AAV8-hSyn-DIO-hM3D(Gq)-mCherry
DOI: 10.1038/s41593-025-02078-y
Resource: RRID:Addgene_44361
Curator: @olekpark
SciCrunch record: RRID:Addgene_44361
AAV8-hSyn-DIO-hM3D(Gq)-mCherry
DOI: 10.1038/s41593-025-02078-y
Resource: RRID:Addgene_44361
Curator: @olekpark
SciCrunch record: RRID:Addgene_44361
125148
DOI: 10.1038/s41588-025-02368-y
Resource: None
Curator: @olekpark
SciCrunch record: RRID:Addgene_125148
99373
DOI: 10.1038/s41588-025-02368-y
Resource: RRID:Addgene_99373
Curator: @olekpark
SciCrunch record: RRID:Addgene_99373
62205
DOI: 10.1038/s41588-025-02368-y
Resource: RRID:Addgene_62205
Curator: @olekpark
SciCrunch record: RRID:Addgene_62205
RRID:SCR_024672
DOI: 10.1038/s41588-025-02368-y
Resource: MapMyCells (RRID:SCR_024672)
Curator: @scibot
SciCrunch record: RRID:SCR_024672
102930
DOI: 10.1038/s41556-025-01774-y
Resource: RRID:Addgene_102930
Curator: @olekpark
SciCrunch record: RRID:Addgene_102930
MSP1E3D1
DOI: 10.1038/s41467-025-65037-y
Resource: RRID:Addgene_20066
Curator: @olekpark
SciCrunch record: RRID:Addgene_20066
p1E3D1
DOI: 10.1038/s41467-025-65037-y
Resource: RRID:Addgene_20066
Curator: @olekpark
SciCrunch record: RRID:Addgene_20066
87306
DOI: 10.1038/s41467-025-64742-y
Resource: RRID:Addgene_87306
Curator: @olekpark
SciCrunch record: RRID:Addgene_87306
121675
DOI: 10.1038/s41467-025-64742-y
Resource: RRID:Addgene_121675
Curator: @olekpark
SciCrunch record: RRID:Addgene_121675
114471
DOI: 10.1038/s41467-025-64742-y
Resource: RRID:Addgene_114471
Curator: @olekpark
SciCrunch record: RRID:Addgene_114471
50459
DOI: 10.1038/s41467-025-64742-y
Resource: RRID:Addgene_50459
Curator: @olekpark
SciCrunch record: RRID:Addgene_50459
44361
DOI: 10.1038/s41467-025-64742-y
Resource: RRID:Addgene_44361
Curator: @olekpark
SciCrunch record: RRID:Addgene_44361
40755
DOI: 10.1038/s41420-025-02789-y
Resource: RRID:Addgene_40755
Curator: @olekpark
SciCrunch record: RRID:Addgene_40755
60505
DOI: 10.1038/s41419-025-07952-y
Resource: RRID:Addgene_60505
Curator: @olekpark
SciCrunch record: RRID:Addgene_60505
80900
DOI: 10.1038/s41419-025-07952-y
Resource: RRID:Addgene_80900
Curator: @olekpark
SciCrunch record: RRID:Addgene_80900
83480
DOI: 10.1038/s41419-025-07952-y
Resource: RRID:Addgene_83480
Curator: @olekpark
SciCrunch record: RRID:Addgene_83480
11813
DOI: 10.1038/s41419-025-07952-y
Resource: RRID:Addgene_11813
Curator: @olekpark
SciCrunch record: RRID:Addgene_11813
12259
DOI: 10.1007/s12672-025-03889-y
Resource: RRID:Addgene_12259
Curator: @olekpark
SciCrunch record: RRID:Addgene_12259
Addgene_52961
DOI: 10.1007/s12672-025-03889-y
Resource: RRID:Addgene_52961
Curator: @scibot
SciCrunch record: RRID:Addgene_52961
Addgene_12260
DOI: 10.1007/s12672-025-03889-y
Resource: RRID:Addgene_12260
Curator: @scibot
SciCrunch record: RRID:Addgene_12260
Addgene_90237
DOI: 10.1038/s41598-025-23075-y
Resource: RRID:Addgene_90237
Curator: @scibot
SciCrunch record: RRID:Addgene_90237
plasmid_215559
DOI: 10.1038/s41467-025-63986-y
Resource: None
Curator: @scibot
SciCrunch record: RRID:Addgene_215559
plasmid_45605
DOI: 10.1038/s41467-025-63986-y
Resource: RRID:Addgene_45605
Curator: @scibot
