origins of replication. Data analysis clearly shows the enrichment of various properties at the origins, and the manuscript concludes with a very well-presented model that clearly explains the authors' understanding and interpretation of the data.
We sincerely thank you for this positive feedback.
(2) The DNA combing experiment is an excellent orthogonal approach to the SNS-seq data. The authors used the different properties of the two experiments (one giving location information, one giving single-molecule information) well to extract information and contrast the experiments.
Thank you very much for this remark.
(3) The discussion is exemplary, as the authors openly discuss the strengths and weaknesses of the approaches used. Further, the discussion serves its purpose of putting the results in both an evolutionary and a trypanosome-focused context.
Thank you for appreciating our discussion.
Weaknesses:
I have major concerns about the origin of replication sites determined from the SNS-seq data. As a caveat, I want to state that, before reading this manuscript, SNS-seq was unknown to me; hence, some of my concerns might be misplaced.
(1) I do not understand why SNS-seq would create peaks. Replication should originate in one locus, then move outward in both directions until the replication fork moving outward from another origin is encountered. Hence, in an asynchronous population average measurement, I would expect SNS data to be broad regions of + and -, which, taken together, cover the whole genome. Why are there so many regions not covered at all by reads, and why are there such narrow peaks?
Thank you for asking these questions. As you correctly point out, replication forks progress in both directions from their origins and ultimately converge at termination sites. However, the SNS-seq method specifically isolates short nascent strands (SNSs) of 0.5–2.5 kb using a sucrose gradient. These short fragments are generated immediately after origin firing and mark the sites of replication initiation, rather than the entire replicated regions. Consequently: (i) SNS-seq does not capture long replication forks or termination regions, only the immediate vicinity of origins. (ii) The narrow peaks indicate the size of selected SNSs (0.5–2.5 kb) and the fact that many cells initiate replication at the same genomic sites, leading to localized enrichment. (iii) Regions without coverage refer to genomic areas that do not serve as efficient origins in the analyzed cell population. Thus, SNS-seq is designed to map origin positions, but not the entire replicated regions.
(2) I am concerned that up to 96% percent of all peaks are filtered away. If there is so much noise in the data, how can one be sure that the peaks that remain are real? Specifically, if the authors placed the same number of peaks as was measured randomly in intergenic regions, would 4% of these peaks pass the filtering process by chance?
Maintaining the strandness of the sequenced DNA fibres enabled us to filter the peaks, thereby increasing the probability that the filtered peak pairs corresponded to origins. Two SNS peaks must be oriented in a way that reflects the topology of the SNS strands within an active origin: the upstream peak must be on the minus strand and followed by the downstream peak on the plus strand.
As suggested by the reviewer, we tested whether randomly placed plus and minus peaks could reproduce the number of filter-passing peaks using the same bioinformatics workflow. Only 1–6% of random peaks passed the filters, compared with 4–12% in our experimental data, resulting in about 50% fewer selected regions (origins). Moreover, the “origins” from random peaks showed 0% reproducibility across replicates, whereas the experimental data showed 7–64% reproducibility. These results indicate that the retainee peaks are highly unlikely to arise by chance and support the specificity of our approach. Thank you for this suggestion.
(3) There are 3 previous studies that map origins of replication in T. brucei. Devlin et al. 2016, Tiengwe et al. 2012, and Krasiļņikova et al. 2025 (https://doi.org/10.1038/s41467-025-56087-3), all with a different technique: MFA-seq. All three previous studies mostly agree on the locations and number of origins. The authors compared their results to the first two, but not the last study; they found that their results are vastly different from the previous studies (see Supplementary Figure 8A). In their discussion, the authors defend this discrepancy mostly by stating that the discrepancy between these methods has been observed in other organisms. I believe that, given the situation that the other studies precede this manuscript, it is the authors' duty to investigate the differences more than by merely pointing to other organisms. A conclusion should be reached on why the results are different, e.g., by orthogonally validating origins absent in the previous studies.
The MFA-seq data for T. brucei were published in two studies by McCulloch’s group: Tiengwe et al. (2012) using TREU927 PCF cells, and Devlin et al. (2016) using PCF and BSF Lister427 cells. In Krasilnikova et al. (2025), previously published MFA-seq data from Devlin et al. were remapped to a new genome assembly without generating new MFA-seq data, which explains why we did not include that comparison.
Clarifying the differences between MFA-seq and our stranded SNS-seq data is essential. MFA-seq and SNS-seq interrogate different aspects of replication. SNS-seq is a widely used, high-resolution method for mapping individual replication origins, whereas MFA-seq detects replicated regions by comparing DNA copy number between S and G1 phases. MFA-seq identified broad replicated regions (0.1–0.5 Mb) that were interpreted by McCulloch’s group as containing a single origin. We disagree with this interpretation and consider that there are multiple origins in each broad peaks; theoretical considerations of replication timing indicate that far more origins are required for complete genome duplication during the short S-phase. Once this assumption is reconsidered, MFA-seq and SNS-seq results become complementary: MFA-seq identifies replicated regions, while SNS-seq pinpoints individual origins within those regions. Our analysis revealed that up to 50% of the origins detected by stranded SNS-seq were located within the broad MFA peaks. This pattern—broad MFA-seq regions containing multiple initiation sites—has also recently been found in Leishmania by McCulloch’s team using nanopore sequencing (PMID: 26481451). Nanopore sequencing showed numerous initiation sites within MFA-seq regions and additional numerous sites outside these regions in asynchronous cells, consistent with what we observed using stranded SNS-seq in T. brucei. We will expand our discussion and conclude that the discrepancy arises from methodological differences and interpretation. The two approaches provide complementary insights into replication dynamics, rather than ‘vastly different’ results.
We recognize the importance of validating our results in future using an alternative mapping method and functional assays. However, it is important to emphasize that stranded SNS-seq is an origin mapping technique with a very high level of resolution. This technique can detect regions between two divergent SNS peaks, which should represent regions of DNA replication initiation. At present, no alternative technique has been developed that can match this level of resolution.
(4) Some patterns that were identified to be associated with origins of replication, such as G-quadruplexes and nucleosomes phasing, are known to be biases of SNS-seq (see Foulk et al. Characterizing and controlling intrinsic biases of lambda exonuclease in nascent strand sequencing reveals phasing between nucleosomes and G-quadruplex motifs around a subset of human replication origins. Genome Res. 2015;25(5):725-735. doi:10.1101/gr.183848.114).
It is important to note that the conditions used in our study differ significantly from those applied in the Foulk et al. Genome Res. 2015. We used SNS isolation and enzymatic treatments as described in previous reports (Cayrou, C. et al. Genome Res, 2015 and Cayrou, C et al. Methods, 2012). Here, we enriched the SNS by size on a sucrose gradient and then treated this SNS-enriched fraction with high amounts of repeated λ-exonuclease treatments (100u for 16h at 37oC - see Methods). In contrast, Foulk et al. used sonicated total genomic DNA for origin mapping, without enrichment of SNS on a sucrose gradient as we did, and then they performed a λ-exonuclease treatment. A previous study (Cayrou, C. et al. Genome Res, 2015, Figure S2, which can be found at https://genome.cshlp.org/content/25/12/1873/suppl/DC1) has shown that complete digestion of G4-rich DNA sequences is achieved under the conditions we used.
Furthermore, the SNS depleted control (without RNA) was included in our experimental approach. This control represents all molecules that are difficult to digest with lambda exonuclease, including G4 structures. Peak calling was performed against this background control, with the aim of removing false positive peaks resulting from undigested DNA structures. We explained better this step in the revised manuscript.
The key benefit of our study is that the orientation of the enrichments (peaks) remains consistent throughout the sequencing process. We identified an enrichment of two divergent strands synthesised on complementary strands containing G4s. These two divergent strands themselves do not, however, contain G4s (see Fig. 8 for the model). Therefore, the enriched molecules detected in our study do not contain G4s. They are complementary to the strands enriched with G4s. This means that the observed enrichment of
G4s cannot be an artefact of the enzymatic treatments used in this study. We added this part in the discussion of the revised manuscript.
We also performed an additional control which is not mentioned in the manuscript. In parallel with replicating cells, we isolated the DNA from the stationary phase of growth, which primarily contains non-replicating cells. Following the three λ-exonuclease treatments, there was insufficient DNA remaining from the stationary phase cells to prepare the libraries for sequencing. This control strongly indicated that there was little to no contaminating DNA present with the SNS molecules after λ-exonuclease enrichment.
factors that influence the time on task.
So how can one accurately estimate with these factors?
Food shopping45 minutes30 minutes Drive to work20 minutes20 minutes
I feel like these times will always fluctuate depending in what time of day it is, the weather and your demographics. Its very difficult to put a time on these. And if the person is like me, a store browser, the time will increase immensely OR i will always go over my estimated time. I guess this is why this challenge is important, to ensure we are actively monitoring our times.
If it is going to take you five hours to study for a final exam, you can plan to spread it over five days, with an hour each night, or you can plan on two hours one night and three hours the next.
Its always better to break up studying/learning into small increments. It helps keep the focus going also gives the brain and eyes a rest.
You might receive feedback from more than one reader as you share different stages of your revised draft. In this situation, you may receive feedback from readers who do not understand the assignment or who lack your involvement with and enthusiasm for it. These differing opinions most commonly occur when students ask people outside the classroom to review their writing.
This paragraph explains that when you share your draft with multiple readers, you may get different opinions because not everyone understands the assignment or has the same interest in your topic. This is especially common when students ask people outside the class to review their writing, so writers should consider feedback carefully and focus on what fits the assignment requirements.
By then the clouds had broken up, and below them we could see a big green patch that Mr. Watanabe said was a Japanese Self-Defense Forces training ground and some of the 117 golf courses that lie at the base of the mountain.
I like how this section focuses on the different aspects of Japanese culture present at Mount Fuji (the contrasting imagery of the golf course vs military training grounds). Also, this compliments the imagery of the last paragraph with the mix of many different tourists and cultures mashed together in that small space. It really does well to highlight the conflict between modernity and traditional cultural practices previously mentioned in this piece.
As he chanted and banged on a small brass drum, the rain began to patter and a gust flicked the water in the trees onto the ground.
I like the inclusion of this sensory image here, especially because the previous paragraph had mentioned the religious prevalence of the surrounding area (including the imagery here about the weather/rain/wind implies a validity to the previously mentioned spirituality that the author does not directly state, thus giving the reader a chance to form their own opinions/connections).
He shifted in his seat. Everything he said sounded measured and elegant
I do like the inclusion of how the tone of his voice and the intonation of his speaking patterns. It helps to further characterize the people mentioned during this trip and also couples well with the author's great imagery/visual descriptions.
was starting to wonder how much of the Japanese devotion to climbing Mount Fuji is abstract and conceptual and how much of it involves the material experience of putting on shoes and walking.
I feel like this sentence is accurately getting at and spelling out the author's "point" or purpose in writing this essay (which seems extremely difficult as a travel blog to find some deeper meaning rather than just summarizing the experience). I also feel that this sentence summarizes the contrast that was built up in the previous paragraphs (Mount Fuji as a conventional tourist attraction vs Mount Fuji as a significant cultural icon–but then I suppose this juxtaposition could be related to all of the common/well-known landmarks across the world, thus making this experience seem more relatable to readers).
I was a material climber but I had been won over to the conceptual side.
Love this line. It ties into the idea of materialism and tourism disrupting the beauty of nature. But this is someone who converted to a conceptualist even though the original intent was to do it for tourist-like reasons.
Japan to commit suicide — every year several dozen bodies are recovered in it — and it is one of the most popular places to headquarter a religion
It's an interesting decision to put these facts side by side. One extreme to another.
“I believe I should learn to be more patient.”
The descriptions of the scenes and the people the author talks to are so well done. The dialogue adds to it as well. When I read this part, I could picture Mr. Watanabe in my head and could almost hear him talking.
and with it came millions of visitors by tour bus and subsequently tons of trash
The author keeps returning to this idea about how industrialization and tourism has ruined the natural parts of our world. By showing other people the beauty of the mountain, they've gone too far to the point of it becoming exploitation. The act of showing is also killing.
“I don’t climb it because if I were on the mountain I couldn’t see it.”
That's an interesting thought process. To me, it makes sense because the pollution is already so bad to the point where you can't see the full mountain. He cares more about preserving the image than conquering the landmark. When you're on the mountain, you can't see it, so the motivation to climb must be to see what everything looks like from that perspective.
but anyway then a man went climbing Mount Fuji
I'm hoping the long sentences are just done on purpose now that I'm reading more. I'm reading this and feeling the same way I do whenever a child tries to tell me what they did on the weekend. A few times feels purposeful, but the lack of a break makes my head spin.
backyard of a Shinto shrine that sits next to a Tokyo fire station and across the street from a grocery store where you can buy sake
I kind of like how long this opening sentence is. I feel like the run on sentence captures how elaborate of a scene it truly is. There's a lot to look at.
Your main research question should be substantial enough to form the guiding principle of your paper—but focused enough to guide your research. A strong research question requires you not only to find information but also to put together different pieces of information, interpret and analyze them, and figure out what you think. As you consider potential research questions, ask yourself whether they would be too hard or too easy to answer.
A strong research question should be focused but meaningful, and it should require analysis and interpretation—not just finding basic information.
Just as you need a recipe, ingredients, and proper tools to cook a delicious meal, you also need a plan, resources, and adequate time to create a good written composition. In other words, writing is a process that requires following steps and using strategies to accomplish your goals.
The author explains that writing is a process, like cooking, and it takes planning, resources, time, and strategies to produce a good final result.
Purpose. The reason the writer composes the paragraph. Tone. The attitude the writer conveys about the paragraph’s subject. Audience. The individual or group whom the writer intends to address
Purpose is why the writer writes, tone is the writer’s attitude, and audience is who the writer is writing for.
In college, your instructors are under the impression that you understand the material, and they would like you to deal critically with the material. For that reason, figuring out the academic purpose of an assignment is important.
This section explains that college instructors expect students to already understand the course material and to think critically about it. Because of that, it’s important to identify the academic purpose of an assignment so you can respond in the way the instructor expects.
Greater scrutiny of Child Find practices may therefore be warranted in states that have high rates of poverty and low rates of special education enrollment, such as Alabama, Arkansas, Georgia, and Louisiana
These states often have fewer resources and less funding to invest in special education assessments and programs, despite having higher levels of poverty that are associated with greater need for special education services. I am also curious about the factors that contribute to the relationship between higher poverty levels and increased rates of special education identification.
TEA did not identify all children with disabilities in need of special education, thereby not complying with the Child Find requirement.
The data collection tool itself appears to be flawed. It is hard to fathom that an entire states education system would rely on a tool that is biased and ineffective and one that overlooks a large population of students with disabilities, particularly students of color.
eLife Assessment
This study offers valuable insights into how humans detect and adapt to regime shifts, highlighting dissociable contributions of the frontoparietal network and ventromedial prefrontal cortex to sensitivity to signal diagnosticity and transition probabilities. The combination of an innovative instructed-probability task, Bayesian behavioural modeling, and model-based fMRI analyses provides a solid foundation for the main claims; however, major interpretational limitations remain, particularly a potential confound between posterior switch probability and time in the neuroimaging results. At the behavioural level, reliance on explicitly instructed conditional probabilities leaves open alternative explanations that complicate attribution to a single computational mechanism, such that clearer disambiguation between competing accounts and stronger control of temporal and representational confounds would further strengthen the evidence.
Reviewer #1 (Public review):
Summary:
The study examines human biases in a regime-change task, in which participants have to report the probability of a regime change in the face of noisy data. The behavioral results indicate that humans display systematic biases, in particular, overreaction in stable but noisy environments and underreaction in volatile settings with more certain signals. fMRI results suggest that a frontoparietal brain network is selectively involved in representing subjective sensitivity to noise, while the vmPFC selectively represents sensitivity to the rate of change.
Strengths:
The study relies on a task that measures regime-change detection primarily based on descriptive information about the noisiness and rate of change. This distinguishes the study from prior work using reversal-learning or change-point tasks in which participants are required to learn these parameters from experiences. The authors discuss these differences comprehensively.
The study uses a simple Bayes-optimal model combined with model fitting, which seems to describe the data well. The model is comprehensively validated.
The authors apply model-based fMRI analyses that provide a close link to behavioral results, offering an elegant way to examine individual biases.
Weaknesses:
The authors have adequately addressed my prior concerns.
Reviewer #3 (Public review):
This study concerns how observers (human participants) detect changes in the statistics of their environment, termed regime shifts. To make this concrete, a series of 10 balls are drawn from an urn that contains mainly red or mainly blue balls. If there is a regime shift, the urn is changed over (from mainly red to mainly blue) at some point in the 10 trials. Participants report their belief that there has been a regime shift as a % probability. Their judgement should (mathematically) depend on the prior probability of a regime shift (which is set at one of three levels) and the strength of evidence (also one of three levels, operationalized as the proportion of red balls in the mostly-blue urn and vice versa). Participants are directly instructed of the prior probability of regime shift and proportion of red balls, which are presented on-screen as numerical probabilities. The task therefore differs from most previous work on this question in that probabilities are instructed rather than learned by observation, and beliefs are reported as numerical probabilities rather than being inferred from participants' choice behaviour (as in many bandit tasks, such as Behrens 2007 Nature Neurosci).
The key behavioural finding is that participants over-estimate the prior probability of regime change when it is low, and under estimate it when it is high; and participants over-estimate the strength of evidence when it is low and under-estimate it when it is high. In other words participants make much less distinction between the different generative environments than an optimal observer would. This is termed 'system neglect'. A neuroeconomic-style mathematical model is presented and fit to data.
Functional MRI results how that strength of evidence for a regime shift (roughly, the surprise associated with a blue ball from an apparently red urn) is associated with activity in the frontal-parietal orienting network. Meanwhile at time-points where the probability of a regime shift is high, there is activity in another network including vmPFC. Both networks show individual differences effects, such that people who were more sensitive to strength of evidence and prior probability show more activity in the frontal-parietal and vmPFC-linked networks respectively.
Strengths
(1) The study provides a different task for looking at change-detection and how this depends on estimates of environmental volatility and sensory evidence strength, in which participants are directly and precisely informed of the environmental volatility and sensory evidence strength rather than inferring them through observation as in most previous studies
(2) Participants directly provide belief estimates as probabilities rather than experimenters inferring them from choice behaviour as in most previous studies
(3) The results are consistent with well-established findings that surprising sensory events activate the frontal-parietal orienting network whilst updating of beliefs about the word ('regime shift') activates vmPFC.
Weaknesses
(1) The use of numerical probabilities (both to describe the environments to participants, and for participants to report their beliefs) may be problematic because people are notoriously bad at interpreting probabilities presented in this way, and show poor ability to reason with this information (see Kahneman's classic work on probabilistic reasoning, and how it can be improved by using natural frequencies). Therefore the fact that, in the present study, people do not fully use this information, or use it inaccurately, may reflect the mode of information delivery.
In the response to this comment the authors have pointed out their own previous work showing that system neglect can occur even when numerical probabilities are not used. This is reassuring but there remains a large body of classic work showing that observers do struggle with conditional probabilities of the type presented in the task.
(2) Although a very precise model of 'system neglect' is presented, many other models could fit the data.
For example, you would get similar effects due to attraction of parameter estimates towards a global mean - essentially application of a hyper-prior in which the parameters applied by each participant in each block are attracted towards the experiment-wise mean values of these parameters. For example, the prior probability of regime shift ground-truth values [0.01, 0.05, 0.10] are mapped to subjective values of [0.037, 0.052, 0.069]; this would occur if observers apply a hyper-prior that the probability of regime shift is about 0.05 (the average value over all blocks). This 'attraction to the mean' is a well-established phenomenon and cannot be ruled out with the current data (I suppose you could rule it out by comparing to another dataset in which the mean ground-truth value was different).
More generally, any model in which participants don't fully use the numerical information they were given would produce apparent 'system neglect'. Four qualitatively different example reasons are: 1. Some individual participants completely ignored the probability values given. 2. Participants did not ignore the probability values given, but combined them with a hyperprior as above. 3. Participants had a reporting bias where their reported beliefs that a regime-change had occurred tend to be shifted towards 50% (rather than reporting 'confident' values such 5% or 95%). 4. Participants underweighted probability outliers, resulting in underweighting of evidence in the 'high signal diagnosticity' environment (10.1016/j.neuron.2014.01.020 )
In summary I agree that any model that fits the data would have to capture the idea that participants don't differentiate between the different environments as much as they should, but I think there are a number of qualitatively different reasons why they might do this - of which the above are only examples - hence I find it problematic that the authors present the behaviour as evidence for one extremely specific model.
(3) Despite efforts to control confounds in the fMRI study, including two control experiments, I think some confounds remain.
For example, a network of regions is presented as correlating with the cumulative probability that there has been a regime shift in this block of 10 samples (Pt). However, regardless of the exact samples shown, Pt always increases with sample number (as by the time of later samples, there have been more opportunities for a regime shift)? To control for this the authors include, in a supplementary analysis, an 'intertemporal prior.' I would have preferred to see the results of this better-controlled analysis presented in the main figure. From the tables in the SI it is very difficult to tell how the results change with the includion of the control regressors.
On the other hand, two additional fMRI experiments are done as control experiments and the effect of Pt in the main study is compared to Pt in these control experiments. Whilst I admire the effort in carrying out control studies, I can't understand how these particular experiment are useful controls. For example, in experiment 3 participants simply type in numbers presented on the screen - how can we even have an estimate of Pt from this task?
(4) The Discussion is very long, and whilst a lot of related literature is cited, I found it hard to pin down within the discussion, what the key contributions of this study are. In my opinion it would be better to have a short but incisive discussion highlighting the advances in understanding that arise from the current study, rather than reviewing the field so broadly.
Author response:
The following is the authors’ response to the current reviews
eLife Assessment
This study offers valuable insights into how humans detect and adapt to regime shifts, highlighting dissociable contributions of the frontoparietal network and ventromedial prefrontal cortex to sensitivity to signal diagnosticity and transition probabilities. The combination of an innovative instructed-probability task, Bayesian behavioural modeling, and model-based fMRI analyses provides a solid foundation for the main claims; however, major interpretational limitations remain, particularly a potential confound between posterior switch probability and time in the neuroimaging results. At the behavioural level, reliance on explicitly instructed conditional probabilities leaves open alternative explanations that complicate attribution to a single computational mechanism, such that clearer disambiguation between competing accounts and stronger control of temporal and representational confounds would further strengthen the evidence.
Thank you. In this revision, we will focus on addressing Reviewer 3’s concern on the potential confound between posterior probability and time in neuroimaging results. First, we will present whole-brain results of subjects’ probability estimates (their subjective posterior probability of switch) after controlling for the effect of time on probability of switch (the intertemporal prior). Second, we will compare the effect of probability estimates (Pt) on vmPFC and ventral striatum activity—which we found to correlate with Pt—with and without including intertemporal prior in the GLM. Third, to address Reviewer 3’s comment that from the Tables of activation in the supplement vmPFC and ventral striatum cannot be located, we will add slice-by-slice image of the whole-brain results on Pt in the Supplemental Information in addition to the Tables of Activation.
Public Reviews:
Reviewer #1 (Public review):<br /> Summary:
The study examines human biases in a regime-change task, in which participants have to report the probability of a regime change in the face of noisy data. The behavioral results indicate that humans display systematic biases, in particular, overreaction in stable but noisy environments and underreaction in volatile settings with more certain signals. fMRI results suggest that a frontoparietal brain network is selectively involved in representing subjective sensitivity to noise, while the vmPFC selectively represents sensitivity to the rate of change.
Strengths:
The study relies on a task that measures regime-change detection primarily based on descriptive information about the noisiness and rate of change. This distinguishes the study from prior work using reversal-learning or change-point tasks in which participants are required to learn these parameters from experiences. The authors discuss these differences comprehensively.
The study uses a simple Bayes-optimal model combined with model fitting, which seems to describe the data well. The model is comprehensively validated.
The authors apply model-based fMRI analyses that provide a close link to behavioral results, offering an elegant way to examine individual biases.
Weaknesses:
The authors have adequately addressed my prior concerns.
Thank you for reviewing our paper and providing constructive comments that helped us improve our paper.
Reviewer #3 (Public review):
Thank you again for reviewing the manuscript. In this revision, we will focus on addressing your concern on the potential confound between posterior probability and time in neuroimaging results. First, we will present whole-brain results of subjects’ probability estimates (Pt, their subjective posterior probability of switch) after controlling for the effect of time on probability of switch (the intertemporal prior). Second, we will compare the effect of probability estimates (Pt) on vmPFC and ventral striatum activity—which we found to correlate with Pt—with and without including intertemporal prior in the GLM. These results will be summarized in a new figure (Figure 4).
Finally, to address that you were not able to locate vmPFC and ventral striatum from the Tables of activation, we will add slice-by-slice image of the whole-brain results on Pt in the supplement in addition to the Tables of Activation.