SciCrunch record: RRID:Addgene_45605
plasmid_117261
DOI: 10.1007/s11274-025-04598-y
Resource: RRID:Addgene_117261
Curator: @scibot
SciCrunch record: RRID:Addgene_117261
plasmid_117262
DOI: 10.1007/s11274-025-04598-y
Resource: None
Curator: @scibot
SciCrunch record: RRID:Addgene_117262
Hola muy buenas noches quisiera poder pedirles respetuosamente el acceder al documento y obtenerlo descargado
Devine SM, O’Geen AT, Larsen RE, Dahlke HE, Liu H, Jin Y, Dahlgren RA. 2019. Microclimateforage growth linkages in a Mediterranean catchment in California’s Central Coast.Ecohydrology. 12(8):e2156
JP
12,260
DOI: 10.1186/s13024-025-00889-y
Resource: RRID:Addgene_12260
Curator: @olekpark
SciCrunch record: RRID:Addgene_12260
Addgene_96
DOI: 10.1186/s13024-025-00889-y
Resource: None
Curator: @olekpark
SciCrunch record: RRID:Addgene_96808
Addgene_12
DOI: 10.1186/s13024-025-00889-y
Resource: None
Curator: @olekpark
SciCrunch record: RRID:Addgene_12259
212936
DOI: 10.1186/s12931-025-03354-y
Resource: RRID:Addgene_212936
Curator: @olekpark
SciCrunch record: RRID:Addgene_212936
1015
DOI: 10.1186/s12929-025-01178-y
Resource: RRID:Addgene_1015
Curator: @olekpark
SciCrunch record: RRID:Addgene_1015
12259
DOI: 10.1038/s42255-025-01358-y
Resource: RRID:Addgene_12259
Curator: @olekpark
SciCrunch record: RRID:Addgene_12259
12260
DOI: 10.1038/s42255-025-01358-y
Resource: RRID:Addgene_12260
Curator: @olekpark
SciCrunch record: RRID:Addgene_12260
83356
DOI: 10.1038/s42255-025-01358-y
Resource: RRID:Addgene_83356
Curator: @olekpark
SciCrunch record: RRID:Addgene_83356
50054
DOI: 10.1038/s42255-025-01358-y
Resource: None
Curator: @olekpark
SciCrunch record: RRID:Addgene_50054
98291
DOI: 10.1038/s42255-025-01358-y
Resource: RRID:Addgene_98291
Curator: @olekpark
SciCrunch record: RRID:Addgene_98291
52961
DOI: 10.1038/s42255-025-01358-y
Resource: RRID:Addgene_52961
Curator: @olekpark
SciCrunch record: RRID:Addgene_52961
79823
DOI: 10.1038/s41586-025-09520-y
Resource: RRID:Addgene_79823
Curator: @olekpark
SciCrunch record: RRID:Addgene_79823
66810
DOI: 10.1038/s41586-025-09520-y
Resource: RRID:Addgene_66810
Curator: @olekpark
SciCrunch record: RRID:Addgene_66810
48138
DOI: 10.1038/s41586-025-09520-y
Resource: RRID:Addgene_48138
Curator: @olekpark
SciCrunch record: RRID:Addgene_48138
Addgene
DOI: 10.1038/s41523-025-00779-y
Resource: Addgene (RRID:SCR_002037)
Curator: @olekpark
SciCrunch record: RRID:SCR_002037
19131
DOI: 10.1038/s41467-025-65706-y
Resource: None
Curator: @olekpark
SciCrunch record: RRID:Addgene_19131
171098
DOI: 10.1038/s41467-025-65706-y
Resource: None
Curator: @olekpark
SciCrunch record: RRID:Addgene_171098
42230
DOI: 10.1038/s41467-025-65706-y
Resource: RRID:Addgene_42230
Curator: @olekpark
SciCrunch record: RRID:Addgene_42230
46569
DOI: 10.1038/s41467-025-65706-y
Resource: RRID:Addgene_46569
Curator: @olekpark
SciCrunch record: RRID:Addgene_46569
179392
DOI: 10.1038/s41467-025-65706-y
Resource: None
Curator: @olekpark
SciCrunch record: RRID:Addgene_179392
psPAX2
DOI: 10.1038/s41467-025-63700-y
Resource: RRID:Addgene_12260
Curator: @olekpark
SciCrunch record: RRID:Addgene_12260
pMD2.G