This study concerns how observers (human participants) detect changes in the statistics of their environment, termed regime shifts. To make this concrete, a series of 10 balls are drawn from an urn that contains mainly red or mainly blue balls. If there is a regime shift, the urn is changed over (from mainly red to mainly blue) at some point in the 10 trials. Participants report their belief that there has been a regime shift as a % probability. Their judgement should (mathematically) depend on the prior probability of a regime shift (which is set at one of three levels) and the strength of evidence (also one of three levels, operationalized as the proportion of red balls in the mostly-blue urn and vice versa). Participants are directly instructed of the prior probability of regime shift and proportion of red balls, which are presented on-screen as numerical probabilities. The task therefore differs from most previous work on this question in that probabilities are instructed rather than learned by observation, and beliefs are reported as numerical probabilities rather than being inferred from participants' choice behaviour (as in many bandit tasks, such as Behrens 2007 Nature Neurosci).
The key behavioural finding is that participants over-estimate the prior probability of regime change when it is low, and under estimate it when it is high; and participants over-estimate the strength of evidence when it is low and under-estimate it when it is high. In other words participants make much less distinction between the different generative environments than an optimal observer would. This is termed 'system neglect'. A neuroeconomic-style mathematical model is presented and fit to data.
Functional MRI results how that strength of evidence for a regime shift (roughly, the surprise associated with a blue ball from an apparently red urn) is associated with activity in the frontal-parietal orienting network. Meanwhile at time-points where the probability of a regime shift is high, there is activity in another network including vmPFC. Both networks show individual differences effects, such that people who were more sensitive to strength of evidence and prior probability show more activity in the frontal-parietal and vmPFC-linked networks respectively.
Strengths
(1) The study provides a different task for looking at change-detection and how this depends on estimates of environmental volatility and sensory evidence strength, in which participants are directly and precisely informed of the environmental volatility and sensory evidence strength rather than inferring them through observation as in most previous studies
(2) Participants directly provide belief estimates as probabilities rather than experimenters inferring them from choice behaviour as in most previous studies
(3) The results are consistent with well-established findings that surprising sensory events activate the frontal-parietal orienting network whilst updating of beliefs about the word ('regime shift') activates vmPFC.
Weaknesses
(1) The use of numerical probabilities (both to describe the environments to participants, and for participants to report their beliefs) may be problematic because people are notoriously bad at interpreting probabilities presented in this way, and show poor ability to reason with this information (see Kahneman's classic work on probabilistic reasoning, and how it can be improved by using natural frequencies). Therefore the fact that, in the present study, people do not fully use this information, or use it inaccurately, may reflect the mode of information delivery.
In the response to this comment the authors have pointed out their own previous work showing that system neglect can occur even when numerical probabilities are not used. This is reassuring but there remains a large body of classic work showing that observers do struggle with conditional probabilities of the type presented in the task.
Thank you. Yes, people do struggle with conditional probabilities in many studies. However, as our previous work suggested (Massey and Wu, 2005), system-neglect was likely not due to response mode (having to enter probability estimates or making binary predictions, and etc.).
(2) Although a very precise model of 'system neglect' is presented, many other models could fit the data.
For example, you would get similar effects due to attraction of parameter estimates towards a global mean - essentially application of a hyper-prior in which the parameters applied by each participant in each block are attracted towards the experiment-wise mean values of these parameters. For example, the prior probability of regime shift ground-truth values [0.01, 0.05, 0.10] are mapped to subjective values of [0.037, 0.052, 0.069]; this would occur if observers apply a hyper-prior that the probability of regime shift is about 0.05 (the average value over all blocks). This 'attraction to the mean' is a well-established phenomenon and cannot be ruled out with the current data (I suppose you could rule it out by comparing to another dataset in which the mean ground-truth value was different).
We thank the reviewer for this comment. We do not disagree that there are alternative models that can describe over- and underreactions seen in the dataset. However, we do wish to point out that since we began with the normative Bayesian model, the natural progression in case the normative model fails to capture data is to modify the starting model. It is under this context that we developed the system-neglect model. It was a simple extension (a parameterized version) of the normative Bayesian model.
Regarding the hyperprior idea, even if the participants have a hyperprior, there has to be some function that describes/implements attraction to the mean. Having a hyperprior itself does not imply attraction to this hyperprior. We therefore were not sure why the hyperprior itself can produce attraction to the mean.
We do look further into the possibility of attraction to the mean. First, as suggested by the reviewer, we looked into another dataset with different mean ground-truth value. In Massey and Wu (2005), the transition probabilities were [0.02 0.05 0.1 0.2], which is different from the current study [0.01 0.05 0.1], and there they also found over- and underreactions as well. Second, we reason that for the attraction to the mean idea to work subjects need to know the mean of the system parameters. This would take time to develop because we did not tell subjects about the mean. If this is caused by attraction to the mean, subjects’ behavior would be different in the early stage of the experiment where they had little idea about the mean, compared with the late stage of the experiment where they knew about the mean. We will further analyze and compare participants’ data at the beginning of the experiment with data at the end of the experiment.
More generally, any model in which participants don't fully use the numerical information they were given would produce apparent 'system neglect'. Four qualitatively different example reasons are: 1. Some individual participants completely ignored the probability values given. 2. Participants did not ignore the probability values given, but combined them with a hyperprior as above. 3. Participants had a reporting bias where their reported beliefs that a regime-change had occurred tend to be shifted towards 50% (rather than reporting 'confident' values such 5% or 95%). 4. Participants underweighted probability outliers, resulting in underweighting of evidence in the 'high signal diagnosticity' environment (10.1016/j.neuron.2014.01.020 )
We thank the reviewer for pointing out these potential explanations. Again, we do not disagree that any model in which participants don’t fully use numerical information they were given would produce system neglect. It is hard to separate ‘not fully using numerical information’ from ‘lack of sensitivity to the numerical information’. We will respond in more details to the four example reasons later.
In summary I agree that any model that fits the data would have to capture the idea that participants don't differentiate between the different environments as much as they should, but I think there are a number of qualitatively different reasons why they might do this - of which the above are only examples - hence I find it problematic that the authors present the behaviour as evidence for one extremely specific model.
Again, we do not disagree with the reviewer on the modeling statement. However, we also wish to point out that the system-neglect model we had is a simple extension of the normative Bayesian model. Had we gone to a non-Bayesian framework, we would have faced the criticism of why we simply do not consider a simple extension of the starting model. In response, we will add a section in Discussion summarizing our exchange on this matter.
(3) Despite efforts to control confounds in the fMRI study, including two control experiments, I think some confounds remain.
For example, a network of regions is presented as correlating with the cumulative probability that there has been a regime shift in this block of 10 samples (Pt). However, regardless of the exact samples shown, Pt always increases with sample number (as by the time of later samples, there have been more opportunities for a regime shift)? To control for this the authors include, in a supplementary analysis, an 'intertemporal prior.' I would have preferred to see the results of this better-controlled analysis presented in the main figure. From the tables in the SI it is very difficult to tell how the results change with the includion of the control regressors.
Thank you. In response, we will add a new figure, now Figure 4, showing the results of Pt and delta Pt from GLM-2 where we added the intertemporal prior as a regressor to control for temporal confounds. We compared Pt and delta Pt results in vmPFC and ventral striatum between GLM-1 and GLM-2. We also will show the results of intertemporal prior on vmPFC and ventral striatum under GLM-2.
On the other hand, two additional fMRI experiments are done as control experiments and the effect of Pt in the main study is compared to Pt in these control experiments. Whilst I admire the effort in carrying out control studies, I can't understand how these particular experiment are useful controls. For example, in experiment 3 participants simply type in numbers presented on the screen - how can we even have an estimate of Pt from this task?
We thank the reviewer for this comment. On the one hand, the effect of Pt we see in brain activity can be simply due to motor confounds and the purpose of Experiment 3 was to control for them. Our question was, if subjects saw the similar visual layout and were just instructed to press buttons to indicate two-digit numbers, would we observe the vmPFC, ventral striatum, and the frontoparietal network like what we did in the main experiment (Experiment 1)?
On the other hand, the effect of Pt can simply reflect probability estimates of that the current regime is the blue regime, and therefore not particularly about change detection. In Experiment 2, we tested that idea, namely whether what we found about Pt was unique to change detection. In Experiment 2, subjects estimated the probability that the current regime is the blue regime (just as they did in Experiment 1) except that there were no regime shifts involved. In other words, it is possible that the regions we identified were generally associated with probability estimation and not particularly about probability estimates of change. We used Experiment 2 to examine whether this were true.
To make the purpose of the two control experiments clearer, we updated the paragraph describing the control experiments on page 9:
“To establish the neural representations for regime-shift estimation, we performed three fMRI experiments ( subjects for each experiment, 90 subjects in total). Experiment 1 was the main experiment, while Experiments 2 to 3 were control experiments that ruled out two important confounds (Fig. 1E). The control experiments were designed to clarify whether any effect of subjects’ probability estimates of a regime shift, , in brain activity can be uniquely attributed to change detection. Here we considered two major confounds that can contribute to the effect of . First, since subjects in Experiment 1 made judgments about the probability that the current regime is the blue regime (which corresponded to probability of regime change), the effect of did not particularly have to do with change detection. To address this issue, in Experiment 2 subjects made exactly the same judgments as in Experiment 1 except that the environments were stationary (no transition from one regime to another was possible), as in Edwards (1968) classic “bookbag-and-poker chip” studies. Subjects in both experiments had to estimate the probability that the current regime is the blue regime, but this estimation corresponded to the estimates of regime change only in Experiment 1. Therefore, activity that correlated with probability estimates in Experiment 1 but not in Experiment 2 can be uniquely attributed to representing regime-shift judgments. Second, the effect of can be due to motor preparation and/or execution, as subjects in Experiment 1 entered two-digit numbers with button presses to indicate their probability estimates. To address this issue, in Experiment 3 subjects performed a task where they were presented with two-digit numbers and were instructed to enter the numbers with button presses. By comparing the fMRI results of these experiments, we were therefore able to establish the neural representations that can be uniquely attributed to the probability estimates of regime-shift.”
To further make sure that the probability-estimate signals in Experiment 1 were not due to motor confounds, we implemented an action-handedness regressor in the GLM, as we described below on page 19:
“Finally, we note that in GLM-1, we implemented an “action-handedness” regressor to directly address the motor-confound issue, that higher probability estimates preferentially involved right-handed responses for entering higher digits. The action-handedness regressor was parametric, coding -1 if both finger presses involved the left hand (e.g., a subject pressed “23” as her probability estimate when seeing a signal), 0 if using one left finger and one right finger (e.g., “75”), and 1 if both finger presses involved the right hand (e.g., “90”). Taken together, these results ruled out motor confounds and suggested that vmPFC and ventral striatum represent subjects’ probability estimates of change (regime shifts) and belief revision.”
(4) The Discussion is very long, and whilst a lot of related literature is cited, I found it hard to pin down within the discussion, what the key contributions of this study are. In my opinion it would be better to have a short but incisive discussion highlighting the advances in understanding that arise from the current study, rather than reviewing the field so broadly.
Thank you. We thank the reviewer for pushing us to highlight the key contributions. In response, we added a paragraph at the beginning of Discussion to better highlight our contributions:
“In this study, we investigated how humans detect changes in the environments and the neural mechanisms that contribute to how we might under- and overreact in our judgments. Combining a novel behavioral paradigm with computational modeling and fMRI, we discovered that sensitivity to environmental parameters that directly impact change detection is a key mechanism for under- and overreactions. This mechanism is implemented by distinct brain networks in the frontal and parietal cortices and in accordance with the computational roles they played in change detection. By introducing the framework in system neglect and providing evidence for its neural implementations, this study offered both theoretical and empirical insights into how systematic judgment biases arise in dynamic environments.”
**Recommendations for the authors:
Reviewer #3 (Recommendations for the authors):**
Thank you for pointing out the inclusion of the intertemporal prior in glm2, this seems like an important control that would address my criticism. Why not present this better-controlled analysis in the main figure, rather than the results for glm1 which has no effective control of the increasing posterior probability of a reversal with time?
Thank you for this suggestion. We added a new figure (Figure 4) that showed results from GLM-2. In this new figure, we showed whole-brain results on Pt and delta Pt, ROI results of vmPFC and ventral striatum on Pt, delta Pt, and intertemporal prior.
The reason we kept results from GLM-1 (Figure 3) was primarily because we wanted to compare the effect of Pt between experiments under identical GLM. In other words, the regressors in GLM-1 was identical across all 3 experiments. In Experiments 1 and 2, Pt and delta Pt were respectively probability estimates and belief updates that current regime was the Blue regime. In Experiment 3, Pt and delta Pt were simply the number subjects were instructed to press (Pt) and change in number between successive periods (delta Pt).
As a further point I could not navigate the tables of fMRI activations in SI and recommend replacing or supplementing these with images. For example I cannot actually find a vmPFC or ventral striatum cluster listed for the effect of Pt in GLM1 (version in table S1), which I thought were the main results? Beyond that, comparing how much weaker (or not) those results are when additional confound regressors are included in GLM2 seems impossible.
The vmPFC and ventral striatum were part of the cluster labeled as Central Opercular cortex. In response, we will provide information about coordinates on the local maxima within the cluster. We will also add slice-by-slice images showing the effect of Pt.
The following is the authors’ response to the original reviews
eLife Assessment
This study offers valuable insights into how humans detect and adapt to regime shifts, highlighting distinct contributions of the frontoparietal network and ventromedial prefrontal cortex to sensitivity to signal diagnosticity and transition probabilities. The combination of an innovative task design, behavioral modeling, and model-based fMRI analyses provides a solid foundation for the conclusions; however, the neuroimaging results have several limitations, particularly a potential confound between the posterior probability of a switch and the passage of time that may not be fully controlled by including trial number as a regressor. The control experiments intended to address this issue also appear conceptually inconsistent and, at the behavioral level, while informing participants of conditional probabilities rather than requiring learning is theoretically elegant, such information is difficult to apply accurately, as shown by well-documented challenges with conditional reasoning and base-rate neglect. Expressing these probabilities as natural frequencies rather than percentages may have improved comprehension. Overall, the study advances understanding of belief updating under uncertainty but would benefit from more intuitive probabilistic framing and stronger control of temporal confounds in future work.
We thank the editors for the assessment and we appreciate your efforts in reviewing the paper. The editors added several limitations in the assessment based on the new reviewer 3 in this round, which we would like to clarify below.
With regard to temporal confounds, we clarified in the main text and response to Reviewer 3 that we had already addressed the potential confound between posterior probability of a switch and passage of time in GLM-2 with the inclusion of intertemporal prior. After adding intertemporal prior in the GLM, we still observed the same fMRI results on probability estimates. In addition, we did two other robustness checks, which we mentioned in the manuscript.
With regard to response mode (probability estimation rather than choice or indicating natural frequencies), we wish to point out that the in previous research by Massey and Wu (2005), which the current study was based on, the concern of participants showing system-neglect tendencies due to the mode of information delivery, namely indicating beliefs through reporting probability estimates rather than through choice or other response mode was addressed. Massy and Wu (2005, Study 3) found the same biases when participants performed a choice task that did not require them to indicate probability estimates.
With regard to the control experiments, the control experiments in fact were not intended to address the confounds between posterior probability and passage of time. Rather, they aimed to address whether the neural findings were unique to change detection (Experiment 2) and to address visual and motor confounds (Experiment 3). These and the results of the control experiments were mentioned on page 18-19.
We also wish to highlight that we had performed detailed model comparisons after reviewer 2’s suggestions. Although reviewer 2 was unable to re-review the manuscript, we believe this provides insight into the literature on change detection. See “Incorporating signal dependency into system-neglect model led to better models for regime-shift detection” (p.27-30). The model comparison showed that system-neglect models that incorporate signal dependency are better models than the original system-neglect model in describing participants probability estimates. This suggests that people respond to change-consistent and change-inconsistent signals differently when judging whether the regime had changed. This was not reported in previous behavioral studies and was largely inspired by the neural finding on signal dependency in the frontoparietal cortex. It indicates that neural findings can provide novel insights into computational modeling of behavior.
To better highlight and summarize our key contributions, we added a paragraph at the beginning of Discussion:
“In this study, we investigated how humans detect changes in the environments and the neural mechanisms that contribute to how we might under- and overreact in our judgments. Combining a novel behavioral paradigm with computational modeling and fMRI, we discovered that sensitivity to environmental parameters that directly impact change detection is a key mechanism for under- and overreactions. This mechanism is implemented by distinct brain networks in the frontal and parietal cortices and in accordance with the computational roles they played in change detection. By introducing the framework in system neglect and providing evidence for its neural implementations, this study offered both theoretical and empirical insights into how systematic judgment biases arise in dynamic environments.”
Public Reviews:
Reviewer #1 (Public review):
Summary:
The study examines human biases in a regime-change task, in which participants have to report the probability of a regime change in the face of noisy data. The behavioral results indicate that humans display systematic biases, in particular, overreaction in stable but noisy environments and underreaction in volatile settings with more certain signals. fMRI results suggest that a frontoparietal brain network is selectively involved in representing subjective sensitivity to noise, while the vmPFC selectively represents sensitivity to the rate of change.
Strengths:
- The study relies on a task that measures regime-change detection primarily based on descriptive information about the noisiness and rate of change. This distinguishes the study from prior work using reversal-learning or change-point tasks in which participants are required to learn these parameters from experiences. The authors discuss these differences comprehensively.
- The study uses a simple Bayes-optimal model combined with model fitting, which seems to describe the data well. The model is comprehensively validated.
- The authors apply model-based fMRI analyses that provide a close link to behavioral results, offering an elegant way to examine individual biases.
We thank the reviewer for the comments.
Weaknesses:
The authors have adequately addressed most of my prior concerns.
We thank the reviewer for recognizing our effort in addressing your concerns.
My only remaining comment concerns the z-test of the correlations. I agree with the non-parametric test based on bootstrapping at the subject level, providing evidence for significant differences in correlations within the left IFG and IPS.
However, the parametric test seems inadequate to me. The equation presented is described as the Fisher z-test, but the numerator uses the raw correlation coefficients (r) rather than the Fisher-transformed values (z). To my understanding, the subtraction should involve the Fisher z-scores, not the raw correlations.
More importantly, the Fisher z-test in its standard form assumes that the correlations come from independent samples, as reflected in the denominator (which uses the n of each independent sample). However, in my opinion, the two correlations are not independent but computed within-subject. In such cases, parametric tests should take into account the dependency. I believe one appropriate method for the current case (correlated correlation coefficients sharing a variable [behavioral slope]) is explained here:
Meng, X.-l., Rosenthal, R., & Rubin, D. B. (1992). Comparing correlated correlation coefficients. Psychological Bulletin, 111(1), 172-175. https://doi.org/10.1037/0033-2909.111.1.172
It should be implemented here:
Diedenhofen B, Musch J (2015) cocor: A Comprehensive Solution for the Statistical Comparison of Correlations. PLoS ONE 10(4): e0121945. https://doi.org/10.1371/journal.pone.0121945
My recommendation is to verify whether my assumptions hold, and if so, perform a test that takes correlated correlations into account. Or, to focus exclusively on the non-parametric test.
In any case, I recommend a short discussion of these findings and how the authors interpret that some of the differences in correlations are not significant.
Thank you for the careful check. Yes. This was indeed a mistake from us. We also agree that the two correlations are not independent. Therefore, we modified the test that accounts for dependent correlations by following Meng et al. (1992) suggested by the reviewer. We updated in the Methods section on p.56-57:
“In the parametric test, we adopted the approach of Meng et al. (1992) to statistically compare the two correlation coefficients. This approach specifically tests differences between dependent correlation coefficients according to the following equation
Where N is the number of subjects, z<sub>ri</sub> is the Fisher z-transformed value of r<sub>i</sub>,(r<sub>1</sub> = r<sub>blue</sub> and r<sub>2</sub> = r<sub>red</sub>), and r<sub>x</sub> is the correlation between the neural sensitivity at change-consistent signals and change-inconsistent signals. The computation of h is based on the following equations
Where 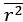 is the mean of the
is the mean of the 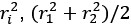 , and f should be set to 1 if > 1.”
, and f should be set to 1 if > 1.”
We updated on the Results section on p.29:
“Since these correlation coefficients were not independent, we compared them using the test developed in Meng et al. (1992) (see Methods). We found that among the five ROIs in the frontoparietal network, two of them, namely the left IFG and left IPS, the difference in correlation was significant (one-tailed z test; left IFG: z = 1.8908, p = 0.0293; left IPS: z = 2.2584, p = 0.0049). For the remaining three ROIs, the difference in correlation was not significant (dmPFC: z = 0.9522, p = 0.1705; right IFG: z = 0.9860, p = 0.1621; right IPS: z = 1.4833, p = 0.0690).”
We added a Discussion on these results on p.41:
“Interestingly, such sensitivity to signal diagnosticity was only present in the frontoparietal network when participants encountered change-consistent signals. However, while most brain areas within this network responded in this fashion, only the left IPS and left IFG showed a significant difference in coding individual participants’ sensitivity to signal diagnosticity between change-consistent and change-inconsistent signals. Unlike the left IPS and left IFG, we observed in dmPFC a marginally significant correlation with behavioral sensitivity at change-inconsistent signals as well. Together, these results indicate that while different brain areas in the frontoparietal network responded similarly to change-consistent signals, there was a greater degree of heterogeneity in responding to change-inconsistent signals.”
Reviewer #3 (Public review):
This study concerns how observers (human participants) detect changes in the statistics of their environment, termed regime shifts. To make this concrete, a series of 10 balls are drawn from an urn that contains mainly red or mainly blue balls. If there is a regime shift, the urn is changed over (from mainly red to mainly blue) at some point in the 10 trials. Participants report their belief that there has been a regime shift as a % probability. Their judgement should (mathematically) depend on the prior probability of a regime shift (which is set at one of three levels) and the strength of evidence (also one of three levels, operationalized as the proportion of red balls in the mostly-blue urn and vice versa). Participants are directly instructed of the prior probability of regime shift and proportion of red balls, which are presented on-screen as numerical probabilities. The task therefore differs from most previous work on this question in that probabilities are instructed rather than learned by observation, and beliefs are reported as numerical probabilities rather than being inferred from participants' choice behaviour (as in many bandit tasks, such as Behrens 2007 Nature Neurosci).
The key behavioural finding is that participants over-estimate the prior probability of regime change when it is low, and under estimate it when it is high; and participants over-estimate the strength of evidence when it is low and under-estimate it when it is high. In other words participants make much less distinction between the different generative environments than an optimal observer would. This is termed 'system neglect'. A neuroeconomic-style mathematical model is presented and fit to data.
Functional MRI results how that strength of evidence for a regime shift (roughly, the surprise associated with a blue ball from an apparently red urn) is associated with activity in the frontal-parietal orienting network. Meanwhile, at time-points where the probability of a regime shift is high, there is activity in another network including vmPFC. Both networks show individual differences effects, such that people who were more sensitive to strength of evidence and prior probability show more activity in the frontal-parietal and vmPFC-linked networks respectively.
We thank the reviewer for the overall descriptions of the manuscript.
Strengths
(1) The study provides a different task for looking at change-detection and how this depends on estimates of environmental volatility and sensory evidence strength, in which participants are directly and precisely informed of the environmental volatility and sensory evidence strength rather than inferring them through observation as in most previous studies
(2) Participants directly provide belief estimates as probabilities rather than experimenters inferring them from choice behaviour as in most previous studies
(3) The results are consistent with well-established findings that surprising sensory events activate the frontal-parietal orienting network whilst updating of beliefs about the word ('regime shift') activates vmPFC.
Thank you for these assessments.
Weaknesses
(1) The use of numerical probabilities (both to describe the environments to participants, and for participants to report their beliefs) may be problematic because people are notoriously bad at interpreting probabilities presented in this way, and show poor ability to reason with this information (see Kahneman's classic work on probabilistic reasoning, and how it can be improved by using natural frequencies). Therefore the fact that, in the present study, people do not fully use this information, or use it inaccurately, may reflect the mode of information delivery.
We appreciate the reviewer’s concern on this issue. The concern was addressed in Massey and Wu (2005) as participants performed a choice task in which they were not asked to provide probability estimates (Study 3 in Massy and Wu, 2005). Instead, participants in Study 3 were asked to predict the color of the ball before seeing a signal. This was a more intuitive way of indicating his or her belief about regime shift. The results from the choice task were identical to those found in the probability estimation task (Study 1 in Massey and Wu). We take this as evidence that the system-neglect behavior the participants showed was less likely to be due to the mode of information delivery.
(2) Although a very precise model of 'system neglect' is presented, many other models could fit the data.
For example, you would get similar effects due to attraction of parameter estimates towards a global mean - essentially application of a hyper-prior in which the parameters applied by each participant in each block are attracted towards the experiment-wise mean values of these parameters. For example, the prior probability of regime shift ground-truth values [0.01, 0.05, 0.10] are mapped to subjective values of [0.037, 0.052, 0.069]; this would occur if observers apply a hyper-prior that the probability of regime shift is about 0.05 (the average value over all blocks). This 'attraction to the mean' is a well-established phenomenon and cannot be ruled out with the current data (I suppose you could rule it out by comparing to another dataset in which the mean ground-truth value was different).
We thank the reviewer for this comment. It is true that the system-neglect model is not entirely inconsistent with regression to the mean, regardless of whether the implementation has a hyper prior or not. In fact, our behavioral measure of sensitivity to transition probability and signal diagnosticity, which we termed the behavioral slope, is based on linear regression analysis. In general, the modeling approach in this paper is to start from a generative model that defines ideal performance and consider modifying the generative model when systematic deviations in actual performance from the ideal is observed. In this approach, a generative Bayesian model with hyper priors would be more complex to begin with, and a regression to the mean idea by itself does not generate a priori predictions.
More generally, any model in which participants don't fully use the numerical information they were given would produce apparent 'system neglect'. Four qualitatively different example reasons are: 1. Some individual participants completely ignored the probability values given. 2. Participants did not ignore the probability values given, but combined them with a hyperprior as above. 3. Participants had a reporting bias where their reported beliefs that a regime-change had occurred tend to be shifted towards 50% (rather than reporting 'confident' values such 5% or 95%). 4. Participants underweighted probability outliers resulting in underweighting of evidence in the 'high signal diagnosticity' environment (10.1016/j.neuron.2014.01.020)
In summary I agree that any model that fits the data would have to capture the idea that participants don't differentiate between the different environments as much as they should, but I think there are a number of qualitatively different reasons why they might do this - of which the above are only examples - hence I find it problematic that the authors present the behaviour as evidence for one extremely specific model.
Thank you for raising this point. The modeling principle we adopt is the following. We start from the normative model—the Bayesian model—that defined what normative behavior should look like. We compared participants’ behavior with the Bayesian model and found systematic deviations from it. To explain those systematic deviations, we considered modeling options within the confines of the same modeling framework. In other words, we considered a parameterized version of the Bayesian model, which is the system-neglect model and examined through model comparison the best modeling choice. This modeling approach is not uncommon in economics and psychology. For example, Kahneman and Tversky adopted this approach when proposing prospect theory, a modification of expected utility theory where expected utility theory can be seen as one specific model for how utility of an option should be computed.
(3) Despite efforts to control confounds in the fMRI study, including two control experiments, I think some confounds remain.
For example, a network of regions is presented as correlating with the cumulative probability that there has been a regime shift in this block of 10 samples (Pt). However, regardless of the exact samples shown, doesn't Pt always increase with sample number (as by the time of later samples, there have been more opportunities for a regime shift)? Unless this is completely linear, the effect won't be controlled by including trial number as a co-regressor (which was done).
Thank you for raising this concern. Yes, Pt always increases with sample number regardless of evidence (seeing change-consistent or change-inconsistent signals). This is captured by the ‘intertemporal prior’ in the Bayesian model, which we included as a regressor in our GLM analysis (GLM-2), in addition to Pt. In short, GLM-1 had Pt and sample number. GLM-2 had Pt, intertemporal prior, and sample number, among other regressors. And we found that, in both GLM-1 and GLM-2, both vmPFC and ventral striatum correlated with Pt.
To make this clearer, we updated the main text to further clarify this on p.18:
“We examined the robustness of P<sub>t</sub> representations in these two regions in several follow-up analyses. First, we implemented a GLM (GLM-2 in Methods) that, in addition to P<sub>t</sub>, included various task-related variables contributing to P<sub>t</sub> as regressors (Fig. S7 in SI). Specifically, to account for the fact that the probability of regime change increased over time, we included the intertemporal prior as a regressor in GLM-2. The intertemporal prior is the natural logarithm of the odds in favor of regime shift in the t-th period, 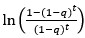 where q is transition probability and t = 1,…,10 is the period (see Eq. 1 in Methods). It describes normatively how the prior probability of change increased over time regardless of the signals (blue and red balls) the subjects saw during a trial. Including it along with P<sub>t</sub> would clarify whether any effect of P<sub>t</sub> can otherwise be attributed to the intertemporal prior. Second, we implemented a GLM that replaced P<sub>t</sub> with the log odds of P<sub>t</sub>, ln (P<sub>t</sub>/(1-P<sub>t</sub>)) (Fig. S8 in SI). Third, we implemented a GLM that examined separately on periods when change-consistent (blue balls) and change-inconsistent (red balls) signals appeared (Fig. S9 in SI). Each of these analyses showed the same pattern of correlations between P<sub>t</sub> and activation in vmPFC and ventral striatum, further establishing the robustness of the P<sub>t</sub> findings.”
where q is transition probability and t = 1,…,10 is the period (see Eq. 1 in Methods). It describes normatively how the prior probability of change increased over time regardless of the signals (blue and red balls) the subjects saw during a trial. Including it along with P<sub>t</sub> would clarify whether any effect of P<sub>t</sub> can otherwise be attributed to the intertemporal prior. Second, we implemented a GLM that replaced P<sub>t</sub> with the log odds of P<sub>t</sub>, ln (P<sub>t</sub>/(1-P<sub>t</sub>)) (Fig. S8 in SI). Third, we implemented a GLM that examined separately on periods when change-consistent (blue balls) and change-inconsistent (red balls) signals appeared (Fig. S9 in SI). Each of these analyses showed the same pattern of correlations between P<sub>t</sub> and activation in vmPFC and ventral striatum, further establishing the robustness of the P<sub>t</sub> findings.”
On the other hand, two additional fMRI experiments are done as control experiments and the effect of Pt in the main study is compared to Pt in these control experiments. Whilst I admire the effort in carrying out control studies, I can't understand how these particular experiment are useful controls. For example in experiment 3 participants simply type in numbers presented on the screen - how can we even have an estimate of Pt from this task?
We thank the reviewer for this comment. On the one hand, the effect of Pt we see in brain activity can be simply due to motor confounds and the purpose of Experiment 3 was to control for them. Our question was, if subjects saw the similar visual layout and were just instructed to press buttons to indicate two-digit numbers, would we observe the vmPFC, ventral striatum, and the frontoparietal network like what we did in the main experiment (Experiment 1)?
On the other hand, the effect of Pt can simply reflect probability estimates of that the current regime is the blue regime, and therefore not particularly about change detection. In Experiment 2, we tested that idea, namely whether what we found about Pt was unique to change detection. In Experiment 2, subjects estimated the probability that the current regime is the blue regime (just as they did in Experiment 1) except that there were no regime shifts involved. In other words, it is possible that the regions we identified were generally associated with probability estimation and not particularly about probability estimates of change. We used Experiment 2 to examine whether this were true.
To make the purpose of the two control experiments clearer, we updated the paragraph describing the control experiments on page 9:
“To establish the neural representations for regime-shift estimation, we performed three fMRI experiments (n\=30 subjects for each experiment, 90 subjects in total). Experiment 1 was the main experiment, while Experiments 2 to 3 were control experiments that ruled out two important confounds (Fig. 1E). The control experiments were designed to clarify whether any effect of subjects’ probability estimates of a regime shift, P<sub>t</sub>, in brain activity can be uniquely attributed to change detection. Here we considered two major confounds that can contribute to the effect of . First, since subjects in Experiment 1 made judgments about the probability that the current regime is the blue regime (which corresponded to probability of regime change), the effect of P<sub>t</sub> did not particularly have to do with change detection. To address this issue, in Experiment 2 subjects made exactly the same judgments as in Experiment 1 except that the environments were stationary (no transition from one regime to another was possible), as in Edwards (1968) classic “bookbag-and-poker chip” studies. Subjects in both experiments had to estimate the probability that the current regime is the blue regime, but this estimation corresponded to the estimates of regime change only in Experiment 1. Therefore, activity that correlated with probability estimates in Experiment 1 but not in Experiment 2 can be uniquely attributed to representing regime-shift judgments. Second, the effect of P<sub>t</sub> can be due to motor preparation and/or execution, as subjects in Experiment 1 entered two-digit numbers with button presses to indicate their probability estimates. To address this issue, in Experiment 3 subjects performed a task where they were presented with two-digit numbers and were instructed to enter the numbers with button presses. By comparing the fMRI results of these experiments, we were therefore able to establish the neural representations that can be uniquely attributed to the probability estimates of regime-shift.”
To further make sure that the probability-estimate signals in Experiment 1 were not due to motor confounds, we implemented an action-handedness regressor in the GLM, as we described below on page 19:
“Finally, we note that in GLM-1, we implemented an “action-handedness” regressor to directly address the motor-confound issue, that higher probability estimates preferentially involved right-handed responses for entering higher digits. The action-handedness regressor was parametric, coding -1 if both finger presses involved the left hand (e.g., a subject pressed “23” as her probability estimate when seeing a signal), 0 if using one left finger and one right finger (e.g., “75”), and 1 if both finger presses involved the right hand (e.g., “90”). Taken together, these results ruled out motor confounds and suggested that vmPFC and ventral striatum represent subjects’ probability estimates of change (regime shifts) and belief revision.”
(4) The Discussion is very long, and whilst a lot of related literature is cited, I found it hard to pin down within the discussion, what the key contributions of this study are. In my opinion it would be better to have a short but incisive discussion highlighting the advances in understanding that arise from the current study, rather than reviewing the field so broadly.
Thank you. We thank the reviewer for pushing us to highlight the key contributions. In response, we added a paragraph at the beginning of Discussion to better highlight our contributions:
“In this study, we investigated how humans detect changes in the environments and the neural mechanisms that contribute to how we might under- and overreact in our judgments. Combining a novel behavioral paradigm with computational modeling and fMRI, we discovered that sensitivity to environmental parameters that directly impact change detection is a key mechanism for under- and overreactions. This mechanism is implemented by distinct brain networks in the frontal and parietal cortices and in accordance with the computational roles they played in change detection. By introducing the framework in system neglect and providing evidence for its neural implementations, this study offered both theoretical and empirical insights into how systematic judgment biases arise in dynamic environments.”
Recommendations for the authors:
Reviewer #3 (Recommendations for the authors):
Many of the figures are too tiny - the writing is very small, as are the pictures of brains. I'd suggest adjusting these so they will be readable without enlarging.
Thank you. We apologize for the poor readability of the figures. We had enlarged the figures (Fig. 5 in particular) and their font size to make them more readable.
After reviewing Figure 1 and the descriptions of various types of writing assignments, watch the following video about the writing process. No matter what type of assignment you are writing, it will be important for you to follow a writing process: a series of steps a writer takes to complete a writing task. Making use of a writing process ensures that you stay organized and focused while allowing you to break up a larger assignment into several distinct tasks.
This paragraph explains that writing uses a process to stay organized, but it isn’t always linear. Writing is recursive, so writers often revisit steps to revise and improve their work.
With what the visitor has been immersed in thus far, it is easy to see how the real and the fake become one.
another reaction of hyper-reality.
When hearing the two “German” men speak, it almost feels as if the conversation is real and taking place now.
a reaction to hyper-reality.
His main argument is that when visitors come to these locations, what they are seeing is the real and the fake becoming ambiguous and hard to differentiate.
This is umberto's argument
plethora
what does this word mean
There may be sophistry in all this; but the condition of a slave confuses all principles ofmorality, and, in fact, renders the practice of them impossible.
To me, this seems like she and the slave community do what they can, if there is a chance of some improvement in their condition. This is regardless of what "morals" are broken. I found it especially moving that she says the practice of morals is impossible due to their condition. I've never thought of this perspective before!
For example, you update your status on a social networking site with the awareness of who will digitally follow the post. If you want to brag about a good grade, you may write the post to please family members. If you want to describe a funny moment, you may write with your friends’ senses of humor in mind. Even at work, you send e-mails with an awareness of an unintended receiver who could intercept the message
This section explains that writers must consider their audience when choosing content and tone. What we write should match who will read it, so the message is appropriate and interesting for that audience.
Tone identifies a speaker’s attitude toward a subject or another person. You may pick up a person’s tone of voice fairly easily in conversation. A friend who tells you about her weekend may speak excitedly about a fun skiing trip. An instructor who means business may speak in a low, slow voice to emphasize her serious mood. Or, a coworker who needs to let off some steam after a long meeting may crack a sarcastic joke.
Tone shows attitude, and it can be recognized through the way someone speaks, like excited, serious, or sarcastic.
Once you’ve decided on the most appropriate voice and style for your document, you will want to continue enhancing your writing to engage your reader. The writing process requires many steps, and in order to ensure you have created a style that meets the needs of both the assignment and your reader, spend some time enhancing your word choices, developing your descriptions, and clarifying your sentence constructions.
This section says good writing takes revision, including improving word choice, description, and sentence clarity to better engage the reader.
They valued astronomy mainly for its practical usefulness in making maps and calendars rather than for its larger philosophical implications.
I think this is interesting because it shows that the Ottoman Empire did not reject science, but used it differently than Europeans did. Instead of focusing on how astronomy might challenge religious or philosophical beliefs, Ottomans focused on what was useful for everyday needs. This helps explain why ideas like a sun centered universe did not cause major controversy there the way they did in Europe.
In Marx’s view the coming of socialism — a society without classes or class conflict — was not simply a good idea; it was inevitable, inscribed in the laws of historical development.
This line stands out because Marx is saying socialism is not just a political belief, but something that history will naturally lead to. He treats social change like a scientific process driven by class conflict, not by individual choices or moral arguments. This shows a big shift away from Enlightenment ideas that focused on reason and independent individuals as the main drivers of progress.
Thus, the earth was no longer unique or at the obvious center of God’s attention.
This line shows why the Scientific Revolution was so unsettling for people at the time. If the earth was no longer at the center of the universe, it challenged the idea that humans held a special place in God’s plan. I think this explains why new scientific ideas caused fear and resistance, not just because they changed science, but because they forced people to rethink religion and humanity’s role in the universe.
“there is no Hindu; there is no Muslim; only God.”
This line stands out because it pushes back against the idea that religious labels define who people are. Guru Nanak seems to be saying that faith matters more than whether someone is Hindu or Muslim. At a time when religion shaped social life and identity, this would have been a powerful and challenging idea that helped Sikhism grow into its own tradition.
eLife Assessment
This article reports an algorithm for inferring the presence of synaptic connection between neurons based on naturally occurring spiking activity of a neuronal network. One key improvement is to combine self-supervised and synthetic approaches to learn to focus on features that generalize to the conditions of the observed network. This valuable contribution is currently supported by incomplete evidence.
Reviewer #1 (Public review):
Summary:
The authors proposed a new method to infer connectivity from spike trains whose main novelty relies on their approach to mitigate the problem of model mismatch. The latter arises when the inference algorithm is trained or based on a model that does not accurately describe the data. They propose combining domain adaptation with a deep neural architecture and in an architecture called DeepDAM. They apply DeepDAM to an in vivo ground-truth dataset previously recorded in mouse CA1, show that it performs better than methods without domain adaptation, and evaluate its robustness. Finally, they show that their approach can also be applied to a different problem i.e., inferring biophysical properties of individual neurons.
Strengths:
(1) The problem of inferring connectivity from extracellular recording is a very timely one: as the yield of silicon probes steadily increases, the number of simultaneously recorded pairs does so quadratically, drastically increasing the possibility of detecting connected pairs.
(2) Using domain adaptation to address model mismatch is a clever idea, and the way the authors introduced it into the larger architecture seems sensible.
(3) The authors clearly put a great effort into trying to communicate the intuitions to the reader.
Weaknesses:
(1) The validation of the approach is incomplete: due to its very limited size, the single ground-truth dataset considered does not provide a sufficient basis to draw a strong conclusion. While the authors correctly note that this is the only dataset of its kind, the value of this validation is limited compared to what could be done by carefully designing in silico experiments.
(2) Surprisingly, the authors fail to compare their method to the approach originally proposed for the data they validate on (English et al., 2017).
(3) The authors make a commendable effort to study the method's robustness by pushing the limits of the dataset. However, the logic of the robustness analysis is often unclear, and once again, the limited size of the dataset poses major limitations to the authors.
(4) The lack of details concerning both the approach and the validation makes it challenging for the reader to establish the technical soundness of the study.
Although in the current form this study does not provide enough basis to judge the impact of DeepDAM in the broader neuroscience community, it nevertheless puts forward a valuable and novel idea: using domain adaptation to mitigate the problem of model mismatch. This approach might be leveraged in future studies and methods to infer connectivity.
Reviewer #2 (Public review):
The article is very well written, and the new methodology is presented with care. I particularly appreciated the step-by-step rationale for establishing the approach, such as the relationship between K-means centers and the various parameters. This text is conveniently supported by the flow charts and t-SNE plots. Importantly, I thought the choice of state-of-the-art method was appropriate and the choice of dataset adequate, which together convinced me in believing the large improvement reported. I thought that the crossmodal feature-engineering solution proposed was elegant and seems exportable to other fields. Here are a few notes.<br /> While the validation data set was well chosen and of high quality, it remains a single dataset and also remains a non-recurrent network. The authors acknowledge this in the discussion, but I wanted to chime in to say that for the method to be more than convincing, it would need to have been tested on more datasets. It should be acknowledged that the problem becomes more complicated in a recurrent excitatory network, and thus the method may not work as well in the cortex or in CA3.
While the data is shown to work in this particular dataset (plus the two others at the end), I was left wondering when the method breaks. And it should break if the models are sufficiently mismatched. Such a question can be addressed using synthetic-synthetic models. This was an important intuition that I was missing, and an important check on the general nature of the method that I was missing.
While the choice of state-of-the-art is good in my opinion, I was looking for comments on the methods prior to that. For instance, methods such based on GLMs have been used by the Pillow, Paninski, and Truccolo groups. I could not find a decent discussion of these methods in the main text and thought that both their acknowledgement and rationale for dismissing were missing.
While most of the text was very clear, I thought that page 11 was odd and missing much in terms of introductions. Foremost is the introduction of the dataset, which is never really done. Page 11 refers to 'this dataset', while the previous sentence was saying that having such a dataset would be important and is challenging. The dataset needs to be properly described: what's the method for labeling, what's the brain area, what were the spike recording methodologies, what is meant by two labeling methodologies, what do we know about the idiosyncrasies of the particular network the recording came from (like CA1 is non-recurrent, so which connections)? I was surprised to see 'English et al.' cited in text only on page 13 since their data has been hailed from the beginning.
Further elements that needed definition are the Nsyn and i, which were not defined in the cortex of Equation 2-3: I was not sure if it referred to different samples or different variants of the synthetic model. I also would have preferred having the function f defined earlier, as it is defined for Equation 3, but appears in Equation 2.
When the loss functions are described, it would be important to define 'data' and 'labels' here. This machine learning jargon has a concrete interpretation in this context, and making this concrete would be very important for the readership.
While I appreciated that there was a section on robustness, I did not find that the features studied were the most important. In this context, I was surprised that the other datasets were relegated to supplementary, as these appeared more relevant.
Some of the figures have text that is too small. In particular, Figure 2 has text that is way too small. It seemed to me that the pseudo code could stand alone, and the screenshot of the equations did not need to be repeated in a figure, especially if their size becomes so small that we can't even read them.
Author response:
General Response
We thank the reviewers for their positive assessment of our work and for acknowledging the timeliness of the problem and the novelty of using domain adaptation to address model mismatch. We appreciate the constructive feedback regarding validation and clarity. In the revised manuscript, we will address these points as follows:
(1) Systematic Validation: We will design and perform systematic in silico experiments to evaluate the method beyond the single in vivo dataset , including robustness tests regarding recording length and network synchrony.
(2) Recurrent Networks & Failure Analysis: We will test our method on synthetic datasets generated from highly recurrent networks and analyze exactly when the method breaks as a function of mismatch magnitude.
(3) Method Comparisons: We will report the Matthews Correlation Coefficient (MCC) for the approach by English et al. (2017) and expand our comparison and discussion of GLM-based methods.
(4) Clarifications: We will rigorously define the dataset details (labeling, recording methodology), mathematical notation, and machine learning terminology ('data', 'labels').
(5) Discussion of Limitations: We will explicitly discuss the challenges and limitations inherent in generalizing to more recurrently connected regions.
Below are our more detailed responses:
Public Reviews:
Reviewer #1 (Public review):
Weaknesses:
(1) The validation of the approach is incomplete: due to its very limited size, the single ground-truth dataset considered does not provide a sufficient basis to draw a strong conclusion. While the authors correctly note that this is the only dataset of its kind, the value of this validation is limited compared to what could be done by carefully designing in silico experiments.
We thank the reviewer for acknowledging the scarcity of suitable in vivo ground-truth datasets and the limitations this poses. We agree that additional validation is necessary to draw strong conclusions. In the revised manuscript, we will systematically design and perform in silico experiments for evaluations beyond the single in vivo dataset.
(2) Surprisingly, the authors fail to compare their method to the approach originally proposed for the data they validate on (English et al., 2017).
We agree that this is an essential comparison. We will report the Matthews Correlation Coefficient (MCC) result of the approach by English et al. (2017) on the spontaneous period of the recording.
(3) The authors make a commendable effort to study the method's robustness by pushing the limits of the dataset. However, the logic of the robustness analysis is often unclear, and once again, the limited size of the dataset poses major limitations to the authors.
We appreciate the reviewer recognizing our initial efforts to evaluate robustness. In our original draft, we tested recording length, network model choices, and analyzed failure cases. However, we agree that the limited real data restricts the scope of these tests. To address this, we will perform more systematic robustness tests on the newly generated synthetic datasets in the revised version, allowing us to evaluate performance under a wider range of conditions.
(4) The lack of details concerning both the approach and the validation makes it challenging for the reader to establish the technical soundness of the study.
We will revise the manuscript thoroughly to better present the methodology of our framework and the validation pipelines. We will ensure that the figures and text clearly articulate the technical details required to assess the soundness of the study.
Although in the current form this study does not provide enough basis to judge the impact of DeepDAM in the broader neuroscience community, it nevertheless puts forward a valuable and novel idea: using domain adaptation to mitigate the problem of model mismatch. This approach might be leveraged in future studies and methods to infer connectivity.
We thank the reviewer again for acknowledging the novelty and importance of our work.
Reviewer #2 (Public review):
While the validation data set was well chosen and of high quality, it remains a single dataset and also remains a non-recurrent network. The authors acknowledge this in the discussion, but I wanted to chime in to say that for the method to be more than convincing, it would need to have been tested on more datasets. It should be acknowledged that the problem becomes more complicated in a recurrent excitatory network, and thus the method may not work as well in the cortex or in CA3.
We will carefully revise our text to specifically discuss this limitation and the challenges inherent in generalizing to more recurrently connected regions. Furthermore, to empirically address this concern, we will test our method extensively on synthetic datasets generated from highly recurrent networks to quantify performance in these regimes.
While the data is shown to work in this particular dataset (plus the two others at the end), I was left wondering when the method breaks. And it should break if the models are sufficiently mismatched. Such a question can be addressed using synthetic-synthetic models. This was an important intuition that I was missing, and an important check on the general nature of the method that I was missing.
We thank the reviewer for this insight regarding the general nature of the method. While we previously analyzed failure cases regarding strong covariation and low spike counts, we agree that a systematic analysis of mismatch magnitude is missing. Building on our planned experiments with synthetic data, we will analyze and discuss exactly when the method breaks as a function of the mismatch magnitude between datasets.
While the choice of state-of-the-art is good in my opinion, I was looking for comments on the methods prior to that. For instance, methods such based on GLMs have been used by the Pillow, Paninski, and Truccolo groups. I could not find a decent discussion of these methods in the main text and thought that both their acknowledgement and rationale for dismissing were missing.
As the reviewer noted, we extensively compared our method with a GLM-based method (GLMCC) and CoNNECT, whose superiority over other GLM-based methods, such as extend GLM method (Ren et al., 2020, J Neurophysiol), have already been demonstrated in their papers (Endo et al., Sci Rep, 2021). However, we acknowledge that the discussion of the broader GLM literature was insufficient. To make the comparison more thorough, we will conduct comparisons with additional GLM-based methods and include a detailed discussion of these approaches.
Endo, D., Kobayashi, R., Bartolo, R., Averbeck, B. B., Sugase-Miyamoto, Y., Hayashi, K., ... & Shinomoto, S. (2021). A convolutional neural network for estimating synaptic connectivity from spike trains. Scientific Reports, 11(1), 12087.
Ren, N., Ito, S., Hafizi, H., Beggs, J. M., & Stevenson, I. H. (2020). Model-based detection of putative synaptic connections from spike recordings with latency and type constraints. Journal of Neurophysiology, 124(6), 1588-1604.
While most of the text was very clear, I thought that page 11 was odd and missing much in terms of introductions. Foremost is the introduction of the dataset, which is never really done. Page 11 refers to 'this dataset', while the previous sentence was saying that having such a dataset would be important and is challenging. The dataset needs to be properly described: what's the method for labeling, what's the brain area, what were the spike recording methodologies, what is meant by two labeling methodologies, what do we know about the idiosyncrasies of the particular network the recording came from (like CA1 is non-recurrent, so which connections)? I was surprised to see 'English et al.' cited in text only on page 13 since their data has been hailed from the beginning.
Further elements that needed definition are the Nsyn and i, which were not defined in the cortex of Equation 2-3: I was not sure if it referred to different samples or different variants of the synthetic model. I also would have preferred having the function f defined earlier, as it is defined for Equation 3, but appears in Equation 2.
When the loss functions are described, it would be important to define 'data' and 'labels' here. This machine learning jargon has a concrete interpretation in this context, and making this concrete would be very important for the readership.
We thank the reviewer for these constructive comments on the writing. We will clarify the introduction of the dataset (labeling method, brain area, recording methodology) and ensure all mathematical terms (such as Nsyn, i, and function f) and machine learning terminology (definitions of 'data' and 'labels' in this context) are rigorously defined upon first use in the revised manuscript.
While I appreciated that there was a section on robustness, I did not find that the features studied were the most important. In this context, I was surprised that the other datasets were relegated to supplementary, as these appeared more relevant.