DOI: 10.1038/s41467-025-63700-y
Resource: RRID:Addgene_12259
Curator: @olekpark
SciCrunch record: RRID:Addgene_12259
162076
DOI: 10.1038/s41467-025-63592-y
Resource: RRID:Addgene_162076
Curator: @olekpark
SciCrunch record: RRID:Addgene_162076
162075
DOI: 10.1038/s41467-025-63592-y
Resource: RRID:Addgene_162075
Curator: @olekpark
SciCrunch record: RRID:Addgene_162075
Plasmid_50005
DOI: 10.1038/s44172-025-00364-y
Resource: RRID:Addgene_50005
Curator: @scibot
SciCrunch record: RRID:Addgene_50005
RRID:IMSR_JAX
DOI: 10.7554/eLife.100248
Resource: None
Curator: @olekpark
SciCrunch record: RRID:IMSR_JAX:000664
plasmid_6541359
DOI: 10.1038/s42003-025-08647-y
Resource: None
Curator: @olekpark
SciCrunch record: RRID:Addgene_65413
plasmid_3295636
DOI: 10.1038/s42003-025-08647-y
Resource: None
Curator: @olekpark
SciCrunch record: RRID:Addgene_32956
plasmid_2717358
DOI: 10.1038/s42003-025-08647-y
Resource: None
Curator: @olekpark
SciCrunch record: RRID:Addgene_27173
disminuir las creencias irracionales expuestas anteriormente, reemplazándolas por pensamientos alternativos que sean más adaptativos y permitan modificar la interpretación negativa que tiene R. de sí mismo y de su relación con otros. Disminuir el exceso de horas laborales para así, sustituirlas mediante actividades de ocio. Así como, desarrollar un adecuado manejo de la expresión emocional y un estilo de afrontamiento más funcional.
Objetivos
Existe una ley interna en la naturaleza a la que ningún ser vivo puede escapar. El cuerpo biológico nace, crece, madura y después decae hasta morir. Algunos pensadores, como Oswald Splenger, cayeron en el error de aplicar este mismo proceso a las sociedades humanas. Como respuesta a esta visión del desarrollo civilizatorio que le llevó a Spengler a escribir su famosa obra «La decadencia de Occidente», autores como Lewis Mumford o Waldo Frank defendieron que las comunidades orgánicas presentan una forma parabólica, siempre abierta y cambiante. El término elegido por Mumford para definir este proceso fue el de «equilibrio dinámico»
¿Es una comunidad orgánica un símil de una [[cibernética]] positiva y abierta? ¿Es posible pensarlo desde ahí?
El reto que tenemos ante nosotros, la revolución esperada, es el triunfo de la visión orgánica. Este momento llegará, según Waldo Frank, cuando el hombre, «que durante dilatadas épocas ha empleado todos sus órganos individuales y colectivos para el bienestar del yo, empíricamente considerado, aprenda que este yo, así cuidado y así servido, pierde su salud: que por su bienestar debe esforzarse en ser un integrador dentro de un todo metafísicamente fuera de él». En resumidas cuentas, nuestra misión futura consiste en la reordenación de los tres componentes del yo: el ego social, el ego somático y el yo cósmico. Este último, el espíritu, con capacidad infinita para elevarse, tiene que ocupar el lugar central, hoy día monopolizado por el ego somático, dando lugar al egoísmo e individualismo reinante. Este proceso de reacondicionamiento interno está todavía en sus primeras etapas y aparece fugazmente en ocasiones puntuales que calificamos de «revolucionarias».