Robustness is an important aspect of our framework to demonstrate its applicability to real experimental scenarios. We specifically analyzed how synchrony between neurons, the number of recorded spikes and the choice of the network influence the performance of our method. We also agree that these aspects are limited by the one dataset we evaluated on. Therefore, we will test the robustness of our method more systematically on synthetic datasets.
With more extensive analysis on synthetic datasets, we believe that the results on inferring biophysical properties of single neuron and microcircuit models remain in the supplement, such that the main figures focus purely on synaptic connectivity inference.
Some of the figures have text that is too small. In particular, Figure 2 has text that is way too small. It seemed to me that the pseudo code could stand alone, and the screenshot of the equations did not need to be repeated in a figure, especially if their size becomes so small that we can't even read them.
We will remove the pseudo-code and equations from Figure 2 to improve readability. The pseudo-code will be presented as a distinct box in the main text.
Organizations
It would be interesting to look now - almost 20 years after this declaration - at to what extent these 387 organisations have changed their practice to be more in line with the principles of higher education.
The paper begins with an explanation of my approach toward analyzing the media coverage of the release of “Where That Came From,” followed by a mostly chronological overview of the reporting about its production and release. After this descriptive overview, I will analyze four themes that run through the media coverage: the promise of “giving back his voice” to Travis, the way in which the relationship between human and technological labor is presented in this story, its framing as a human-interest story, and finally, its broader implications in terms of the strategic and financial interests that are at play.
method -- author's method described
When you interact with your classmates in your English class and learn the language of writing processes, for example, determining your rhetorical situation, you are participating in a specific discourse community
This helped me understand that even an English class is a discourse community because it has its own language, writing processes, and expectations. It explains why we have to follow certain writing rules.
Many students think contacting their instructor shows that they weren’t paying attention or that they are the only student did not understand something, so they often keep quiet and go on trying to do work that they do not understand. Other students think that their teacher is their own private tutor, so they email or message the teacher several times a day to ask questions that likely have answers in the syllabus and in the learning module instructions.
This stood out to me because many students avoid contacting instructors out of fear of looking like they weren’t paying attention. I think asking questions is better than struggling in silence
The main purpose of communication whether it be email, text, tweet, blog, discussion, presentation, written assignment, or speech is always to help the receiver(s) of the message understand the idea that the sender of the message is trying to share
The main purpose of communication is to help the receiver understand the idea being shared. This matters in college because clear messages prevent confusion with assignments and expectations
the parameters will have been “picked” or “fixed” already.
why not write the test error as \mathcal{E}(h, \theta) = ... and then say for a fixed $\theta \in \Theta$ instead of burying the dependence on \theta? This is partially addressed in the note but it might make the note more intuitive. Also, \Theta is typically reserved for parameter space and \theta are instantiations of the parameters. Since we are not adopting this convention making we can introduce notation for a fixed \Theta and still incorporate it
where Θ stands for all the parameters in our model (i.e., all possible choices over parameters).
Why not place this above where we first introduce this notation (i.e. for training error)?
Note: This response was posted by the corresponding author to Review Commons. The content has not been altered except for formatting.
Learn more at Review Commons
In our manuscript, we describe a role for the nuclear mRNA export factor UAP56 (a helicase) during metamorphic dendrite and presynapse pruning in flies. We characterize a UAP56 ATPase mutant and find that it rescues the pruning defects of a uap56 mutant. We identify the actin severing enzyme Mical as a potentially crucial UAP56 mRNA target during dendrite pruning and show alterations at both the mRNA and protein level. Finally, loss of UAP56 also causes presynapse pruning defects with actin abnormalities. Indeed, the actin disassembly factor cofilin is required for pruning specifically at the presynapse.
We thank the reviewers for their constructive comments, which we tried to address experimentally as much as possible. To summarize briefly, while all reviewers saw the results as interesting (e. g., Reviewer 3's significance assessment: "Understanding how post-transcriptional events are linked to key functions in neurons is important and would be of interest to a broad audience") and generally methodologically strong, they thought that our conclusions regarding the potential specificity of UAP56 for Mical mRNA was not fully covered by the data. To address this criticism, we added more RNAi analyses of other mRNA export factors and rephrased our conclusions towards a more careful interpretation, i. e., we now state that the pruning process is particularly sensitive to loss of UAP56. In addition, reviewer 1 had technical comments regarding some of our protein and mRNA analyses. We added more explanations and an additional control for the MS2/MCP system. Reviewers 2 and 3 wanted to see a deeper characterization of the ATPase mutant provided. We generated an additional UAP56 mutant transgene, improved our analyses of UAP56 localization, and added a biochemical control experiment. We hope that our revisions make our manuscript suitable for publication.
This section is mandatory. *Please insert a point-by-point reply describing the revisions that were already carried out and included in the transferred manuscript. *
Comments by reviewer 1.
Major comments
1.
For Figure 4, the MS2/MCP system is not quantitative. Using this technique, it is impossible to determine how many RNAs are located in each "dot". Each of these dots looks quite large and likely corresponds to some phase-separated RNP complex where multiple RNAs are stored and/or transported. Thus, these data do not support the conclusion that Mical mRNA levels are reduced upon UAP56 knockdown. A good quantitative microscopic assay would be something like smFISH. Additinally, the localization of Mical mRNA dots to dendrites is not convincing as it looks like regions where there are dendritic swellings, the background is generally brighter.
Our response
We indeed found evidence in the literature that mRNPs labeled with the MS2/MCP or similar systems form condensates (Smith et al., JCB 2015). Unfortunately, smFISH is not established for this developmental stage and would likely be difficult due to the presence of the pupal case. To address whether the Mical mRNPs in control and UAP56 KD neurons are comparable, we characterized the MCP dots in the respective neurons in more detail and found that their sizes did not differ significantly between control and UAP56 KD neurons. To facilitate interpretability, we also increased the individual panel sizes and include larger panels that only show the red (MCP::RFP) channel. We think these changes improved the figure. Thanks for the insight.
Changes introduced: Figure 5 (former Fig. 4): Increased panel size for MCP::RFP images, left out GFP marker for better visibility. Added new analysis of MCP::RFP dot size (new Fig. 5 I).
1.
Alternatively, levels of Mical mRNA could be verified by qPCR in the laval brain following pan-neuronal UAP56 knockdown or in FACS-sorted fluorescently labeled da sensory neurons. Protein levels could be analyzed using a similar approach.
Our response
We thank the reviewer for this comment. Unfortunately, these experiments are not doable as neuron-wide UAP56 KD is lethal (see Flybase entry for UAP56). From our own experience, FACS-sorting of c4da neurons would be extremely difficult as the GFP marker fluorescence intensity of UAP56 KD neurons is weak - this would likely result in preferential sorting of subsets of neurons with weaker RNAi effects. In addition, FACS-sorting whole neurons would not discriminate between nuclear and cytoplasmic mRNA.
The established way of measuring protein content in the Drosophila PNS system is immunofluorescence with strong internal controls. In our case, we also measured Mical fluorescence intensity of neighboring c1da neurons that do not express the RNAi and show expression levels as relative intensities compared to these internal controls. This procedure rules out the influence of staining variation between samples and is used by other labs as well.
1.
In Figure 5, the authors state that Mical expression could not be detected at 0 h APF. The data presented in Fig. 5C, D suggest the opposite as there clearly is some expression. Moreover, the data shown in Fig. 5D looks significantly brighter than the Orco dsRNA control and appears to localize to some type of cytoplasmic granule. So the expression of Mical does not look normal.
Our response
We thank the reviewer for this comment. In the original image in Fig. 5 C, the c4da neuron overlaps with the dendrite from a neighboring PNS neuron (likely c2da or c3da). The latter neuron shows strong Mical staining. We agree that this image is confusing and exchanged this image for another one from the same genotype.
Changes introduced: Figure 5 L (former Fig. 5 C): Exchanged panel for image without overlap from other neuron.
1.
Sufficient data are not presented to conclude any specificity in mRNA export pathways. Data is presented for one export protein (UAP56) and one putative target (Mical). To adequately assess this, the authors would need to do RNA-seq in UAP56 mutants.
Our response
We thank the reviewer for this comment. To address this, we tested whether knockdown of three other mRNA export factors (NXF1, THO2, THOC5) causes dendrite pruning defects, which was not the case (new Fig. S1). While these data are consistent with specific mRNA export pathways, we agree that they are not proof. We therefore toned down our interpretation and removed the conclusion about specificity. Instead, we now use the more neutral term "increased sensibility (to loss of UAP56)".
Changes introduced: Added new Figure S1: RNAi analyses of NXF1, THO2 and THOC5 in dendrite pruning. Introduced concluding sentence at the end of first Results paragraph: We conclude that c4da neuron dendrite pruning is particularly sensitive to loss of UAP56. (p. 6)
1.
In summary, better quantitative assays should be used in Figures 4 and 5 in order to conclude the expression levels of either mRNA or protein. In its current form, this study demonstrates the novel finding that UAP56 regulates dendrite and presynaptic pruning, potentially via regulation of the actin cytoskeleton. However, these data do not convincingly demonstrate that UAP56 controls these processes by regulating of Mical expression and defintately not by controlling export from the nucleus.
Our response
We hope that the changes we introduced above help clarify this.
1.
While there are clearly dendrites shown in Fig. 1C', the cell body is not readily identifiable. This makes it difficult to assess attachment and suggests that the neuron may be dying. This should be replaced with an image that shows the soma.
Our response
We thank the reviewer for this comment. Changes introduced: we replaced the picture in the panel with one where the cell body is more clearly visible.
1.
The level of knockdown in the UAS56 RNAi and P element insertion lines should be determined. It would be useful to mention the nature of the RNAi lines (long/short hairpin). Some must be long since Dcr has been co-expressed. Another issue raised by this is the potential for off-target effects. shRNAi lines would be preferable because these effects are minimized.
Our response
We thank the reviewer for this comment. Assessment of knockdown efficiency is a control to make sure the manipulations work the way they are intended to. As mRNA isolation from Drosophila PNS neurons is extremely difficult, RNAi or mutant phenotypes in this system are controlled by performing several independent manipulations of the same gene. In our case, we used two independent RNAi lines (both long hairpins from VDRC/Bloomington and an additional insertion of the VDRC line, see Table S1) as well as a mutant P element in a MARCM experiment, i. e., a total of three independent manipulations that all cause pruning defects, and the VDRC RNAi lines do not have any predicted OFF targets (not known for the Bloomington line). If any of these manipulations would not have matched, we would have generated sgRNA lines for CRISPR to confirm.
Minor comments:
1.
The authors should explain what EB1:GFP is marking when introduced in the text.
Our response
We thank the reviewer for this comment. Changes introduced: we explain the EB1::GFP assay in the panel with one where the cell body is more clearly visible.
1.
The da neuron images throughout the figures could be a bit larger.
Our response
We thank the reviewer for this comment. Changes introduced: we changed the figure organization to be able to use larger panels:
the pruning analysis of the ATPase mutations (formerly Fig. 2) is now its own figure (Figure 3).
we increased the panel sizes of the MCP::RFP images (Figure 5 A - I, formerly Fig. 4).
Reviewer #1 (Significance (Required)):
Strengths:
The methodology used to assess dendrite and presynaptic prunings are strong and the phenotypic analysis is conclusive.
Our response
We thank the reviewer for this comment.
Weakness:
The evidence demonstrating that UAP56 regulates the expression of Mical is unconvincing. Similarly, no data is presented to show that there is any specificity in mRNA export pathways. Thus, these major conclusions are not adequately supported by the data.
Our response
We hope the introduced changes address this comment.
__Reviewer #2 (Evidence, reproducibility and clarity (Required)): __
In this paper, the authors describe dendrite pruning defects in c4da neurons in the DEXD box ATPase UAP56 mutant or in neuronal RNAi knockdown. Overexpression UAP56::GFP or UAP56::GFPE194Q without ATPase activity can rescue dendrite pruning defects in UAP56 mutant. They further characterized the mis-localization of UAP56::GFPE194Q and its binding to nuclear export complexes. Both microtubules and the Ubiquitin-proteasome system are intact in UAP56RNAi neurons. However, they suggest a specific effect on MICAL mRNA nuclear export shown by using the MS2-MCP system., resulting in delay of MICAL protein expression in pruned neurons. Furthermore, the authors show that UAP56 is also involved in presynaptic pruning of c4da neuros in VNC and Mica and actin are also required for actin disassembly in presynapses. They propose that UAP56 is required for dendrite and synapse pruning through actin regulation in Drosophila. Following are my comments.
Major comments
1.
The result that UAP56::GFPE194Q rescues the mutant phenotype while the protein is largely mis-localized suggests a novel mechanism or as the authors suggested rescue from combination of residual activities. The latter possibility requires further support, which is important to support the role mRNA export in dendrite and pre-synapse pruning. One approach would be to examine whether other export components like REF1, and NXF1 show similar mutant phenotypes. Alternatively, depleting residual activity like using null mutant alleles or combining more copies of RNAi transgenes could help.
Our response
We thank the reviewer for this comment. We agree that the mislocalization phenotype is interesting and could inform further studies on the mechanism of UAP56. To further investigate this and to exclude that this could represent a gain-of-function due to the introduced mutation, we made and characterized a new additional transgene, UAP56::GFP E194A. This mutant shows largely the same phenotypes as E194Q, with enhanced interactions with Ref1 and partial mislocalization to the cytoplasm. In addition, we tested whether knockdown of THO2, THOC5 or NXF1 causes pruning defects (no).
Changes introduced:
added new Figure S1: RNAi analyses of NXF1, THO2 and THOC5 in dendrite pruning.
made and characterized a new transgene UAP56 E194A (new Fig. 2 B, E, E', 3 C, C', E, F).
1.
The localization of UAP56::GFP (and E194Q) should be analyzed in more details. It is not clear whether the images in Fig. 2A and 2B are from confocal single sections or merged multiple sections. The localization to the nuclear periphery of UAP56::GFP is not clear, and the existence of the E194Q derivative in both nucleus and cytosol (or whether there is still some peripheral enrichment) is not clear if the images are stacked.
Our response
We thank the reviewer for this comment. It is correct that the profiles in the old Figure 2 were from single confocal sections from the displayed images. As it was difficult to create good average profiles with data from multiple neurons, we now introduce an alternative quantification based on categories (nuclear versus dispersed) which includes data from several neurons for each genotype, including the new E194A transgene (new Fig 3 G). Upon further inspection, the increase at the nuclear periphery was not always visible and may have been a misinterpretation. We therefore removed this statement.
Changes introduced:
1.
The Ub-VV-GFP is a new reagent, and its use to detect active proteasomal degradation is by the lack of GFP signals, which could be also due to the lack of expression. The use of Ub-QQ-GFP cannot confirm the expression of Ub-VV-GFP. The proteasomal subunit RPN7 has been shown to be a prominent component in the dendrite pruning pathway (Development 149, dev200536). Immunostaining using RPN7 antibodies to measure the RPN expression level could be a direct way to address the issue whether the proteasomal pathway is affected or not.
Our response
We thank the reviewer for this comment. We agree that it is wise to not only introduce a positive control for the Ub-VV-GFP sensor (the VCP dominant-negative VCP QQ), but also an independent control. As mutants with defects in proteasomal degradation accumulate ubiquitinated proteins (see, e. g., Rumpf et al., Development 2011), we stained controls and UAP56 KD neurons with antibodies against ubiquitin and found that they had similar levels (new Fig. S3).
Changes introduced:
1.
Using the MS2/MCP system to detect the export of MICAL mRNA is a nice approach to confirm the UAP56 activity; lack of UAP56 by RNAi knockdown delays the nuclear export of MS2-MICAL mRNA. The rescue experiment by UAS transgenes could not be performed due to the UAS gene dosage, as suggested by the authors. However, this MS2-MICAL system is also a good assay for the requirement of UAP56 ATPase activity (absence in the E194Q mutant) in this process. Could authors use the MARCM (thus reduce the use of UAS-RNAi transgene) for the rescue experiment? Also, the c4da neuronal marker UAS-CD8-GFP used in Fig4 could be replaced by marker gene directly fused to ppk promoter, which can save a copy of UAS transgene. The results from the rescue experiment would test the dependence of ATPase activity in nuclear export of MICAL mRNA.
Our response
We thank the reviewer for this comment. This is a great idea but unfortunately, this experiment was not feasible due to the (rare) constraints of Drosophila genetics. The MARCM system with rescue already occupies all available chromosomes (X: FLPase, 2nd: FRT, GAL80 + mutant, 3rd: GAL4 + rescue construct), and we would have needed to introduce three additional ones (MCP::RFP and two copies of unmarked genomic MICAL-MS2, all on the third chromosome) that would have needed to be introduced by recombination. Any Drosophilist will see that this is an extreme, likely undoable project :-(
1.
The UAP56 is also involved in presynaptic pruning through regulating actin assembly, and the authors suggest that Mical and cofilin are involved in the process. However, direct observation of lifeact::GFP in Mical or cofilin RNAi knockdown is important to support this conclusion.
Our response
We thank the reviewer for this comment. In response, we analyzed the lifeact::GFP patterns of control and cofilin knockdown neurons and found that loss of cofilin also leads to actin accumulation (new Fig. 7 I, J).
Changes introduced:
Minor comments:
1.
RNA localization is important for dendrite development in larval stages (Brechbiel JL, Gavis ER. Curr Biol. 20;18(10):745-750). Yet, the role of UAP56 is relatively specific and shown only in later-stage pruning. It would need thorough discussion.
Our response
We thank reviewer 2 for this comment. We added the following paragraph to the discussion: "UAP56 has also been shown to affect cytoplasmic mRNA localization in Drosophila oocytes (Meignin and Davis, 2008), opening up the possibility that nuclear mRNA export and cytoplasmic transport are linked. It remains to be seen whether this also applies to dendritic mRNA transport (Brechbiel and Gavis, 2008)." (p.13)
1.
Could authors elaborate on the possible upstream regulators that might be involved, as described in "alternatively, several cofilin upstream regulators have been described (Rust, 2015) which might also be involved in presynapse pruning and subject to UAP56 regulation" in Discussion?
Our response
We thank reviewer 2 for this comment. In the corresponding paragraph, we cite as example now that cofilin is regulated by Slingshot phosphatases and LIM kinase (p.14).
1.
In Discussion, the role of cofilin in pre- and post-synaptic processes was described. The role of Tsr/Cofilin regulating actin behaviors in dendrite branching has been described in c3da and c4da neurons (Nithianandam and Chien, 2018 and other references) should be included in Discussion.
Our response
We thank reviewer 2 for this comment. In response we tested whether cofilin is required for dendrite pruning and found that this, in contrast to Mical, is not the case (new Fig. S6). We cite the above paper in the corresponding results section (p.12).
Changes introduced:
new cofilin dendrite pruning analysis (new Fig. S6).
added cofilin reference in Results.
1.
The authors speculate distinct actin structures have to be disassembled in dendrite and presynapse pruning in Discussion. What are the possible actin structures in both sites could be elaborated.
Our response
We thank reviewer 2 for this comment. In response, we specify in the Discussion: "As Mical is more effective in disassembling bundled F-actin than cofilin (Rajan et al., 2023), it is interesting to speculate that such bundles are more prevalent in dendrites than at presynapses." (p14)
Reviewer #2 (Significance (Required)):
The study initiated a genetic screen for factors involved in a dendrite pruning system and reveals the involvement of nuclear mRNA export is an important event in this process. They further identified the mRNA of the actin disassembly factor MICAL is a candidate substrate in the exporting process. This is consistent with previous finding that MICAL has to be transcribed and translated when pruning is initiated. As the presynapses of the model c4da neuron in this study is also pruned, the dependence on nuclear export and local actin remodeling were also shown. Thus, this study has added another layer of regulation (the nuclear mRNA export) in c4da neuronal pruning, which would be important for the audience interested in neuronal pruning. The study is limited for the confusing result whether ATPase activity of the exporting factor is required.
Reviewer #3 (Evidence, reproducibility and clarity (Required)):
Summary: In the manuscript by Frommeyer, Gigengack et al. entitled "The UAP56 mRNA Export Factor is Required for Dendrite and Synapse Pruning via Actin Regulation in Drosophila" the authors surveyed a number of RNA export/processing factors to identify any required for efficient dendrite and/or synapse pruning. They describe a requirement for a general poly(A) RNA export factor, UAP56, which functions as an RNA helicase. They also study links to aspects of actin regulation.
Overall, while the results are interesting and the impact of loss of UAP56 on the pruning is intriguing, some of the data are overinterpreted as presented. The argument that UAP56 may be specific for the MICAL RNA is not sufficiently supported by the data presented. The two stories about poly(A) RNA export/processing and the actin regulation seem to not quite be connected by the data presented. The events are rather distal within the cell, making connecting the nuclear events with RNA to events at the dendrites/synapse challenging.
Our response
We thank reviewer 3 for this comment. To address this, we tested whether knockdown of three other mRNA export factors (NXF1, THO2, THOC5) causes dendrite pruning defects, which was not the case (new Fig. S1). While these data are consistent with specific mRNA export pathways, we agree that they are not proof. We therefore toned down our interpretation and removed the conclusion about specificity. Instead, we now use the more neutral term "increased sensibility (to loss of UAP56)".
We agree that it is a little hard to tie cofilin to UAP56, as we currently have no evidence that cofilin levels are affected by loss of UAP56, even though both seem to affect lifeact::GFP in a similar way (new Fig. 7 I, J). However, a dysregulation of cofilin can also occur through dysregulation of upstream cofilin regulators such as Slingshot and LIM kinase, making such a relationship possible.
Changes introduced:
added new Figure S1: RNAi analyses of NXF1, THO2 and THOC5 in dendrite pruning.
introduced concluding sentence at the end of first Results paragraph: "We conclude that c4da neuron dendrite pruning is particularly sensitive to loss of UAP56." (p. 6)
add new lifeact::GFP analysis of cofilin KD (new Fig. I, J).
identify potential other targets from the literature in the Discussion (Slingshot phosphatases and LIM kinase, p.14).
There are a number of specific statements that are not supported by references. See, for example, these sentences within the Introduction- "Dysregulation of pruning pathways has been linked to various neurological disorders such as autism spectrum disorders and schizophrenia. The cell biological mechanisms underlying pruning can be studied in Drosophila." The Drosophila sentence is followed by some specific examples that do include references. The authors also provide no reference to support the variant that they create in UAP56 (E194Q) and whether this is a previously characterized fly variant or based on an orthologous protein in a different system. If so, has the surprising mis-localization been reported in another system?
Our response
We thank reviewer 3 for this comment. We added the following references on pruning and disease:
1) Howes, O.D., Onwordi, E.C., 2023. The synaptic hypothesis of schizophrenia version III: a master mechanism. Mol. Psychiatry 28, 1843-1856.
2) Tang, G., et al., 2014. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron 83, 1131-43.
To better introduce the E194 mutations, we explain the position of the DECD motif in the Walker B domain, give the corresponding residues in the human and yeast homologues and cite papers demonstrating the importance of this residue for ATPase activity:
3) Saguez, C., et al., 2013. Mutational analysis of the yeast RNA helicase Sub2p reveals conserved domains required for growth, mRNA export, and genomic stability. RNA 19:1363-71.
4) Shen, J., et al., 2007. Biochemical Characterization of the ATPase and Helicase Activity of UAP56, an Essential Pre-mRNA Splicing and mRNA Export Factor. J. Biol. Chem. 282, P22544-22550.
We are not aware of other studies looking at the relationship between the UAP56 ATPase and its localization. Thank you for pointing this out!
Specific Comments:
Specific Comment 1: Figure 1 shows the impact of loss of UAP56 on neuron dendrite pruning. The experiment employs both two distinct dsRNAs and a MARCM clone, providing confidence that there is a defect in pruning upon loss of UAP56. As the authors mention screening against 92 genes that caused splicing defects in S2 cells, inclusion of some examples of these genes that do not show such a defect would enhance the argument for specificity with regard to the role of UAP56. This control would be in addition to the more technical control that is shown, the mCherry dsRNA.
Our response
We thank reviewer 3 for this comment. To address this, we included the full list of screened genes with their phenotypic categorization regarding pruning (103 RNAi lines targeting 64 genes) as Table S1. In addition, we also tested four RNAi lines targeting the nuclear mRNA export factors Nxf1, THO2 and THOC5 which do not cause dendrite pruning defects (Fig. S1).
Changes introduced:
added RNAi screen results as a list in Table S1.
added new Figure S1: RNAi analyses of NXF1, THO2 and THOC5 in dendrite pruning.
Specific Comment 2: Later the authors demonstrate a delay in the accumulation of the Mical protein, so if they assayed these pruning events at later times, would the loss of UAP56 cause a delay in these events as well? Such a correlation would enhance the causality argument the authors make for Mical levels and these pruning events.
Our response
We thank reviewer 3 for this comment. Unfortunately, this is somewhat difficult to assess, as shortly after the 18 h APF timepoint, the epidermal cells that form the attachment substrate for c4da neuron dendrites undergo apoptosis. Where assessed (e. g., Wang et al., 2017, Development) 144: 1851–1862), this process, together with the reduced GAL4 activity of our ppk-GAL4 during the pupal stage (our own observations), eventually leads to pruning, but the causality cannot be easily attributed anymore. We therefore use the 18 h APF timepoint essentially as an endpoint assay.
Specific Comment 3: Figure 2 provides data designed to test the requirement for the ATPase/helicase activity of UAP56 for these trimming events. The first observation, which is surprising, is the mislocalization of the variant (E194Q) that the authors generate. The data shown does not seem to indicate how many cells the results shown represent as a single image and trace is shown the UAP56::GFP wildtype control and the E194Q variant.
Our response
We thank reviewer 3 for this comment. It is correct that the traces shown are from single confocal sections. To better display the phenotypic penetrance, we now added a categorical analysis that shows that the UAP56 E194Q mutant is completely mislocalized in the majority of cells assessed (and the newly added E194A mutant in a subset of cells).
Changes introduced:
__Specific Comment 4: __Given the rather surprising finding that the ATPase activity is not required for the function of UAP56 characterized here, the authors do not provide sufficient references or rationale to support the ATPase mutant that they generate. The E194Q likely lies in the Walker B motif and is equivalent to human E218Q, which can prevent proper ATP hydrolysis in the yeast Sub2 protein. There is no reference to support the nature of the variant created here.
Our response
We thank reviewer 3 for this comment. To better introduce the E194 mutations, we explain the position of the DECD motif in the Walker B domain, give the corresponding residues in the human and yeast homologues (Sub2) and cite papers demonstrating the importance of this residue for ATPase activity:
1) Saguez, C., et al., 2013. Mutational analysis of the yeast RNA helicase Sub2p reveals conserved domains required for growth, mRNA export, and genomic stability. RNA 19:1363-71.
2) Shen, J., et al., 2007. Biochemical Characterization of the ATPase and Helicase Activity of UAP56, an Essential Pre-mRNA Splicing and mRNA Export Factor. J. Biol. Chem. 282, P22544-22550.
__Specific Comment 5: __Given the surprising results, the authors could have included additional variants to ensure the change has the biochemical effect that the authors claim. Previous studies have defined missense mutations in the ATP-binding site- K129A (Lysine to Alanine): This mutation, in both yeast Sub2 and human UAP56, targets a conserved lysine residue that is critical for ATP binding. This prevents proper ATP binding and consequently impairs helicase function. There are also missense mutations in the DEAD-box motif, (Asp-Glu-Ala-Asp) involved in ATP binding and hydrolysis. Mutations in this motif, such as D287A in yeast Sub2 (corresponding to D290A in human UAP56), can severely disrupt ATP hydrolysis, impairing helicase activity. In addition, mutations in the Walker A (GXXXXGKT) and Walker B motifs are can impair ATP binding and hydrolysis in DEAD-box helicases. Missense mutations in these motifs, like G137A (in the Walker A motif), can block ATP binding, while E218Q (in the Walker B motif)- which seems to be the basis for the variant employed here- can prevent proper ATP hydrolysis.
Our response
We thank reviewer 3 for this comment. Our cursory survey of the literature suggested that mutations in the Walker B motif are the most specific as they still preserve ATP binding and their effects have not well been characterized overall. In addition, these mutations can create strong dominant-negatives in related helicases (e. g., Rode et al., 2018 Cell Reports, our lab). To better characterize the role of the Walker B motif in UAP56, we generated and characterized an alternative mutant, UAP56 E194A. While the E194A variant does not show the same penetrance of localization phenotypes as E194Q, it also is partially mislocalized, shows stronger binding to Ref1 and also rescues the uap56 mutant phenotypes without an obvious dominant-negative effect, thus confirming our conclusions regarding E194Q.
Changes introduced:
__Specific Comment 6: __The co-IP results shown in Figure 2C would also seem to have multiple potential interpretations beyond what the authors suggest, an inability to disassemble a complex. The change in protein localization with the E194Q variant could impact the interacting proteins. There is no negative control to show that the UAP56-E194Q variant is not just associated with many, many proteins. Another myc-tagged protein that does not interact would be an ideal control.
Our response
We thank reviewer 3 for this comment. To address this comment, we tried to co-IP UAP56 wt or UAP56 E194Q with a THO complex subunit THOC7 (new Fig. S2). The results show that neither UAP56 variant can co-IP THOC7 under our conditions (likely because the UAP56/THO complex intermediate during mRNA export is disassembled in an ATPase-independent manner (Hohmann et al., Nature 2025)).
Changes introduced:
__Specific Comment 7: __With regard to Figure 3, the authors never define EB1::GFP in the text of the Results, so a reader unfamiliar with this system has no idea what they are seeing. Reading the Materials and Methods does not mitigate this concern as there is only a brief reference to a fly line and how the EB1::GFP is visualized by microscopy. This makes interpretation of the data presented in Figure 3A-C very challenging.
Our response
We thank reviewer 3 for pointing this out. We added a description of the EB1::GFP analysis in the corresponding Results section (p.8).
__Specific Comment 8: __The data shown for MICAL MS2 reporter localization in Figure 4 is nice, but is also fully expected on many former studies analyzing loss of UAP56 or UAP56 hypomorphs in different systems. While creating the reporter is admirable, to make the argument that MICAL localization is in some way preferentially impacted by loss of UAP56, the authors would need to examine several other transcripts. As presented, the authors can merely state that UAP56 seems to be required for the efficient export of an mRNA transcript, which is predicted based on dozens of previous studies dating back to the early 2000s.
Our response
Firstly, thank you for commenting on the validity of the experimental approach! The primary purpose of this experiment was to test whether the mechanism of UAP56 during dendrite pruning conforms with what is known about UAP56's cellular role - which it apparently does. We also noted that our statements regarding the specificity of UAP56 for Mical over other transcripts are difficult. While our experiments would be consistent with such a model, they do not prove it. We therefore toned down the corresponding statements (e. g., the concluding sentence at the end of first Results paragraphis now: "We conclude that c4da neuron dendrite pruning is particularly sensitive to loss of UAP56." (p. 6)).
Minor (and really minor) points:
In the second sentence of the Discussion, the word 'developing' seems to be mis-typed "While a general inhibition of mRNA export might be expected to cause broad defects in cellular processes, our data in develoing c4da neurons indicate that loss of UAP56 mainly affects pruning mechanisms related to actin remodeling."
Sentence in the Results (lack of page numbers makes indicating where exactly a bit tricky)- "We therefore reasoned that Mical expression could be more challenging to c4da neurons." This is a complete sentence as presented, yet, if something is 'more something'- the thing must be 'more than' something else. Presumably, the authors mean that the length of the MICAL transcript could make the processing and export of this transcript more challenging than typical fly transcripts (raising the question of the average length of a mature transcript in flies?).
Our response
Thanks for pointing these out. The typo is fixed, page numbers are added. We changed the sentence to: "Because of the large size of its mRNA, we reasoned that MICAL gene expression could be particularly sensitive to loss of export factors such as UAP56." (p.9) We hope this is more precise language-wise.
Reviewer #3 (Significance (Required)):
Understanding how post-transcriptional events are linked to key functions in neurons is important and would be of interest to a broad audience.
Note: This preprint has been reviewed by subject experts for Review Commons. Content has not been altered except for formatting.
Learn more at Review Commons
Summary:
In the manuscript by Frommeyer, Gigengack et al. entitled "The UAP56 mRNA Export Factor is Required for Dendrite and Synapse Pruning via Actin Regulation in Drosophila" the authors surveyed a number of RNA export/processing factors to identify any required for efficient dendrite and/or synapse pruning. They describe a requirement for a general poly(A) RNA export factor, UAP56, which functions as an RNA helicase. They also study links to aspects of actin regulation.
Overall, while the results are interesting and the impact of loss of UAP56 on the pruning is intriguing, some of the data are overinterpreted as presented. The argument that UAP56 may be specific for the MICAL RNA is not sufficiently supported by the data presented. The two stories about poly(A) RNA export/processing and the actin regulation seem to not quite be connected by the data presented. The events are rather distal within the cell, making connecting the nuclear events with RNA to events at the dendrites/synapse challenging.
There are a number of specific statements that are not supported by references. See, for example, these sentences within the Introduction- "Dysregulation of pruning pathways has been linked to various neurological disorders such as autism spectrum disorders and schizophrenia. The cell biological mechanisms underlying pruning can be studied in Drosophila." The Drosophila sentence is followed by some specific examples that do include references. The authors also provide no reference to support the variant that they create in UAP56 (E194Q) and whether this is a previously characterized fly variant or based on an orthologous protein in a different system. If so, has the surprising mis-localization been reported in another system?
Specific Comments:
Figure 1 shows the impact of loss of UAP56 on neuron dendrite pruning. The experiment employs both two distinct dsRNAs and a MARCM clone, providing confidence that there is a defect in pruning upon loss of UAP56. As the authors mention screening against 92 genes that caused splicing defects in S2 cells, inclusion of some examples of these genes that do not show such a defect would enhance the argument for specificity with regard to the role of UAP56. This control would be in addition to the more technical control that is shown, the mCherry dsRNA. Later the authors demonstrate a delay in the accumulation of the Mical protein, so if they assayed these pruning events at later times, would the loss of UAP56 cause a delay in these events as well? Such a correlation would enhance the causality argument the authors make for Mical levels and these pruning events.
Figure 2 provides data designed to test the requirement for the ATPase/helicase activity of UAP56 for these trimming events. The first observation, which is surprising, is the mislocalization of the variant (E194Q) that the authors generate. The data shown does not seem to indicate how many cells the results shown represent as a single image and trace is shown the UAP56::GFP wildtype control and the E194Q variant.
Given the rather surprising finding that the ATPase activity is not required for the function of UAP56 characterized here, the authors do not provide sufficient references or rationale to support the ATPase mutant that they generate. The E194Q likely lies in the Walker B motif and is equivalent to human E218Q, which can prevent proper ATP hydrolysis in the yeast Sub2 protein. There is no reference to support the nature of the variant created here.
Given the surprising results, the authors could have included additional variants to ensure the change has the biochemical effect that the authors claim. Previous studies have defined missense mutations in the ATP-binding site- K129A (Lysine to Alanine): This mutation, in both yeast Sub2 and human UAP56, targets a conserved lysine residue that is critical for ATP binding. This prevents proper ATP binding and consequently impairs helicase function. There are also missense mutations in the DEAD-box motif, (Asp-Glu-Ala-Asp) involved in ATP binding and hydrolysis. Mutations in this motif, such as D287A in yeast Sub2 (corresponding to D290A in human UAP56), can severely disrupt ATP hydrolysis, impairing helicase activity. In addition, mutations in the Walker A (GXXXXGKT) and Walker B motifs are can impair ATP binding and hydrolysis in DEAD-box helicases. Missense mutations in these motifs, like G137A (in the Walker A motif), can block ATP binding, while E218Q (in the Walker B motif)- which seems to be the basis for the variant employed here- can prevent proper ATP hydrolysis.
The co-IP results shown in Figure 2C would also seem to have multiple potential interpretations beyond what the authors suggest, an inability to disassemble a complex. The change in protein localization with the E194Q variant could impact the interacting proteins. There is no negative control to show that the UAP56-E194Q variant is not just associated with many, many proteins. Another myc-tagged protein that does not interact would be an ideal control.
With regard to Figure 3, the authors never define EB1::GFP in the text of the Results, so a reader unfamiliar with this system has no idea what they are seeing. Reading the Materials and Methods does not mitigate this concern as there is only a brief reference to a fly line and how the EB1::GFP is visualized by microscopy. This makes interpretation of the data presented in Figure 3A-C very challenging. The data shown for MICAL MS2 reporter localization in Figure 4 is nice, but is also fully expected on many former studies analyzing loss of UAP56 or UAP56 hypomorphs in different systems. While creating the reporter is admirable, to make the argument that MICAL localization is in some way preferentially impacted by loss of UAP56, the authors would need to examine several other transcripts. As presented, the authors can merely state that UAP56 seems to be required for the efficient export of an mRNA transcript, which is predicted based on dozens of previous studies dating back to the early 2000s.
Minor (and really minor) points:
In the second sentence of the Discussion, the word 'developing' seems to be mis-typed "While a general inhibition of mRNA export might be expected to cause broad defects in cellular processes, our data in develoing c4da neurons indicate that loss of UAP56 mainly affects pruning mechanisms related to actin remodeling."
Sentence in the Results (lack of page numbers makes indicating where exactly a bit tricky)- "We therefore reasoned that Mical expression could be more challenging to c4da neurons." This is a complete sentence as presented, yet, if something is 'more something'- the thing must be 'more than' something else. Presumably, the authors mean that the length of the MICAL transcript could make the processing and export of this transcript more challenging than typical fly transcripts (raising the question of the average length of a mature transcript in flies?).
Understanding how post-transcriptional events are linked to key functions in neurons is important and would be of interest to a broad audience.
Note: This preprint has been reviewed by subject experts for Review Commons. Content has not been altered except for formatting.
Learn more at Review Commons
In this paper, the authors describe dendrite pruning defects in c4da neurons in the DEXD box ATPase UAP56 mutant or in neuronal RNAi knockdown. Overexpression UAP56::GFP or UAP56::GFPE194Q without ATPase activity can rescue dendrite pruning defects in UAP56 mutant. They further characterized the mis-localization of UAP56::GFPE194Q and its binding to nuclear export complexes. Both microtubules and the Ubiquitin-proteasome system are intact in UAP56RNAi neurons. However, they suggest a specific effect on MICAL mRNA nuclear export shown by using the MS2-MCP system., resulting in delay of MICAL protein expression in pruned neurons. Furthermore, the authors show that UAP56 is also involved in presynaptic pruning of c4da neuros in VNC and Mica and actin are also required for actin disassembly in presynapses. They propose that UAP56 is required for dendrite and synapse pruning through actin regulation in Drosophila. Following are my comments.
Major comments
The result that UAP56::GFPE194Q rescues the mutant phenotype while the protein is largely mis-localized suggests a novel mechanism or as the authors suggested rescue from combination of residual activities. The latter possibility requires further support, which is important to support the role mRNA export in dendrite and pre-synapse pruning. One approach would be to examine whether other export components like REF1, and NXF1 show similar mutant phenotypes. Alternatively, depleting residual activity like using null mutant alleles or combining more copies of RNAi transgenes could help.
The localization of UAP56::GFP (and E194Q) should be analyzed in more details. It is not clear whether the images in Fig. 2A and 2B are from confocal single sections or merged multiple sections. The localization to the nuclear periphery of UAP56::GFP is not clear, and the existence of the E194Q derivative in both nucleus and cytosol (or whether there is still some peripheral enrichment) is not clear if the images are stacked.
The Ub-VV-GFP is a new reagent, and its use to detect active proteasomal degradation is by the lack of GFP signals, which could be also due to the lack of expression. The use of Ub-QQ-GFP cannot confirm the expression of Ub-VV-GFP. The proteasomal subunit RPN7 has been shown to be a prominent component in the dendrite pruning pathway (Development 149, dev200536). Immunostaining using RPN7 antibodies to measure the RPN expression level could be a direct way to address the issue whether the proteasomal pathway is affected or not.
Using the MS2/MCP system to detect the export of MICAL mRNA is a nice approach to confirm the UAP56 activity; lack of UAP56 by RNAi knockdown delays the nuclear export of MS2-MICAL mRNA. The rescue experiment by UAS transgenes could not be performed due to the UAS gene dosage, as suggested by the authors. However, this MS2-MICAL system is also a good assay for the requirement of UAP56 ATPase activity (absence in the E194Q mutant) in this process. Could authors use the MARCM (thus reduce the use of UAS-RNAi transgene) for the rescue experiment? Also, the c4da neuronal marker UAS-CD8-GFP used in Fig4 could be replaced by marker gene directly fused to ppk promoter, which can save a copy of UAS transgene. The results from the rescue experiment would test the dependence of ATPase activity in nuclear export of MICAL mRNA.
The UAP56 is also involved in presynaptic pruning through regulating actin assembly, and the authors suggest that Mical and cofilin are involved in the process. However, direct observation of lifeact::GFP in Mical or cofilin RNAi knockdown is important to support this conclusion.
Minor comments
RNA localization is important for dendrite development in larval stages (Brechbiel JL, Gavis ER. Curr Biol. 20;18(10):745-750). Yet, the role of UAP56 is relatively specific and shown only in later-stage pruning. It would need thorough discussion.
Could authors elaborate on the possible upstream regulators that might be involved, as described in "alternatively, several cofilin upstream regulators have been described (Rust, 2015) which might also be involved in presynapse pruning and subject to UAP56 regulation" in Discussion?
In Discussion, the role of cofilin in pre- and post-synaptic processes was described. The role of Tsr/Cofilin regulating actin behaviors in dendrite branching has been described in c3da and c4da neurons (Nithianandam and Chien, 2018 and other references) should be included in Discussion.
The authors speculate distinct actin structures have to be disassembled in dendrite and presynapse pruning in Discussion. What are the possible actin structures in both sites could be elaborated.
The study initiated a genetic screen for factors involved in a dendrite pruning system and reveals the involvement of nuclear mRNA export is an important event in this process. They further identified the mRNA of the actin disassembly factor MICAL is a candidate substrate in the exporting process. This is consistent with previous finding that MICAL has to be transcribed and translated when pruning is initiated. As the presynapses of the model c4da neuron in this study is also pruned, the dependence on nuclear export and local actin remodeling were also shown. Thus, this study has added another layer of regulation (the nuclear mRNA export) in c4da neuronal pruning, which would be important for the audience interested in neuronal pruning. The study is limited for the confusing result whether ATPase activity of the exporting factor is required.
Note: This preprint has been reviewed by subject experts for Review Commons. Content has not been altered except for formatting.
Learn more at Review Commons
Summary:
This manuscript by Frommeyer et al. explores the role of the helicase and regulator of nuclear export, UAP56, in the control of dendrite and presynaptic pruning in Drosophila larval da sensory neurons. The authors present evidence showing that UAP56 regulates these processes via the actin cytoskeleton and suggest that this is occurs by controlling the expression of the actin severing enzyme, Mical.
Major comments:
The most signficant issue with the manuscript is that some of the major conclusions are not supported by the data. Additional experiment would need to be completed in order support these claims. These (and other) major comments are as follows:
For Figure 4, the ms2/MCP system is not quantitative. Using this technique, it is impossible to determine how many RNAs are located in each "dot". Each of these dots looks quite large and likely corresponds to some phase-separated RNP complex where multiple RNAs are stored and/or transported. Thus, these data do not support the conclusion that Mical mRNA levels are reduced upon UAP56 knockdown. A good quantitative microscopic assay would be something like smFISH. Additinally, the localization of Mical mRNA dots to dendrites is not convincing as it looks like regions where there are dendritic swellings, the background is generally brighter.
Alternatively, levels of Mical mRNA could be verified by qPCR in the laval brain following pan-neuronal UAP56 knockdown or in FACS-sorted fluorescently labeled da sensory neurons. Protein levels could be analyzed using a similar approach.
In Figure 5, the authors state that Mical expression could not be detected at 0 h APF. The data presented in Fig. 5C, D suggest the opposite as there clearly is some expression. Moreover, the data shown in Fig. 5D looks significantly brighter than the Orco dsRNA control and appears to localize to some type of cytoplasmic granule. So the expression of Mical does not look normal.
Sufficient data are not presented to conclude any specificity in mRNA export pathways. Data is presented for one export protein (UAP56) and one putative target (Mical). To adequately assess this, the authors would need to do RNA-seq in UAP56 mutants.
In summary, better quantitative assays should be used in Figures 4 and 5 in order to conclude the expression levels of either mRNA or protein. In its current form, this study demonstrates the novel finding that UAP56 regulates dendrite and presynaptic pruning, potentially via regulation of the actin cytoskeleton. However, these data do not convincingly demonstrate that UAP56 controls these processes by regulating of Mical expression and defintately not by controlling export from the nucleus.
While there are clearly dendrites shown in Fig. 1C', the cell body is not readily identifiable. This makes it difficult to assess attachment and suggests that the neuron may be dying. This should be replaced with an image that shows the soma.
The level of knockdown in the UAS56 RNAi and P element insertion lines should be determined. It would be useful to mention the nature of the RNAi lines (long/short hairpin). Some must be long since Dcr has been co-expressed. Another issue raised by this is the potential for off-target effects. shRNAi lines would be preferable because these effects are minimized.
Minor comments:
The authors should explain what EB1:GFP is marking when introduced in the text.
The da neuron images througout the figures could be a bit larger.
Strengths:
The methodology used to assess dendrite and presynaptic prunings are strong and the phenotypic analysis is conclusive.
Weakness:
The evidence demonstrating that UAP56 regulates the expression of Mical is unconvincing. Similarly, no data is presented to show that there is any specificity in mRNA export pathways. Thus, these major conclusions are not adequately supported by the data.
Advance:
The findings that UAP56 regulate dendrite and synaptic pruning are novel. As is its specific regulation of the actin cytoskeleton. These findings are restricted to a phenotypic analysis and do not show that it is not simply due to the disruption of general mRNA export.
Audience:
In its current form the manuscript whould be of interest to an audience who specializes in the study of RNA binding proteins in the control of neurodevelopment. This would include scientists who work in invertebrate and vertebrate model systems.
My expertise:
My lab uses Drosophila to study the role of RNA binding proteins in neurodevelopment and neurodegeneration. Currently, we use flies as a model to better understand the molecular pathogenesis of neurodevelopmenal disorders such as FXS and ASD.
The pipeline runs from women as the original “computers” in the 1940s, through the masculinization of computing that pushed women into typing pools and administrative support, through the automation of those roles, to AI assistants today automating what remains: scheduling, reminding, organizing, emotional management.
There is a line from computers in the original sense, to typing pools, admin support, to automation to AI.
he connection between AI assistants and human secretaries isn’t metaphorical—it’s genealogical. In a 2020 academic paper titled “Alexa, Tell Me About Your Mother,” researchers Jessa Lingel and Kate Crawford trace the lineage explicitly. Alexa has a mother. Her name was Secretary.
"Alex has a mother. Her name was Secretary"
“Obedient and obliging machines that pretend to be women are entering our homes, cars and offices,” warned UNESCO’s Director for Gender Equality, Saniye Gülser Corat, in the agency’s landmark 2019 report.
Unesco report, Saniye Gülser Corat (dir for gender equality).
This gap between feeling and explanation isn’t mysterious. Neuroscientist Antonio Damasio spent decades documenting what he calls “somatic markers”—physiological responses that guide decision-making before conscious reasoning engages. These signals aren’t primitive or irrational. They’re compressed expertise—the accumulated wisdom of lived experience, speaking faster than language.
Somatic markers, Antonio Damasio physiologically expressed compressed lived experience.
Die Internet-Browser Google Chrome, Microsoft Edge und Safari (Apple) dominieren das Internet. Sie basieren fast alle auf der gleichen Technik (Chromium)
Why do people who should be in-the-know make this mistake? This is one of the weirder variations—usually it's people thinking that Chrome is based on WebKit.
In this instance, they seem to be aware that Chrome isn't using WebKit, but do think that Safari is using "Chromium" (which is neither WebKit, nor Blink, i.e. the browser engine that Chrome is using—Chromium is an open source browser, not a browser engine…).
1. Méthodologie et hypothèses importantes
Doesn't bring to the right section
1. Methodology and significant assumptions
Doesn't bring to the right section
eLife Assessment
This useful paper describes a software tool, "DrosoMating", which allows automated, high-throughput quantification of 6 common metrics of courtship and mating behaviors in Drosophila melanogaster. The validity of the tool is quite convincingly demonstrated by comparing expert human assessments with those made by DrosoMating. The work, however, does not address how DrosoMating compares with or advances on other existing tools for exactly the same purpose, whether it can be used for studies of other Drosophila species, and/or whether finer aspects of courtship response timing - which depend on proximal female signals to the male - could be extracted with more detailed analyses. Some additional statistical analyses would also help further strengthen the authors' current conclusions.
Reviewer #1 (Public review):
Summary:
The study of Drosophila mating behaviors has offered a powerful entry point for understanding how complex innate behaviors are instantiated in the brain. The effectiveness of this behavioral model stems from how readily quantifiable many components of the courtship ritual are, facilitating the fine-scale correlations between the behaviors and the circuits that underpin their implementation. Detailed quantification, however, can be both time-consuming and error-prone, particularly when scored manually. Song et al. have sought to address this challenge by developing DrosoMating, software that facilitates the automated and high-throughput quantification of 6 common metrics of courtship and mating behaviors. Compared to a human observer, DrosoMating matches courtship scoring with high fidelity. Further, the authors demonstrate that the software effectively detects previously described variations in courtship resulting from genetic background or social conditioning. Finally, they validate its utility in assaying the consequences of neural manipulations by silencing Kenyon cells involved in memory formation in the context of courtship conditioning.
Strengths:
(1) The authors demonstrate that for three key courtship/mating metrics, DrosoMating performs virtually indistinguishably from a human observer, with differences consistently within 10 seconds and no statistically significant differences detected. This demonstrates the software's usefulness as a tool for reducing bias and scoring time for analyses involving these metrics.
(2) The authors validate the tool across multiple genetic backgrounds and experimental manipulations to confirm its ability to detect known influences on male mating behavior.
(3) The authors present a simple, modular chamber design that is integrated with DrosoMating and allows for high-throughput experimentation, capable of simultaneously analyzing up to 144 fly pairs across all chambers.
Weaknesses:
(1) DrosoMating appears to be an effective tool for the high-throughput quantification of key courtship and mating metrics, but a number of similar tools for automated analysis already exist. FlyTracker (CalTech), for instance, is a widely used software that offers a similar machine vision approach to quantifying a variety of courtship metrics. It would be valuable to understand how DrosoMating compares to such approaches and what specific advantages it might offer in terms of accuracy, ease of use, and sensitivity to experimental conditions.
(2) The courtship behaviors of Drosophila males represent a series of complex behaviors that unfold dynamically in response to female signals (Coen et al., 2014; Ning et al., 2022; Roemschied et al., 2023). While metrics like courtship latency, courtship index, and copulation duration are useful summary statistics, they compress the complexity of actions that occur throughout the mating ritual. The manuscript would be strengthened by a discussion of the potential for DrosoMating to capture more of the moment-to-moment behaviors that constitute courtship. Even without modifying the software, it would be useful to see how the data can be used in combination with machine learning classifiers like JAABA to better segment the behavioral composition of courtship and mating across genotypes and experimental manipulations. Such integration could substantially expand the utility of this tool for the broader Drosophila neuroscience community.
(3) While testing the software's capacity to function across strains is useful, it does not address the "universality" of this method. Cross-species studies of mating behavior diversity are becoming increasingly common, and it would be beneficial to know if this tool can maintain its accuracy in Drosophila species with a greater range of morphological and behavioral variation. Demonstrating the software's performance across species would strengthen claims about its broader applicability.
Reviewer #2 (Public review):
This paper introduces "DrosoMating," an integrated hardware and software solution for automating the analysis of male Drosophila courtship. The authors aim to provide a low-cost, accessible alternative to expensive ethological rigs by utilizing a custom acrylic chamber and smartphone-based recording. The system focuses on quantifying key temporal metrics-Courtship Index (CI), Copulation Latency (CL), and Mating Duration (MD)-and is applied to behavioral paradigms involving memory mutants (orb2, rut).
The development of open-source behavioral tools is a significant contribution to neuroethology, and the authors successfully demonstrate a system that simplifies the setup for large-scale screens. A major strength of the work is the specific focus on automating Copulation Latency and Mating Duration, metrics that are often labor-intensive to score manually.
However, there are several limitations in the current analysis and validation that affect the strength of the conclusions:
First, the statistical rigor requires substantial improvement. The analysis of multi-group experiments (e.g., comparing four distinct strains or factorial designs with genotype and training) currently relies on multiple independent Student's t-tests. This approach is statistically invalid for these experimental designs as it inflates the family-wise Type I error rate. To support the claims of strain-specific differences or learning deficits, the data must be analyzed using Analysis of Variance (ANOVA) to properly account for multiple comparisons and to explicitly test for interaction effects between genotype and training conditions.
Second, the biological validation using w1118 and y1 mutants entails a potential confound. The authors attribute the low Courtship Index in these strains to courtship-specific deficits. However, both strains are known to exhibit general locomotor sluggishness (due to visual or pigmentation/behavioral defects). Since "following" behavior is likely a component of the Courtship Index, a reduction in this metric could reflect a general motor deficit rather than a specific lack of reproductive motivation. Without controlling for general locomotion, the interpretation of these behavioral phenotypes remains ambiguous.
Third, the benchmarking of the system is currently limited to comparisons against manual scoring. Given that the field has largely adopted sophisticated open-source tracking tools (e.g., Ctrax, FlyTracker, JAABA), the utility of DrosoMating would be better contextualized by comparing its performance - in terms of accuracy, speed, or identity maintenance - against these existing automated standards, rather than solely against human observation.
Finally, the visual presentation of the data hinders the assessment of the system's temporal precision. While the system is designed to capture time-resolved metrics, the results are presented primarily as aggregate bar plots. The absence of behavioral ethograms or raster plots makes it difficult to verify the software's ability to accurately detect specific transitions, such as the exact onset of copulation.
Author response:
Thank you very much for the constructive feedback on our manuscript, "Simple Methods to Acutely Measure Multiple Timing Metrics among Sexual Repertoire of Male Drosophila," and for the opportunity to address the reviewers' comments. We appreciate the time and effort the reviewers have invested in evaluating our work, and we agree that their suggestions will significantly strengthen the manuscript.
We are currently working diligently to address all the concerns raised in the public reviews and recommendations. Below is an outline of the major revisions we plan to implement in the revised version:
(1) Statistical Rigor and Analysis
We acknowledge the statistical limitations pointed out by Reviewer #2. We will re-analyze the multi-group data in Figures 3 and 4 using One-way and Two-way ANOVA with appropriate post-hoc tests (e.g., Tukey's HSD), respectively, to properly account for multiple comparisons and interaction effects between genotype and training conditions.
(2) Comparison with Existing Tools
As suggested by both reviewers, we will provide a detailed comparison of DrosoMating with established automated tracking systems (e.g., FlyTracker, JAABA, Ctrax),and specific use cases where DrosoMating offers distinct advantages in terms of cost, accessibility, and ease of use for high-throughput screening.
(3) Control for Locomotor Activity
To address the potential confound of general locomotor deficits in w1118 and y1 mutants, we will calculate and present general locomotion metrics (e.g., average velocity, total distance traveled) from our tracking data to dissociate motor defects from specific courtship deficits.
(4) Software Capabilities and Cross-Species Applicability
We will clarify how DrosoMating handles fly identification during mating (including occlusion management). We will also discuss or test the software's applicability across different *Drosophila* species, as requested.
(5) Minor Corrections
We will address all textual errors, standardize terminology (e.g., "Mating Duration" vs. "Copulation Duration"), improve figure legibility, and provide complete statistical details for all figures.
We believe these revisions will substantially improve the rigor, clarity, and utility of our manuscript. We aim to resubmit the revised version within the standard timeframe and will ensure the preprint is updated accordingly.
eLife Assessment
This valuable study provides convincing evidence that MgdE, a conserved mycobacterial nucleomodulin, downregulates inflammatory gene transcription by interacting with the histone methyltransferase COMPASS complex and altering histone H3 lysine methylation. This work will interest microbiologists as well as cell and cancer biologists.
Reviewer #1 (Public review):
Summary:
This fundamental study identifies a new mechanism that involves a mycobacterial nucleomodulin manipulation of the host histone methyltransferase COMPASS complex to promote infection. Although other intracellular pathogens are known to manipulate histone methylation, this is the first report demonstrating specific targeting the COMPASS complex by a pathogen. The rigorous experimental design using of state-of-the art bioinformatic analysis, protein modeling, molecular and cellular interaction and functional approaches, culminating with in vivo infection modeling provide convincing, unequivocal evidence that supports the authors claims. This work will be of particular interest to cellular microbiologist working on microbial virulence mechanisms and effectors, specifically nucleomodulins, and cell/cancer biologists that examine COMPASS dysfunction in cancer biology.
Strengths:
(1) The strengths of this study include the rigorous and comprehensive experimental design that involved numerous state-of-the-art approaches to identify potential nucleomodulins, define molecular nucleomodulin-host interactions, cellular nucleomodulin localization, intracellular survival, and inflammatory gene transcriptional responses, and confirmation of the inflammatory and infection phenotype in a small animal model.
(2) The use of bioinformatic, cellular and in vivo modeling that are consistent and support the overall conclusions is a strengthen of the study. In addition, the rigorous experimental design and data analysis including the supplemental data provided, further strengthens the evidence supporting the conclusions.
Weaknesses:
(1) This work could be stronger if the MgdE-COMPASS subunit interactions that negatively impact COMPASS complex function were more well defined. Since the COMPASS complex consists of many enzymes, examining functional impact on each of the components would be interesting.
(2) Examining the impact of WDR5 inhibitors on histone methylation, gene transcription and mycobacterial infection could provide additional rigor and provide useful information related to mechanisms and specific role of WDR5 inhibition on mycobacteria infection.
(3) The interaction between MgdE and COMPASS complex subunit ASH2L is relatively undefined and studies to understand the relationship between WDR5 and ASH2L in COMPASS complex function during infection could provide interesting molecular details that are undefined in this study.
(4) The AlphaFold prediction results for all the nuclear proteins examined could be useful. Since the interaction predictions with COMPASS subunits range from 0.77 for WDR5 and 0.47 for ASH2L, it is not clear how the focus on COMPASS complex over other nuclear proteins was determined.
Comments on revisions:
The authors have addressed the weaknesses that were identified by this reviewer by providing rational explanation and specific references that support the findings and conclusions.
Reviewer #2 (Public review):
Summary:
The manuscript by Chen et al addresses an important aspect of pathogenesis for mycobacterial pathogens, seeking to understand how bacterial effector proteins disrupt the host immune response. To address this question the authors sought to identify bacterial effectors from M. tuberculosis (Mtb) that localize to the host nucleus and disrupt host gene expression as a means of impairing host immune function. Their revised manuscript has strengthened their observations by performing additional experiments with BCG strains expressing tagged MgdE.
Strengths:
The researchers conducted a rigorous bioinformatic analysis to identify secreted effectors containing mammalian nuclear localization signal (NLS) sequences, which formed the basis of quantitative microscopy analysis to identify bacterial proteins that had nuclear targeting within human cells. The study used two complementary methods to detect protein-protein interaction: yeast two-hybrid assays and reciprocal immunoprecipitation (IP). The combined use of these techniques provides strong evidence of interactions between MgdE and SET1 components and suggests the interactions are in fact direct. The authors also carried out rigorous analysis of changes in gene expression in macrophages infected with MgdE mutant BCG. They found strong and consistent effects on key cytokines such as IL6 and CSF1/2, suggesting that nuclear-localized MgdE does in fact alter gene expression during infection of macrophages. The revised manuscript contains additional biochemical analyses of BCG strains expressing tagged MgdE that further supports their microscopy findings.
Weaknesses:
There are some drawbacks in this study that limit the application of the findings to M. tuberculosis (Mtb) pathogenesis. Much of the study relies on transfected/ overexpressed proteins in non-immune cells (HEK293T) or in yeast using 2-hybrid approaches, and pathogenesis is studied using the BCG vaccine strain rather than virulent Mtb. In addition, the magnitude of some of the changes they observe are quite small. However, overall the key findings of the paper - that MgdE interacts with COMPASS and alters gene expression are well-supported.
Comments on revisions:
The authors have performed additional experiments that have addressed several important concerns from the original manuscript and they now include an analysis of BCG strains expressing FLAG-tagged MgdE that supports their model. However here are still a few areas where the data are difficult to interpret or do not support their claims.
Reviewer #3 (Public review):
In this study, Chen L et al. systematically analyzed the mycobacterial nucleomodulins and identified MgdE as a key nucleomodulin in pathogenesis. They found that MgdE enters into host cell nucleus through two nuclear localization signals, KRIR108-111 and RLRRPR300-305, and then interacts with COMPASS complex subunits ASH2L and WDR5 to suppress H3K4 methylation-mediated transcription of pro-inflammatory cytokines, thereby promoting mycobacterial survival.
Comments on revisions:
The authors have adequately addressed previous concerns through additional experimentation. The revised data robustly support the main conclusions, demonstrating that MgdE engages the host COMPASS complex to suppress H3K4 methylation, thereby repressing pro-inflammatory gene expression and promoting mycobacterial survival. This work represents a significant conceptual advance.
Author response:
The following is the authors’ response to the original reviews.
Public Reviews:
Reviewer #1 (Public review):
Summary:
This fundamental study identifies a new mechanism that involves a mycobacterial nucleomodulin manipulation of the host histone methyltransferase COMPASS complex to promote infection. Although other intracellular pathogens are known to manipulate histone methylation, this is the first report demonstrating the specific targeting of the COMPASS complex by a pathogen. The rigorous experimental design using state-of-the art bioinformatic analysis, protein modeling, molecular and cellular interaction, and functional approaches, culminating with in vivo infection modeling, provides convincing, unequivocal evidence that supports the authors' claims. This work will be of particular interest to cellular microbiologists working on microbial virulence mechanisms and effectors, specifically nucleomodulins, and cell/cancer biologists that examine COMPASS dysfunction in cancer biology.
Strengths:
(1) The strengths of this study include the rigorous and comprehensive experimental design that involved numerous state-of-the-art approaches to identify potential nucleomodulins, define molecular nucleomodulin-host interactions, cellular nucleomodulin localization, intracellular survival, and inflammatory gene transcriptional responses, and confirmation of the inflammatory and infection phenotype in a small animal model.
(2) The use of bioinformatic, cellular, and in vivo modeling that are consistent and support the overall conclusions is a strength of the study. In addition, the rigorous experimental design and data analysis, including the supplemental data provided, further strengthen the evidence supporting the conclusions.
Weaknesses:
(1) This work could be stronger if the MgdE-COMPASS subunit interactions that negatively impact COMPASS complex function were better defined. Since the COMPASS complex consists of many enzymes, examining the functional impact on each of the components would be interesting.
We thank the reviewer for this insightful comment. A biochemistry assays could be helpful to interpret the functional impact on each of the components by MgdE interaction. However, the purification of the COMPASS complex could be a hard task itself due to the complexity of the full COMPASS complex along with its dynamic structural properties and limited solubility.
(2) Examining the impact of WDR5 inhibitors on histone methylation, gene transcription, and mycobacterial infection could provide additional rigor and provide useful information related to the mechanisms and specific role of WDR5 inhibition on mycobacterial infection.
We thank the reviewer for the comment. A previous study showed that WIN-site inhibitors, such as compound C6, can displace WDR5 from chromatin, leading to a reduction in global H3K4me3 levels and suppression of immune-related gene expression (Hung et al., Nucleic Acids Res, 2018; Bryan et al., Nucleic Acids Res, 2020). These results closely mirror the functional effects we observed for MgdE, suggesting that MgdE may act as a functional mimic of WDR5 inhibition. This supports our proposed model in which MgdE disrupts COMPASS activity by targeting WDR5, thereby dampening host pro-inflammatory responses.
(3) The interaction between MgdE and COMPASS complex subunit ASH2L is relatively undefined, and studies to understand the relationship between WDR5 and ASH2L in COMPASS complex function during infection could provide interesting molecular details that are undefined in this study.
We thank the reviewer for the comment. In this study, we constructed single and multiple point mutants of MgdE at residues S<sup>80</sup>, D<sup>244</sup>, and H<sup>247</sup> to identify key amino acids involved in its interaction with ASH2L (Figure 5A and B; New Figure S4C). However, these mutations did not interrupt the interaction with MgdE, suggesting that more residues are involved in the interaction.
ASH2L and WDR5 function cooperatively within the WRAD module to stabilize the SET domain and promote H3K4 methyltransferase activity with physiological conditions (Couture and Skiniotis, Epigenetics, 2013; Qu et al., Cell, 2018; Rahman et al., Proc Natl Acad Sci U S A, 2022). ASH2L interacts with RbBP5 via its SPRY domain, whereas WDR5 bridges MLL1 and RbBP5 through the WIN and WBM motifs (Chen et al., Cell Res, 2012; Park et al., Nat Commun, 2019). The interaction status between ASH2L and WDR5 during mycobacterial infection could not be determined in our current study.
(4) The AlphaFold prediction results for all the nuclear proteins examined could be useful. Since the interaction predictions with COMPASS subunits range from 0.77 for WDR5 and 0.47 for ASH2L, it is not clear how the focus on COMPASS complex over other nuclear proteins was determined.
We thank the reviewer for the comment. We employed AlphaFold to predict the interactions between MgdE and the major nuclear proteins. This screen identified several subunits of the SET1/COMPASS complex as high-confidence candidates for interaction with MgdE (Figure S4A). This result is consistent with a proteomic study by Penn et al. which reported potential interactions between MgdE and components of the human SET1/COMPASS complex based on affinity purification-mass spectrometry analysis (Penn et al., Mol Cell, 2018).
Reviewer #2 (Public review):
Summary:
The manuscript by Chen et al addresses an important aspect of pathogenesis for mycobacterial pathogens, seeking to understand how bacterial effector proteins disrupt the host immune response. To address this question, the authors sought to identify bacterial effectors from M. tuberculosis (Mtb) that localize to the host nucleus and disrupt host gene expression as a means of impairing host immune function.
Strengths:
The researchers conducted a rigorous bioinformatic analysis to identify secreted effectors containing mammalian nuclear localization signal (NLS) sequences, which formed the basis of quantitative microscopy analysis to identify bacterial proteins that had nuclear targeting within human cells. The study used two complementary methods to detect protein-protein interaction: yeast two-hybrid assays and reciprocal immunoprecipitation (IP). The combined use of these techniques provides strong evidence of interactions between MgdE and SET1 components and suggests that the interactions are, in fact, direct. The authors also carried out a rigorous analysis of changes in gene expression in macrophages infected with the mgdE mutant BCG. They found strong and consistent effects on key cytokines such as IL6 and CSF1/2, suggesting that nuclear-localized MgdE does, in fact, alter gene expression during infection of macrophages.
Weaknesses:
There are some drawbacks in this study that limit the application of the findings to M. tuberculosis (Mtb) pathogenesis. The first concern is that much of the study relies on ectopic overexpression of proteins either in transfected non-immune cells (HEK293T) or in yeast, using 2-hybrid approaches. Some of their data in 293T cells is hard to interpret, and it is unclear if the protein-protein interactions they identify occur during natural infection with mycobacteria. The second major concern is that pathogenesis is studied using the BCG vaccine strain rather than virulent Mtb. However, overall, the key findings of the paper - that MgdE interacts with SET1 and alters gene expression are well-supported.
We thank the reviewer for the comment. We agree that the ectopic overexpression could not completely reflect a natural status, although these approaches were adopted in many similar experiments (Drerup et al., Molecular plant, 2013; Chen et al., Cell host & microbe, 2018; Ge et al., Autophagy, 2021). Further, the MgdE localization experiment using Mtb infected macrophages will be performed to increase the evidence in the natural infection.
We agree with the reviewer that BCG strain could not fully recapitulate the pathogenicity or immunological complexity of M. tuberculosis infection. We employed BCG as a biosafe surrogate model since it was acceptable in many related studies (Wang et al., Nat Immunol, 2025; Wang et al., Nat Commun, 2017; Péan et al., Nat Commun, 2017; Li et al., J Biol Chem, 2020).
Reviewer #3 (Public review):
In this study, Chen L et al. systematically analyzed the mycobacterial nucleomodulins and identified MgdE as a key nucleomodulin in pathogenesis. They found that MgdE enters into host cell nucleus through two nuclear localization signals, KRIR<sup>108-111</sup> and RLRRPR<sup>300-305</sup>, and then interacts with COMPASS complex subunits ASH2L and WDR5 to suppress H3K4 methylation-mediated transcription of pro-inflammatory cytokines, thereby promoting mycobacterial survival. This study is potentially interesting, but there are several critical issues that need to be addressed to support the conclusions of the manuscript.
(1) Figure 2: The study identified MgdE as a nucleomodulin in mycobacteria and demonstrated its nuclear translocation via dual NLS motifs. The authors examined MgdE nuclear translocation through ectopic expression in HEK293T cells, which may not reflect physiological conditions. Nuclear-cytoplasmic fractionation experiments under mycobacterial infection should be performed to determine MgdE localization.
We thank the reviewer for this insightful comment. In the revised manuscript, we addressed this concern by performing nuclear-cytoplasmic fractionation experiments using M. bovis BCG-infected macrophages to assess the subcellular localization of MgdE (New Figure 2F) (Lines 146–155). Nuclear-cytoplasmic fractionation experiments showed that WT MgdE and the NLS single mutants (MgdE<sup>ΔNLS1</sup> and MgdE<sup>ΔNLS2</sup>) could be detected both in the cytoplasm and in the nucleus, while the double mutant MgdE<sup>ΔNLS1-2</sup> was detectable only in the cytoplasm. These findings strongly indicate that MgdE is capable of translocating into the host cell nucleus during BCG infection, and that this nuclear localization relies on the dual NLS motifs.
(2) Figure 2F: The authors detected MgdE-EGFP using an anti-GFP antibody, but EGFP as a control was not detected in its lane. The authors should address this technical issue.
We thank the reviewer for this question. In the revised manuscript, we have included the uncropped immunoblot images, which clearly show the EGFP band in the corresponding lane. These have been provided in the New Figure 2E.
(3) Figure 3C-3H: The data showing that the expression of all detected genes in 24 h is comparable to that in 4 h (but not 0 h) during WT BCG infection is beyond comprehension. The issue is also present in Figure 7C, Figure 7D, and Figure S7. Moreover, since Il6, Il1β (pro-inflammatory), and Il10 (anti-inflammatory) were all upregulated upon MgdE deletion, how do the authors explain the phenomenon that MgdE deletion simultaneously enhanced these gene expressions?
We thank the reviewer for the comment. A relative quantification method was used in our qPCR experiments to normalize the WT expression levels in Figure 3C–3H, Figure 7C, 7D, and New Figure S6.
The concurrent induction of both types of cytokines likely represents a dynamic host strategy to fine-tune immune responses during infection. This interpretation is supported by previous studies (Podleśny-Drabiniok et al., Cell Rep, 2025; Cicchese et al., Immunological Reviews, 2018).
(4) Figure 5: The authors confirmed the interactions between MgdE and WDR5/ASH2L. How does the interaction between MgdE and WDR5 inhibit COMPASS-dependent methyltransferase activity? Additionally, the precise MgdE-ASH2L binding interface and its functional impact on COMPASS assembly or activity require clarification.
We thank the reviewer for this insightful comment. We cautiously speculate that the MgdE interaction inhibits COMPASS-dependent methyltransferase activity by interfering with the integrity and stability of the COMPASS complex. Accordingly, we have incorporated the following discussion into the revised manuscript (Lines 303-315):
“The COMPASS complex facilitates H3K4 methylation through a conserved assembly mechanism involving multiple core subunits. WDR5, a central scaffolding component, interacts with RbBP5 and ASH2L to promote complex assembly and enzymatic activity (Qu et al., 2018; Wysocka et al., 2005). It also recognizes the WIN motif of methyltransferases such as MLL1, thereby anchoring them to the complex and stabilizing the ASH2L-RbBP5 dimer (Hsu et al., Cell, 2018). ASH2L further contributes to COMPASS activation by interacting with both RbBP5 and DPY30 and by stabilizing the SET domain, which is essential for efficient substrate recognition and catalysis (Qu et al., Cell, 2018; Park et al., Nat Commun, 2019). Our work shows that MgdE binds both WDR5 and ASH2L and inhibits the methyltransferase activity of the COMPASS complex. Site-directed mutagenesis revealed that residues D<sup>224</sup> and H<sup>247</sup> of MgdE are critical for WDR5 binding, as the double mutant MgdE-D<sup>224</sup>A/H<sup>247</sup>A fails to interact with WDR5 and shows diminished suppression of H3K4me3 levels (Figure 5D).”
Regarding the precise MgdE-ASH2L binding interface, we attempted to identify the key interaction site by introducing point mutations into ASH2L. However, these mutations did not disrupt the interaction (Figure 5A and B; New Figure S4C), suggesting that more residues are involved in the interaction.
(5) Figure 6: The authors proposed that the MgdE-regulated COMPASS complex-H3K4me3 axis suppresses pro-inflammatory responses, but the presented data do not sufficiently support this claim. H3K4me3 inhibitor should be employed to verify cytokine production during infection.
We thank the reviewer for the comment. We have now revised the description in lines 220-221 and lines 867-868 "MgdE suppresses host inflammatory responses probably by inhibition of COMPASS complex-mediated H3K4 methylation."
(6) There appears to be a discrepancy between the results shown in Figure S7 and its accompanying legend. The data related to inflammatory responses seem to be missing, and the data on bacterial colonization are confusing (bacterial DNA expression or CFU assay?).
We thank the reviewer for the comment. New Figure S6 specifically addresses the effect of MgdE on bacterial colonization in the spleens of infected mice, which was assessed by quantitative PCR rather than by CFU assay.
We have now revised the legend of New Figure S6 as below (Lines 986-991):
“MgdE facilitates bacterial colonization in the spleens of infected mice. Bacterial colonization was assessed in splenic homogenates from infected mice (as described in Figure 7A) by quantifying bacterial DNA using quantitative PCR at 2, 14, 21, 28, and 56 days post-infection.”
(7) Line 112-116: Please provide the original experimental data demonstrating nuclear localization of the 56 proteins harboring putative NLS motifs.
We thank the reviewer for the comment. We will provide this data in the New Table S3.
Recommendations for the authors:
Reviewer #2 (Recommendations for the authors):
There are a few concerns about specific experiments:
Major Comments:
(1) Questions about the exact constructs used in their microscopy studies and the behavior of their controls. GFP is used as a negative control, but in the data they provide, the GFP signal is actually nuclear-localized (for example, Figure 1c, Figure 2a). Later figures do show other constructs with clear cytoplasmic localization, such as the delta-NLS-MgdE-GFP in Figure 2D. This raises significant questions about how the microscopy images were analyzed and clouds the interpretation of these findings. It is also not clear if their microscopy studies use the mature MdgE, lacking the TAT signal peptide after signal peptidase cleavage (the form that would be delivered into the host cell) or if they are transfecting the pro-protein that still has the TAT signal peptide (a form that would present in the bacterial cell but that would not be found in the host cell). This should be clarified, and if their construct still has the TAT peptide, then key findings such as nuclear localization and NLS function should be confirmed with the mature protein lacking the signal peptide.
We thank the reviewer for this question. EGFP protein can passively diffuse through nuclear pores due to its smaller size (Petrovic et al., Science, 2022; Yaseen et al., Nat Commun, 2015; Bhat et al., Nucleic Acids Res, 2015). However, upon transfection with EGFP-tagged wild-type MdgE and its NLS deletion mutants (MdgE<sup>ΔNLS1</sup>, MdgE<sup>ΔNLS2</sup>, and MdgE<sup>ΔNLS1-2</sup>), we observed significantly stronger nuclear fluorescence in cells expressing wild-type MdgE compared to the EGFP protein. Notably, the MdgE<sup>ΔNLS1-2</sup>-EGFP mutant showed almost no detectable nuclear fluorescence (Figure 2C, D, and E). These results indicate that (i) MdgE-EGFP fusion protein could not enter the nucleus by passive diffusion, and (ii) EGFP does not interfere with the nuclear targeting ability of MdgE.
We did not construct a signal peptide-deleted MgdE for transfection assays. Instead, we performed an infection experiment using recombinant M. bovis BCG strains expressing Flag-tagged wild-type MgdE. The mature MgdE protein (signal peptide cleaved) can be detected in the nucleus fractionation (New Figure 2F), suggesting that the signal peptide does not play a role for the nuclear localization of MgdE.
(2) The localization of MdgE is not shown during actual infection. The study would be greatly strengthened by an analysis of the BCG strain expressing their MdgE-FLAG construct.
We thank the reviewer for the comment. In the revised manuscript, we constructed M. bovis BCG strains expressing FLAG-tagged wild-type MdgE as well as NLS deletion mutants (MdgE<sup>ΔNLS1</sup>, MdgE<sup>ΔNLS2</sup>, and MdgE<sup>ΔNLS1-2</sup>). These strains were used to infect THP-1 cells, and nuclear-cytoplasmic fractionation was performed 24 hours post-infection.
Nuclear-cytoplasmic fractionation experiments showed that WT MgdE and the NLS single mutants could be detected both in the cytoplasm and in the nucleus by immunoblotting, while the double mutant MgdE<sup>ΔNLS1-2</sup> was detectable only in the cytoplasm (New Figure 2F) (Lines 146–155). These findings indicate that MdgE is capable of entering the host cell nucleus during BCG infection, and that this nuclear localization depends on the presence of both its N-terminal and C-terminal NLS motifs.
(3) Their pathogenesis studies suggesting a role for MdgE would be greatly strengthened by studying MdgE in virulent Mtb rather than the BCG vaccine strain. If this is not possible because of technical limitations (such as lack of a BSL3 facility), then at least a thorough discussion of studies that examined Rv1075c/MdgE in Mtb is important. This would include a discussion of the phenotype observed in a previously published study examining the Mtb Rv1075c mutant that showed a minimal phenotype in mice (PMID: 31001637) and would also include a discussion of whether Rv1075c was identified in any of the several in vivo Tn-Seq studies done on Mtb.
We thank the reviewer for this insightful comment. In the revised manuscript, we have incorporated a more thorough discussion of prior studies that examined Rv1075c/MgdE in Mtb, including the reported minimal phenotype of an Mtb MgdE mutant in mice (PMID: 31001637) (Lines 288–294).
In the latest TnSeq studies in M. tuberculosis, Rv1075c/MgdE was not classified as essential for in vivo survival or virulence (James et al., NPJ Vaccines, 2025; Zhang et al., Cell, 2013). However, this absence should not be interpreted as evidence of dispensability since these datasets also failed to identify some well characterized virulence factors including Rv2067c (Singh et al., Nat Commun, 2023), PtpA (Qiang et al., Nat Commun, 2023), and PtpB (Chai et al., Science, 2022) which were demonstrated to be required for the virulence of Mtb.
Minor Comments:
(1) Multiple figures with axes with multiple discontinuities used when either using log-scale or multiple graphs is more appropriate, including 3B, 7A.
We sincerely thank the reviewer for pointing this out. In the revised manuscript, we have updated Figure 3B and Figure 7A.
(2) Figure 1C - Analysis of only nuclear MFI can be very misleading because it is affected by the total expression of each construct. Ratios of nuclear to cytoplasmic MFI are a more rigorous analysis.
We thank the reviewer for this comment. We agree that analyzing the ratio of nuclear to cytoplasmic mean fluorescence intensity (MFI) provides a more rigorous quantification of nuclear localization, particularly when comparing constructs with different expression levels. However, the analysis presented in Figure 1C was intended as a preliminary qualitative screen to identify Tat/SPI-associated proteins with potential nuclear localization, rather than a detailed quantitative assessment.
(3) Figure 5C - Controls missing and unclear interpretation of their mutant phenotype. There is no mock or empty-vector control transfection, and their immunoblot shows a massive increase in total cellular H3K4me3 signal in the bulk population, although their prior transfection data show only a small fraction of cells are expressing MdgE. They also see a massive increase in methylation in cells transfected with the inactive mutant, but the reason for this is unclear. Together, these data raise questions about the specificity of the increasing methylation they observe. An empty vector control should be included, and the phenotype of the mutant explained.
We thank the reviewer for this comment. In the revised manuscript, we transfected HEK293T cells with an empty EGFP vector and performed a quantitative analysis of H3K4me3 levels. The results demonstrated that, at the same time point, cells expressing MdgE showed significantly lower levels of H3K4me3 compared to both the EGFP control and the catalytically inactive mutant MdgE (D<sup>244</sup>A/H<sup>247</sup>A) (New Figure 5D) (Lines 213–216). These findings support the conclusion that MdgE specifically suppresses H3K4me3 levels in cells.
(4) Figure S1A - The secretion assay is lacking a critical control of immunoblotting a cytoplasmic bacterial protein to demonstrate that autolysis is not releasing proteins into the culture filtrate non-specifically - a common problem with secretion assays in mycobacteria.
We thank the reviewer for this comment. To address the concerns, we examined FLAG-tagged MgdE and the secreted antigen Ag85B in the culture supernatants by monitoring the cytoplasmic protein GlpX. The absence of GlpX in the supernatant confirmed that there was no autolysis in the experiment. We could detect MgdE-Flag in the culture supernatant (New Figure S2A), indicating that MgdE is a secreted protein.
(5) The volcano plot of their data shows that the proteins with the smallest p-values have the smallest fold-changes. This is unusual for a transcriptomic dataset and should be explained.
We thank the reviewer for this comment. We are not sure whether the p-value is correlated with fold-change in the transcriptomic dataset. This is probably case by case.
Reviewer #3 (Recommendations for the authors):
There are several minor comments:
(1) Line 104-109: The number of proteins harboring NLS motifs and candidate proteins assigned to the four distinct pathways does not match the data presented in Table S2. Please recheck the details. Figure 1A and B, as well as Figure S1A and B, should also be corrected accordingly.
We thank the reviewer for the comment. We have carefully checked the details and the numbers were confirmed and updated.
(2) Please add the scale bar in all image figures, including Figure 1C, Figure 2D, Figure 5C, Figure 7B, and Figure S2.
We thank the reviewer for this suggestion. We have now added scale bars to all relevant image figures in the revised manuscript, including Figure 1C, New Figure 2C, Figure 5C, Figure 7B, and New Figure S2B.
(3) Please add the molecular marker in all immunoblotting figures, including Figure 2C, Figure 2F, Figure 4B, Figure 4C, Figure 5B, Figure 5D, and Figure S5.
We thank the reviewer for this suggestion. We have now added the molecular marker in all immunoblotting figures in the revised manuscript, including New Figure 2E–F, Figure 4B–C, Figure 5B and D, Figure S2A, New Figure S2E and New Figure S4C.
References
Bryan AF, Wang J, Howard GC, Guarnaccia AD, Woodley CM, Aho ER, Rellinger EJ, Matlock BK, Flaherty DK, Lorey SL, Chung DH, Fesik SW, Liu Q, Weissmiller AM, Tansey WP (2020) WDR5 is a conserved regulator of protein synthesis gene expression Nucleic Acids Res 48:2924-2941.
Chai Q, Yu S, Zhong Y, Lu Z, Qiu C, Yu Y, Zhang X, Zhang Y, Lei Z, Qiang L, Li BX, Pang Y, Qiu XB, Wang J, Liu CH (2022) A bacterial phospholipid phosphatase inhibits host pyroptosis by hijacking ubiquitin Science 378(6616):eabq0132.
Chen C, Nguyen BN, Mitchell G, Margolis SR, Ma D, Portnoy DA (2018) The listeriolysin O PEST-like sequence co-opts AP-2-mediated endocytosis to prevent plasma membrane damage during Listeria infection Cell host & microbe 23: 786-795.
Chen Y, Cao F, Wan B, Dou Y, Lei M (2012) Structure of the SPRY domain of human Ash2L and its interactions with RbBP5 and DPY30 Cell Res 22:598–602.
Cicchese JM, Evans S, Hult C, Joslyn LR, Wessler T, Millar JA, Marino S, Cilfone NA, Mattila JT, Linderman JJ, Kirschner DE (2018) Dynamic balance of pro‐ and anti‐inflammatory signals controls disease and limits pathology Immunological Reviews 285: 147–167.
Couture JF, Skiniotis G (2013) Assembling a COMPASS Epigenetics 8:349-54
Drerup MM, Schlücking K, Hashimoto K, Manishankar P, Steinhorst L, Kuchitsu K, Kudla J (2013) The calcineurin B-like calcium sensors CBL1 and CBL9 together with their interacting protein kinase CIPK26 regulate the Arabidopsis NADPH oxidase RBOHF Molecular plant 6: 559-569.
Ge P, Lei Z, Yu Y, Lu Z, Qiang L, Chai Q, Zhang Y, Zhao D, Li B, Pang Y, Liu C, Wang J (2021) M. tuberculosis PknG Manipulates Host Autophagy Flux to Promote Pathogen Intracellular Survival Autophagy 18: 576–94.
Hung KH, Woo YH, Lin IY, Liu CH, Wang LC, Chen HY, Chiang BL, Lin KI (2018) The KDM4A/KDM4C/NF-κB and WDR5 epigenetic cascade regulates the activation of B cells Nucleic Acids Res 46:5547–5560.
James KS, Jain N, Witzl K, Cicchetti N, Fortune SM, Ioerger TR, Martinot AJ, Carey AF (2025) TnSeq identifies genetic requirements of Mycobacterium tuberculosis for survival under vaccine-induced immunity NPJ Vaccines 10:103.
Li X, Chen L, Liao J, Hui J, Li W, He ZG (2020) A novel stress-inducible CmtR-ESX3-Zn²⁺ regulatory pathway essential for survival of Mycobacterium bovis under oxidative stress J Biol Chem 295:17083–17099.
Park SH, Ayoub A, Lee YT, Xu J, Kim H, Zheng W, Zhang B, Sha L, An S, Zhang Y, Cianfrocco MA, Su M, Dou Y, Cho US (2019) Cryo-EM structure of the human MLL1 core complex bound to the nucleosome Nat Commun 10:5540.
Penn BH, Netter Z, Johnson JR, Von Dollen J, Jang GM, Johnson T, Ohol YM, Maher C, Bell SL, Geiger K (2018) An Mtb-human protein-protein interaction map identifies a switch between host antiviral and antibacterial responses Mol Cell 71:637-648.e5.
Petrovic S, Samanta D, Perriches T, Bley CJ, Thierbach K, Brown B, Nie S, Mobbs GW, Stevens TA, Liu X, Tomaleri GP, Schaus L, Hoelz A (2022) Architecture of the linker-scaffold in the nuclear pore Science 376: eabm9798.
Podleśny-Drabiniok A, Romero-Molina C, Patel T, See WY, Liu Y, Marcora E, Goate AM (2025) Cytokine-induced reprogramming of human macrophages toward Alzheimer's disease-relevant molecular and cellular phenotypes in vitro Cell Rep 44:115909.
Qiang L, Zhang Y, Lei Z, Lu Z, Tan S, Ge P, Chai Q, Zhao M, Zhang X, Li B, Pang Y, Zhang L, Liu CH, Wang J (2023) A mycobacterial effector promotes ferroptosis-dependent pathogenicity and dissemination Nat Commun 14:1430.
Qu Q, Takahashi YH, Yang Y, Hu H, Zhang Y, Brunzelle JS, Couture JF, Shilatifard A, Skiniotis G (2018) Structure and Conformational Dynamics of a COMPASS Histone H3K4 Methyltransferase Complex Cell 174:1117-1126.e12.
Rahman S, Hoffmann NA, Worden EJ, Smith ML, Namitz KEW, Knutson BA, Cosgrove MS, Wolberger C (2022) Multistate structures of the MLL1-WRAD complex bound to H2B-ubiquitinated nucleosome Proc Natl Acad Sci U S A 119:e2205691119.
Sharma G, Upadhyay S, Srilalitha M, Nandicoori VK, Khosla S 2015 The interaction of mycobacterial protein Rv2966c with host chromatin is mediated through non-CpG methylation and histone H3/H4 binding Nucleic Acids Res 43:3922-37.
Singh PR, Dadireddy V, Udupa S, Kalladi SM, Shee S, Khosla S, Rajmani RS, Singh A, Ramakumar S, Nagaraja V (2023) The Mycobacterium tuberculosis methyltransferase Rv2067c manipulates host epigenetic programming to promote its own survival Nat Commun 14:8497.
Wang J, Ge P, Qiang L, Tian F, Zhao D, Chai Q, Zhu M, Zhou R, Meng G, Iwakura Y, Gao GF, Liu CH (2017) The mycobacterial phosphatase PtpA regulates the expression of host genes and promotes cell proliferation Nat Commun 8:244.
Wang J, Li BX, Ge PP, Li J, Wang Q, Gao GF, Qiu XB, Liu CH (2015) Mycobacterium tuberculosis suppresses innate immunity by coopting the host ubiquitin system Nat Immunol 16:237–245
Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD (2005) WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development Cell 121:859-72.
Yaseen I, Kaur P, Nandicoori VK, Khosla S (2015) Mycobacteria modulate host epigenetic machinery by Rv1988 methylation of a non-tail arginine of histone H3 Nat Commun 6:8922.
Zhang L, Kent JE, Whitaker M, Young DC, Herrmann D, Aleshin AE, Ko YH, Cingolani G, Saad JS, Moody DB, Marassi FM, Ehrt S, Niederweis M (2022) A periplasmic cinched protein is required for siderophore secretion and virulence of Mycobacterium tuberculosis Nat Commun 13:2255.
Zhang YJ, Reddy MC, Ioerger TR, Rothchild AC, Dartois V, Schuster BM, Trauner A, Wallis D, Galaviz S, Huttenhower C, Sacchettini JC, Behar SM, Rubin EJ (2013) Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing Cell 155:1296-308.
Figure 1: The coalescent process with four samples. The S (starting) state just signifies that the process begins in state (4,0,0,0). Edges are annotated with the coalescence rates that, in this model represents the transition rates of the model
Hvorfor stopper processen, når vi ender i staten [0, 0, 0, 1]?
Spanish court decision that algorithms used to decide about citizens by government entities, need to be transparant. Presented as landmark ruling with EU consequences, but I'm not sure if this isn't already true for public sector decision making under the AI Act. In NL the national algorithm register and tools like IAMA are aimed at this too, and have been in place for some time, even if not fully implemented yet.
eLife Assessment
This work describes a useful computational tool for automated morphometry of dynamic organelles from microscope images. However, the supporting evidence and novelty of the manuscript as presented are incomplete and could be improved. The work will be of interest to microscopists and bioimage analysts who are non-experts but wish to improve quantitative analysis of cellular structures.
Reviewer #1 (Public review):
Summary:
The authors develop a Python-based analysis framework for cellular organelle segmentation, feature extraction, and analysis for live-cell imaging videos. They demonstrate that their pipeline works for two organelles (mitochondria and lysosomes) and provide a step-by-step overview of the AutoMorphoTrack package.
Strengths:
The authors provide evidence that the package is functional and can provide publication-quality data analysis for mitochondrial and lysosomal segmentation and analysis.
Weaknesses:
(1) I was enthusiastic about the manuscript as a good end-to-end cell/organelle segmentation and quantification pipeline that is open-source, and is indeed useful to the field. However, I'm not certain AutoMorphoTrack fully fulfills this need. It appears to stitch together basic FIJI commands in a Python script that an experienced user can put together within a day. The paper reads as a documentation page, and the figures seem to be individual analysis outputs of a handful of images. Indeed, a recent question on the image.sc forum prompted similar types of analysis and outputs as a simple service to the community, and with seemingly better results and integrated organelle identity tracking (which is necessary in my opinion for live imaging). I believe this is a better fit in the methods section of a broader work. https://forum.image.sc/t/how-to-analysis-organelle-contact-in-fiji-with-time-series-data/116359/5.
(2) The authors do not discuss or compare to any other pipelines that can accomplish similar analyses, such as Imaris, CellProfiler, or integrate options for segmentation, etc., such as CellPose, StarDist.
(3) Although LLM-based chatbot integration seems to have been added for novelty, the authors do not demonstrate in the manuscript, nor provide instructions for making this easy-to-implement, given that it is directed towards users who do not code, presumably.
Reviewer #2 (Public review):
Summary:
AutoMorphoTrack provides an end-to-end workflow for organelle-scale analysis of multichannel live-cell fluorescence microscopy image stacks. The pipeline includes organelle detection/segmentation, extraction of morphological descriptors (e.g., area, eccentricity, "circularity," solidity, aspect ratio), tracking and motility summaries (implemented via nearest-neighbor matching using cKDTree), and pixel-level overlap/colocalization metrics between two channels. The manuscript emphasizes a specific application to live imaging in neurons, demonstrated on iPSC-derived dopaminergic neuronal cultures with mitochondria in channel 0 and lysosomes in channel 1, while asserting adaptability to other organelle pairs.
The tool is positioned for cell biologists, including users with limited programming experience, primarily through two implemented modes of use: (i) a step-by-step Jupyter notebook and (ii) a modular Python package for scripted or batch execution, alongside an additional "AI-assisted" mode that is described as enabling analyses through natural-language prompts.
The motivation and general workflow packaging are clear, and the notebook-plus-modules structure is a reasonable engineering choice. However, in its current form, the manuscript reads more like a convenient assembly of standard methods than a validated analytical tool. Key claims about robustness, accuracy, and scope are not supported by quantitative evidence, and the 'AI-assisted' framing is insufficiently defined and attributes to the tool capabilities that are provided by external LLM platforms rather than by AutoMorphoTrack itself. In addition, several figure, metric, and statistical issues-including physically invalid plots and inconsistent metric definitions-directly undermine trust in the quantitative outputs.
Strengths:
(1) Clear motivation: lowering the barrier for organelle-scale quantification for users who do not routinely write custom analysis code.
(2) Multiple entry points: an interactive notebook together with importable modules, emphasizing editable parameters rather than a fully opaque black box.
(3) End-to-end outputs: automated generation of standardized visualizations and tables that, if trustworthy, could help users obtain quantitative summaries without assembling multiple tools.
Weaknesses:
(1) "AI-assisted / natural-language" functionality is overstated.
The manuscript implies an integrated natural-language interface, but no such interface is implemented in the software. Instead, users are encouraged to use external chatbots to help generate or modify Python code or execute notebook steps. This distinction is not made clearly and risks misleading readers.
(2) No quantitative validation against trusted ground truth.
There is no systematic evaluation of segmentation accuracy, tracking fidelity, or interaction/overlap metrics against expert annotations or controlled synthetic data. Without such validation, accuracy, parameter sensitivity, and failure modes cannot be assessed.
(3) Limited benchmarking and positioning relative to existing tools.
The manuscript does not adequately compare AutoMorphoTrack to established platforms that already support segmentation, morphometrics, tracking, and colocalization (e.g., CellProfiler) or to mitochondria-focused toolboxes (e.g., MiNA, MitoGraph, Mitochondria Analyzer). This is particularly problematic given the manuscript's implicit novelty claims.
(4) Core algorithmic components are basic and likely sensitive to imaging conditions.
Heavy reliance on thresholding and morphological operations raises concerns about robustness across varying SNR, background heterogeneity, bleaching, and organelle density; these issues are not explored.
(5) Multiple figure, metric, and statistical issues undermine confidence.
The most concerning include:<br /> (i) "Circularity (4πA/P²)" values far greater than 1 (Figures 2 and 7, and supplementary figures), which is inconsistent with the stated definition and strongly suggests a metric/label mismatch or computational error.
(ii) A displacement distribution extending to negative values (Figure 3B). This is likely a plotting artifact (e.g., KDE boundary bias), but as shown, it is physically invalid and undermines confidence in the motility analysis.
(iii) Colocalization/overlap metrics that are inconsistently defined and named, with axis ranges and terminology that can mislead (e.g., Pearson r reported for binary masks without clarification).
(iv) Figure legends that do not match the displayed panels, and insufficient reporting of Ns, p-values, sampling units, and statistical assumptions.
Reviewer #3 (Public review):
Summary:
AutoMorphoTrack is a Python package for quantitatively evaluating organelle shape, movement, and colocalization in high-resolution live cell imaging experiments. It is designed to be a beginning-to-end workflow from segmentation through metric graphing, which is easy to implement. The paper shows example results from their images of mitochondria and lysosomes within cultured neurons, demonstrating how it can be used to understand organelle processing.
Strengths:
The text is well-written and easy to follow. I particularly appreciate tables 1 and 2, which clearly define the goals of each module, the tunable parameters, and the input and outputs. I can see how the provided metrics would be useful to other groups studying organelle dynamics. Additionally, because the code is open-source, it should be possible for experienced coders to use this as a backbone and then customize it for their own purposes.
Weaknesses:
Unfortunately, I was not able to install the package to test it myself using any standard install method. This is likely fixable by the authors, but until a functional distribution exists, the utility of this tool is highly limited. I would be happy to re-review this work after this is fixed.
The authors claim that there is "AI-Assisted Execution and Natural-Language Interface". However, this is never defended in any of the figures, and from quickly reviewing the .py files, there does not seem to be any built-in support or interface for this. Without significantly more instructions on how to connect this package to a (free) LLM, along with data to prove that this works reproducibly to produce equivalent results, this section should be removed.
Additionally, I have a few suggestions/questions:
(1) Red-green images are difficult for colorblind readers. I recommend that the authors change all raw microscopy images to a different color combination.
(2) For all of the velocity vs displacement graphs (Figure 3C and subpart G of every supplemental figure), there is a diagonal line clearly defining a minimum limit of detected movement. Is this a feature of the dataset (drift /shakiness /etc) or some sort of minimum movement threshold in the tracking algorithm? This should be discussed in the text.
(3) Integrated Correlation Summary (Figure 5) - Pearson is likely the wrong metric for most of these metric pairs because even interesting relationships may be non-linear. Please replace with Spearman correlation, which is less dependent on linearity.
Author response:
Reviewer #1
We thank the reviewer for their thoughtful and constructive assessment of AutoMorphoTrack and for recognizing its potential utility as an open-source end-to-end workflow for organelle analysis.
(1) Novelty and relationship to existing tools / FIJI workflows
We appreciate this concern and agree that many of the underlying image-processing operations (e.g., thresholding, morphological cleanup, region properties) are well-established. Our goal with AutoMorphoTrack is not to introduce new segmentation algorithms, but rather to provide a curated, reproducible, and extensible end-to-end workflow that integrates segmentation, morphology, tracking, motility, and colocalization into a single, transparent pipeline tailored for live-cell organelle imaging.
While an experienced user could assemble similar analyses ad hoc using FIJI or custom scripts, our contribution lies in:
Unifying these steps into a single workflow with consistent parameterization and outputs
Generating standardized, publication-ready visualizations and tables at each step,
Enabling batch and longitudinal analyses across cells and conditions, and
Lowering the barrier for users who do not routinely write custom analysis code.
We note that the documentation-style presentation of the manuscript is intentional, as it serves both as a methods paper and a practical reference for users implementing the workflow. We agree, however, that the manuscript currently overemphasizes step-by-step execution at the expense of positioning. In revision, we will more explicitly frame AutoMorphoTrack as a workflow integration and usability contribution, rather than a fundamentally new algorithmic advance.
We will also cite and discuss the image.sc example referenced by the reviewer, clarifying conceptual overlap and differences in scope.
(2) Comparison to existing pipelines (Imaris, CellProfiler, CellPose, StarDist)
We agree and thank the reviewer for highlighting this omission. In the revised manuscript, we will expand the related-work and positioning section to explicitly compare AutoMorphoTrack with established commercial (e.g., Imaris) and open-source (e.g., CellProfiler, MiNA, MitoGraph) platforms, as well as learning-based segmentation tools such as CellPose and StarDist.
Rather than claiming superiority, we will clarify trade-offs, emphasizing that AutoMorphoTrack prioritizes:
Transparency and parameter interpretability,
Lightweight dependencies suitable for small live-imaging datasets
Direct integration of morphology, tracking, and colocalization in a single workflow, and
Ease of modification for domain-specific use cases.
(3) AI / chatbot integration
We appreciate this critique and agree that the current description is insufficiently precise. AutoMorphoTrack does not implement a native natural-language interface. Instead, our intent was to convey that the workflow can be executed and modified with assistance from external large language models (LLMs) in a notebook-based environment.
In revision, we will revise this section to:
Clearly distinguish AutoMorphoTrack’s functionality from that of external LLM tools,
Remove any implication of a built-in AI interface, and
Provide concrete, reproducible examples of how non-coding users may interact with the pipeline using natural-language prompts mediated by external tools.
Reviewer #2
We thank the reviewer for their detailed and technically rigorous evaluation. We appreciate the recognition of the workflow’s motivation and structure, and we agree that several aspects of validation, positioning, and quantitative reporting must be strengthened.
(1) AI-assisted / natural-language functionality
We agree with this critique. AutoMorphoTrack does not provide a native natural-language execution layer, and the manuscript currently overstates this aspect. In revision, we will explicitly scope any reference to AI assistance as external, optional support for code generation and parameter editing, with clearly documented examples and stated limitations.
We agree that conflating external LLM capabilities with the software itself risks misleading readers, and we will correct this accordingly.
(2) Lack of quantitative validation
We fully agree that the current manuscript lacks formal quantitative validation. In the revised version, we will add a dedicated validation section including:
Segmentation accuracy compared to expert annotations using overlap metrics (e.g., Dice / IoU),
Tracking fidelity assessed using manually annotated tracks and/or synthetic ground truth,
Sensitivity analyses for key parameters (e.g., thresholding and linking distance), and
Explicit discussion of failure modes and quality-control indicators.
We acknowledge that without such validation, claims of robustness are not sufficiently supported.
(3) Benchmarking and positioning relative to existing tools
We agree and will substantially strengthen AutoMorphoTrack’s benchmarking and positioning relative to existing platforms. Rather than framing novelty algorithmically, we will clarify that the primary contribution is a reproducible, integrated workflow designed specifically for two-organelle live imaging in neurons, with transparent parameters and standardized outputs.
We note that our goal is not to exhaustively benchmark against all available tools, but rather to provide representative comparisons that clarify operating regimes, assumptions, and trade-offs. We will add a comparative table and/or qualitative comparison highlighting strengths, assumptions, and limitations relative to existing tools.
(4) Core algorithms and robustness
We agree that reliance on threshold-based segmentation introduces sensitivity to imaging conditions. In revision, we will:
Explicitly discuss the operating regime and assumptions under which AutoMorphoTrack performs reliably,
Clarify that the framework is modular and can accept alternative segmentation backends, and
Include guidance on when outputs should be treated with caution.
(5) Figure, metric, and statistical issues
We thank the reviewer for identifying several critical issues and agree that these undermine confidence. In revision, we will correct all figure, metric-definition, and reporting inconsistencies, including:
Resolving circularity values exceeding 1 by correcting computation and/or labeling errors,
Revising physically invalid displacement plots and clarifying kernel-density limitations,
Ensuring colocalization metrics are consistently defined, named, and interpreted, with explicit clarification of whether calculations are intensity- or mask-based,
Correcting figure legends to match displayed panels, and
Clearly reporting sample size, sampling units, and statistical assumptions, including handling of multiple comparisons where applicable.
(6) Value-added demonstration
We agree that the manuscript would benefit from a clearer demonstration of value-added use cases. In revision, we will include at least one realistic example showing how AutoMorphoTrack enables a complete, reproducible analysis workflow with reduced setup burden compared to manually assembling multiple tools.
(7) Editorial suggestions
We agree and will streamline the Results section to reduce procedural repetition and focus more on validation, limitations, and quality-control guidance.
Reviewer #3
We thank the reviewer for their positive assessment of clarity and organization, and for the constructive practical feedback.
Installation issues
We appreciate the detailed report of installation failures and acknowledge that the current packaging and distribution are inadequate. Prior to revision, we will:
Fix the package structure to support standard installation methods,
Ensure all required files (e.g., setup configuration, README) are correctly included,
Test installation on clean environments across platforms, and
Correct broken links to notebooks and documentation.
We agree that without a functional installation pathway, the utility of the tool is severely limited.
AI-assisted claims
We agree with the reviewer and echo our responses above. The AI-assisted description will be clarified and appropriately scoped in the revised manuscript.
Additional suggestions
Color accessibility: We will revise all figures to use colorblind-safe palettes.
Velocity–displacement diagonal: We will explicitly explain the origin of this relationship, including whether it reflects dataset properties, tracking assumptions, or minimum detectable motion.
Integrated correlation metric: We agree that Spearman correlation is more appropriate for many of these relationships and will replace Pearson correlations accordingly.
Supplementary movies: We agree that providing raw movies would improve interpretability and will add representative examples as supplementary material.
The 1970s was marked by the onset of the neo-liberal revolution and food commodity production and the accelerated and uneven transformation of small-holder farming in the Global South was part of that. Jason Moore’s work is significant in asking the question whether this was a conjuncture that signaled the tipping point of neoliberalism or if this was part of a more structural/epochal ecological crisis (Moore, 2010a, p. 233). Araghi (2010) has called the onset of the 70s transformatory crisis an indication of the exhaustion of the regime of “cheap ecology”.
1970 neoliberal revoliution and food commodity production
Two themes are important in this analysis. The first is the impact that increased concentration and centralization in the world food market has deepened the trend toward monopolistic and oligopolistic control. This has reinforced the economic power of agribusiness. The lengthening of supply chains under such monopoly control reduces autonomy and independence of especially small food producers increasing vulnerability to world market forces and environmental hazard.The second theme, that has created conditions for the recent food crisis is the penetration of finance capital in the food sector. That has generated increased price volatility and world market oscillations notably because “acceptable” profit margins for equity capital investors are significantly higher than other economic actors (The Economist, 2015). This combination of finance with farming (Fairbairn, 2014) is financial speculation (Clapp, 2014, Isakson, 2014). It is sometimes linked to the boom in biofuels which is estimated to have accounted for 30% of the increase in average grain prices
two important aspects in the definition of food crisis 1 concentration and centralisation of the world food market (increading power of agrobusinesses, monopolies) 2 penetration of finance capital into the food sector
It helps to explore why and how food as a commodity determines people’s uneven access to what is essential for life and the ways in which large social movements like Via Campesina emerged to mobilize small food producers to protest their increased (global) dispossession from the planet’s most important means of production: land
this related to petty commodity producers
the dominant food policy response seems to have been an attempt to forge one inclusive package of measures: there was no attempt to establish an understanding of the differential food needs of different social categories of poor, and the diverse socio-ecological conditions of African countries.
one inclusive package of measures
Economic growth seemed to represent the pre-requisite for long-term food security, and foreign direct investment was the instrument to get out of the crisis.
econ growth -> food security forign dircet investment
The FAO (1986) argued that the growing gap between high rates of demographic growth, and low increases of food production, required the rehabilitation of agriculture with an emphasis on the increase of commercial production. According to the FAO, the crises required “technical adjustments” to pave the way to agricultural investments and agricultural modernization. The productivity emphasis echoed the trade-oriented and market-liberalizing focus of the World Bank that privileged instruments to stabilize food availability
FAO's view, increase food productivity and make technical adjustments, agro modernisation and open to investments. Similar to market liberalisation
The concept of food self-sufficiency has been a central element of a wider strategy aimed at establishing forms of self-centered and endogenous development.
endogenous developments opposing extraversion
Many African states were at a crossroad in the early 1970s. They either accepted policy reform of market liberalization advised by the IFIs that de-regulated production and access to food, or they tried to further define a politics of self-sufficiency. This latter was to be anchored in the control and autonomy of the agri-food national system
market liberalisation vs setting a national system for food
Our argument is that in its present form the notion of food security has been emptied of any critical political content and mainstreamed. This is the result of a long historical process that we explore below.There was an important and diverse antecedent to the mainstream capture of food security and this can be traced to the post WW2 period. This was the view expressed first by Mao Tse Tung, and then by Latin American and subsequently African radical liberation activists, that premised the objective of development on the central need for food self-sufficiency (and productivity).
food security as a goal, development as cenreal for food self sufficiency and productivity
1.2 Integrate Razorpay Shield JS Integrate SHIELD JS and pass session_id in Create a Payment step. JavaScriptcopy<script src="https://checkout.razorpay.com/v1/shield.js"></script> // later, at the time of payment initialisation:const checkout_session_id = await RazorpayShield.getCheckoutSessionId() // pass it to your backend
Instead of this, we need to update the below shield config just for Singapore country - https://docs.google.com/document/d/1FoKeoOz1ODlVJEatojKZBUmt8zVY8-TZLlVWfTf7cas/edit?tab=t.0#heading=h.2qwnt1y19i36
We can discuss this in a call
the Hypothesis extension in Chrome
hehe
Marriage hearse
Kind of an oxymoron? Marriage can be related to the ideals of life, and hearse is related to death.
Chimney-sweepers cry Every blackning Church appalls
Here, there is the contrast of innocent chimney sweepers being placed in dangerous work positions by family and institutions, while those same people go to church and pray for their sins.
I wander thro' each charter'd street, Near where the charter'd Thames does flow.
The repetition of "charter'd" reminds me of the feelings of ownership, a sense of having control vs losing control. The speaker is setting the tone of individualism vs industrialization.
In the Sunday, March 8, 1964 edition of The Plain Dealer Heloise Cruse suggest putting several drops of pure glycerin onto one's dried out typewriter ribbon as a means of rejuvenating them. She recommends letting them soak for at least three days.
https://www.facebook.com/groups/TypewriterCollectors/posts/10163177170344678/

Swift also sold an “All Too Well” typewriter Christmas ornament at the craft store Michaels.
via https://www.404media.co/taylor-swift-typewriter-tortured-poets-department/
Advanced filtering
There is also another functionality that was introduced: "Convert filter to query string". I think it's also worth mentioning it under this paragraph.
We introduced a dedicated request/response pair event for converting filters to query strings:
octane_convert_filter_to_query:
---Direction: Sent from the external action to the client.
---Purpose:
Requests active filter with filter_id to be converted to query strings, associated within the widget identified by panel_id.
---Example payload:
js
{
event_name: 'octane_convert_filter_to_query',
workspace: this.params.workspace,
shared_space: this.params.shared_space,
data: {
panel_id: this.params.panel_id,
filter_id: filterId,
filter: filter,
entity_type: 'work_item'
}
}
and:
octane_filter_converted_to_query:
---Direction:
Sent from the client to the external action as a response to the octane_convert_filter_to_query.
---Purpose:
Provides the filter with filter_id converted to query strings, associated within the widget identified by panel_id.
---Example payload:
js
{
data: {
panel_id: panelId,
filter_id: filterId,
query: query
}
}
'octane_open_advanced_filter'
octane_show_filter_dialog -this is the final event name
data: {
we should also specify 'filter_id' which is unique and it's associated with the filter instance
You send an event requesting that OpenText Core Software Delivery Platform open the Advanced filter dialog box with specified scope and constraint
We support multiple advanced filters within the same widget, and the unique 'filter_id' is how we differentiate between them.
About 90% of the lithogenous sediment in the oceans is though to have come from river discharge, particularly from Asia. Most of this sediment, especially the larger particles, will be deposited and remain fairly close to the coastline, however, smaller clay particles may remain suspended in the water column for long periods of time and may be transported great distances from the source.
If most lithogenous sediment enters the ocean through rivers, how does this affect sediment distribution in areas far from major river systems?
High energy conditions, such as strong currents or waves, usually results in the deposition of only the larger particles as the finer ones will be carried away. Lower energy conditions will allow the smaller particles to settle out and form finer sediments.
This part stood out to me because it shows how sediment characteristics can be used as evidence of past environmental conditions. By looking at grain size, scientists can infer whether an area experienced high energy processes like strong currents or low energy conditions where finer sediments were able to settle.
Benefits of Functions# There are several advantages to creating and using functions in computer programs, such as: Reusing code instead of repeating code: When we find ourselves repeating a set of actions in our program, we end up writing (or copying) the same code multiple times. If we put that repeated code in a function, then we only have to write it once and then use that function in all the places we were repeating the code. Single, standardized definitions: Let’s say we made code that takes a name and tries to split it into a first name and last name, and we have that code copied in several places in our program. Then we realize that our code isn’t handling some last names correctly, like “O’Reilly” and “Del Toro.” If we fix this bug in one of the places the code is copied in our program it still will be broken elsewhere, so we have to find all the places and fix it there. If, on the other hand we had the code to split names in a function, and used that function everywhere else, then we only have to fix the bug inside that one function and our code everywhere is fixed. Code organization: Making functions also can help us organize our code. It lets us give a name to a block of code, and when we use it, those function names can help make the code more understandable. Making code as functions also helps in letting us put those pieces of code in other files or in code libraries, so the file we are working on is smaller and easier to manage.
This explanation clearly shows how functions improve efficiency and clarity in programming by reducing repetition, standardizing logic, and making code easier to read and manage.
Whether written into the margins of texts, integratedinto the print, or digitally superimposed, in what contexts have you encoun-tered annotations written by another person? In what ways have you sharedyour annotations with other readers?
In school, when using class copies of book or even paper copies of articles there would be little notes from someone's own thought process while reading or a teacher's teachings of the text.
Reference:
Preprints. (2024). Traditional ecological knowledge and environmental stewardship. https://doi.org/10.20944/preprints202406.1838.v1
“Environmental stewardship in Indigenous and local communities should not be understood only as a response to poverty or lack of alternatives.”(Preprints,2024)
The key idea of this article is that Environmental stewardship is an intention choice instead of that something people do because of no option. The author challenges the assumption that local people and communities protect the environment because of poverty. In this article Stewardship is presented as a value based practice that is shaped by culture, ethics and responsibility to future generations. By definition and reframing stewardship as purposeful, the author argues that local communities are active decision makers in environmental protection, not a passive participants.
Reference:
Preprints. (2024). Traditional ecological knowledge and environmental stewardship. https://doi.org/10.20944/preprints202406.1838.v1
“Practices rooted in Traditional Ecological Knowledge have contributed to long-term ecosystem resilience in various local contexts.”(Preprints,2024)
This idea can be connected to real world example like indigenous fire management practices . IFMP controlled burns that reduces wildlife risk and maintain ecosystem balance. These practices are based on long term observation of land and seasonal patterns instead of modern emergency responses. In many cases, when these practices ignored, then ecosystems became more vulnerable to environmental damage on large scale. This example shows that how TEK is applied in real world and it produces measurable environmental benefits. This example also supports the article that TEK is not just about theoretical knowledge but also effective practically in managing ecosystems and sustainability.
Reference:
Preprints. (2024). Traditional ecological knowledge and environmental stewardship. https://doi.org/10.20944/preprints202406.1838.v1
“Traditional Ecological Knowledge (TEK) represents a knowledge system grounded in lived experience and intergenerational learning.”(Preprints,2024)
This article connects strongly to our course because in class we will study sustainability and social-ecological systems, In class we already discussed about social ecological system where we learned that Environmental management is not just technical but it's also related to social and historical things. In class. we discussed about how ecosystems and humans influence each other, which is exactly what Traditional ecological knowledge is based on. TEK is defined as generations of observation, trial and error instead of short term scientific studies and policies. This article also connects to discussion about whose knowledge is considered valid in environmental decision making. The course gives the idea that ignoring indigenous people's knowledge leads to ineffective policies and wrong results. This article support the course argument that for effective environmental management multiple ways of knowing the TEK is necessary. This article also reinforces that sustainability is not just about protecting nature but also about respecting culture, people and historical experience.
Reference
Preprints. (2024). Traditional ecological knowledge and environmental stewardship. https://doi.org/10.20944/preprints202406.1838.v1
"Traditional Ecological Knowledge (TEK) refers to the cumulated wisdom, practices, and beliefs concerning their natural environment developed over centuries by indigenous and local communities"(Preprints,2024)
This definition explains that Traditional Ecological Knowledge (TEK) is not just a information about nature but it is a system that includes historical knowledge developed over past generations. The phrase "long term interaction" is important because it shows that TEK knowledge comes from repeated experience with same land over time. TEK focus on continuous observation and adaptation as compared to scientific research which focus on limited timeframes. This definition gave clarification that TEK is structured and intentional, not outdated or random knowledge. TEK challenges modern science that Environmental knowledge only comes from modern science.
In music, it is a simple direction that means that you should repeat the piece of music from the beginning.
"Play it again."
Da capo is an Italian word found in music that is translated as “from the head/beginning”.
"Da Capo" from Italy.
They also break down how prison labor is exploited as a loophole in the 13th amendment which was supposed to abolish slavery, but allowed slave-labor “as punishment for crime.”
wow we are leading history repeat it self by allowing this kinds of this happen. So why are there amendments if we don't respect them ?
In Chapter 2: “The Ongoing Struggle for Ethnic Studies,” Espinoza-Kulick examines how Ethnic Studies was birthed out of struggle, which helps to define this unique discipline.
that makes a lot of sense because I feel that ethnic studies are important to educate us about the struggles and differences of other people culture and backgrounds.
AI systems that empower users, enhancing human capabilities rather than replacing them
My opinion on this may not be popular, however we live in a society that will always choose profitability over employees and so the concept of AI not replacing human jobs is not realistic because it is cheaper for corporations and organizations to use AI than to pay employees.
These nuances are often missed by AI systems that are trained on culturally narrow datasets.
I think that this is something that people often miss when discussing AI. AI is generally seen as "more intelligent than us" and its advice is held to a higher standard and yet it gets things wrong all the time and is an unreliable source when it comes to cultural communication.
but on a psychological level we feel more compelled to do something if we tell someone else.
Most agreed!
Rewarding yourself for the completion of tasks or meeting goals is a good way to avoid procrastination. An example of this would be rewarding yourself with the time to watch a movie you would enjoy after you have finished the things you need to do, rather than using the movie to keep yourself from getting things done.
I agree with the reward system, it works.
Putting aside distractions is one of the primary functions of setting priorities.
I find it less distracting to study at a library vs. at home. At home there are so many distractions and escape routes that can lead to procrastination.
a boost of mental urgency.
Some people work well under pressure, and even perform at their best.
like low physical energy,
Sometimes a quick walk outside or take a few steps around the house, or even do a short 10-15 minute exercise; jump-n-jacks, arm circles, jump rope, sit ups, legs lifts. It gets the blood flowing and combats tiredness.
situation. It could be something as simple as a lack of sleep or improper diet.
Sometimes, before tackling an assignment or task, its better to properly fuel and rest the body and mind.
were a temporary setback (e.g., temporary loss of transportation, temporary loss of housing, an illness that lasted more than a week, etc.)? What could you do for a backup plan if something did happen?
Most of this is not planned, most people just manage as it comes. Its good to have a plan, however so many set-backs can happen, so we plan for every single kind there is? Sounds impossible
from a financial perspective you should do all you can to graduate as soon as you can.
With so much weighing on a college student , I think it not fair to have this thought of finances lingering over your head. Focusing on classes, studying and just surviving is enough.
Many college administrators that work directly with students are aware that a single mishap or a case of poor time management can set into motion a series of events that can seriously jeopardize a student’s success.
I find this a bit extreme, a SINGLE mishap, just one can send ones success crashing and burning ?
self-advocacy serves as a model:.d-undefined, .lh-undefined { background-color: rgba(0, 0, 0, 0.2) !important; }1RaeAnn Gutierrez a sustained attention to how systems of oppression intersect with each other, informed by the knowledge that comes from direct experience. It offers a guide for challenging power and working toward justice
I hadn't considered the power of self-advocacy. I'm glad to have learned about this as a potential pathway to change.
Darden’s rise into the leadership ranks at NASA was largely the result of her own knowledge, experience, and grit
A very inspiring story! I'm glad her experience and skill was rewarded.
intersectionality is essential for this whole book, let’s get a bit more specific. The term was coined by legal theorist Kimberlé Crenshaw in the late 1980s
Glad to become familiar with this term. I've heard it used before but hadn't been explained what it meant up until now.
feminisms, which clearly signals the range of—and, at times, the incompatibilities among—these various strains of feminist activism and political thought
A great point of clarity: feminism has a wide range of meanings, and being aware of this can help facilitate conversations on this topic.
The most outstanding result of this study is that a high UPF intake promotes a more pro-oxidant and pro-inflammatory status in patients with MetS without differences in their anthropometric parameters and biochemical profile.
This shows that inflammation may differ among individuals of different weights, heights, etc. Although obesity/excess fat can increase inflammation, so can several other factors. I wonder how they could limit more of those in future studies.
differences in the levels of endogenous antioxidants and prooxidants
This shows that antioxidant levels and pro-oxidant levels may be independent of one another and not strictly an inverse relationship, interesting
n = 92)
This is a small sample size and may not be representative of the population
Perhaps there were also unspoken rules like “no one talks to Mom before 9 am” or “no one disagrees with John”.
I made a connection during this section, especially the idea that power is usually unspoken but predictable. The examples about the types of rules reminded me of groups that I have seen in school and at work. It is when certain peoples preferences automatically get prioritized even when they dont have an offical supirior title like professor or manager. It connects to the idea that power doesnt come from a title, it can come from personality, populatirty, or confidence. It helped me to better understand how informational power show up everyday settings. It also made me realized how easiy it is to follow this pattern.
I also made a connection to cumulative stress and identity based harm. The sections on micro affirmations and micro interventions stood out because they go beyond just identifying harm and instead focusing on smaller, more practical actions people can take. That makes the topic feel more like action, instead of just theoretical , especially for people in this field.
True Christians are conservative. True Christians are progressive.
This is something I really relate to also. The fact that I am considered myself a Christian doesnt mean I cannot do whatever I desire. I’ve heard this millions of times, and sometimes it bothers me, but then I start thinking, “they don’t know how really I am, they just see the “outside” of me”. And even thought I do try to show my inner me on my outside its hard, because not everyone knows how to read people in a sense where anyone can understand me instantly.
How is it that she could only see junk where I saw my entire life story?
This sentence in itself is very important and impactful. People can judge others hobbies or interests and see their perspective on it has boring or uninteresting. For that person is it something more than just what meets the eyes. It’s something that they enjoy, find meaningful, interesting etc. This just introduces the difference between people. We all have different ideas, interests, etc.
I could relate inthat I felt growing up was certainly not all that it had been built up to be.
The author is disappointed in adulthood especially around the time of hardships.
A Hitchhiker’s Guide to the Galaxy orthe entire Harry Potter series? How could I possibly give up my National Forensics League RuleBook, The Art of War
This shows that fictional and fantastical books hold the same value to the speaker as non-fiction informative books.
to change the discourse. If we can control the discourse, we can contr
Who controls the narrative? How do we control it? Politics feels like a war of optics and the left seems to need a recalibration.
health and well-being. Thus, the poverty that exists in one part of the world is related to the affluence in another part. Similarly, the poverty that exists on one side of town is related to the affluence and
How do we say this? What language do we need to use for people to hear us?
understandings. Culture is what people mak
Period. Culture is what people make it. Period.
People have literally died for education, yet we keep hearing that certain families do not value it
People have literally died for it, that is how important it is. The hypocrisy in the highlighted statement is deafening.
eferred to as "ac
Pulling up the top two results from a quick "academic redshirt" search in the Auririan Library with minimal constraints show the value of academic redshirting and how it is primarily done by white upper-middle class families. ISSN: ISSN-1083-6470 https://www.jstor.org/stable/43773434
mplies, "If you come to school not reading you get treated as if you have no right to be in sc
Following up on what Fagana said, I find it interesting that there is such a dichotamy between "They should learn that at school" vs "They should learn that at home." That type of thinking is a lose-lose mentatlity, and allows both the school and parents to cope out of responsibility. Sex ed for example, I have had many example of parents saying "the school has no right to teach my child about sex ed", and then the parent procedes to NOT teach the child about sex ed....
These Children Don't Have Enough Exposure/Experiences
So provide it
students' parents were less likely to have transportation to travel across the city or that it was not particularly safe for Black people to be found in the school's neighborhood after d
Many schools offer additional "voluntary" support and call it tier 1 support. anything that is voluntary is automatically tier 2 because it is not given to the whole school. Saying you "offered it" is not good enough when some families literally cannot attend for a myriad of reasons. Tier 1 means the ENTIRE school gets access. Voluntary opportunities are opportunites for the privledged.
his sensory barrage can shift the very nature of curiosity, altering not just the depth but also the motivations behind our inquiries.
This section explains how misinformation and too much information can confuse people and affect real curiosity. It shows that not everything online is trustworthy, and people can get stuck in false ideas. This makes me realize that I need to be more careful about what sources I believe and use for school.
There’s a profound difference between merely knowing an answer and understanding its foundations and implications deeply. By offering instant gratification, the internet may inadvertently deter individuals from cultivating deeper, sustained interests and passions.
This part explains that getting instant answers can stop people from thinking deeply. Instead of struggling through a problem, people may just look it up and move on. I agree with this because sometimes I search answers without fully understanding them, especially when studying.
The digital age has, in many ways, transformed the very fabric of learning and inquiry.
This section talks about how the internet gives people unlimited access to knowledge from around the world. IT explains that learning is easier now because we don't have to only rely on books or teachers. I like this point because I use the internet all the time for schoolwork and researching topics I'm interested in.
Join me as we explore both the empowering and potentially stifling effects the internet has on our inherent quest for knowledge.
This introduction explains how the internet has changed the way people look for information and satisfy their curiosity. It shows that humans have always been curious, but now we rely on technology to explore questions faster. I think this is true because most of the time when I'm curious about something, I immediately search it on my phone.