Interesante posibilidad para cruzar con el #TFM y la cuestión del #encantamiento
Después de mucho tiempo dándole vueltas a la cabeza, he llegado a la misma conclusión a la que llegaron Lewis Mumford y su colega Waldo Frank: uno de los asuntos claves en la humanidad y en su modo de organización como sociedad es el eterno conflicto en la visión mecánica y la visión orgánica de la existencia humana y todo lo que con ella se relaciona. La primera de las visiones se relaciona con la máquina, la segunda con la naturaleza. Cada día este eterno conflicto entre mecanicismo y organicismo se aprecia con más claridad. El escenario donde se libra la batalla entre mecanicista y organicista ha sido y es de lo más variado. En arquitectura, Frank Lloyd Wright y Antoni Gaudí frente a Le Corbusier y los representantes del llamado «Estilo Internacional»; en la música, Mozart frente a la música electrónica; el cerebro frente a la inteligencia artificial; el proyecto educativo de Dewey frente a los postulados de Comenius; la pintura de Goya frente a los cuadros de Andy Warhol; la medicina natural frente a la institucional, etc…
[[Lewis Mumford]] y [[Waldo Frank]] sobre el conflicto de la visión mecánica u orgánica.
Términos como organismo, mecanicismo, organización, 15M, democracia, política,…, son las piezas claves del puzzle y una metáfora en sí misma de la idea principal que las une a todas: la relación entre el todo y las partes.
Sobre [[mecanicismo]] y [[organicismo]].
P element allele of vps24, yw;;P w+y+ vps24[EY04708]
DOI: 10.1371/journal.pone.0251184
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: (WB Cat# WBStrain00004840,RRID:WB-STRAIN:WBStrain00004840)
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00004840
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: (WB Cat# WBStrain00000001,RRID:WB-STRAIN:WBStrain00000001)
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00000001
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00006376
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00027365
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00041003
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00007572
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: RRID:WB-STRAIN:WBStrain00027424
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00027424
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00005261
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: (WB Cat# WBStrain00041969,RRID:WB-STRAIN:WBStrain00041969)
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00041969
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: RRID:WB-STRAIN:WBStrain00027363
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00027363
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00022773
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: (WB Cat# WBStrain00040806,RRID:WB-STRAIN:WBStrain00040806)
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00040806
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00022197
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: (WB Cat# WBStrain00024040,RRID:WB-STRAIN:WBStrain00024040)
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00024040
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00005327
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00022768
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00026779
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00022771
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00022185
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: RRID:WB-STRAIN:WBStrain00004310
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00004310
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00031767
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00005487
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: (WB Cat# WBStrain00034065,RRID:WB-STRAIN:WBStrain00034065)
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00034065
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: RRID:WB-STRAIN:WBStrain00000264
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00000264
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00029930
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: RRID:WB-STRAIN:WBStrain00008608
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00008608
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00031144
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: RRID:WB-STRAIN:WBStrain00008309
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00008309
Supplementary Information
DOI: 10.1038/s41467-025-58478-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00029100
Dossier d'Information : La Quête de la Parentalité Idéale
Ce document synthétise une discussion radiophonique sur la notion de "bon parent", explorant les pressions, les doutes et les stratégies qui définissent la parentalité contemporaine.
Il ressort que l'idéal du parent parfait est une source de stress et de culpabilité, largement alimentée par la compétition sociale et un afflux de connaissances scientifiques qui peuvent être à la fois une aide et un fardeau.
Les intervenants s'accordent sur le fait que la parentalité est un exercice d'équilibriste constant, oscillant entre de grands succès et des échecs patents.
Les thèmes centraux incluent le conflit entre le désir de façonner un "enfant idéal" et la nécessité d'accepter l'enfant réel, la difficulté de se défaire de ses propres projections et traumatismes, et la charge mentale disproportionnée qui pèse souvent sur les mères.
La discussion met en lumière le concept de "parent suffisamment bon" de Donald Winnicott, qui valorise non pas la perfection, mais la capacité à répondre aux besoins de l'enfant tout en introduisant une frustration gérable, essentielle à son développement.
Finalement, la parentalité est présentée comme une expérience partagée, où l'échange, la reconnaissance de sa propre faillibilité et la capacité à "réparer" ses erreurs sont plus importants que la poursuite d'un idéal inaccessible.
--------------------------------------------------------------------------------
La question "Qu'est-ce qu'un bon parent ?" a fait l'objet d'une émission sur France Inter, réunissant des chroniqueurs, auteurs et parents pour partager leurs expériences et réflexions.
La discussion, présentée comme une conversation de "praticiens" plutôt que de spécialistes, a exploré les multiples facettes de la parentalité moderne.
Intervenants Principaux :
Nom
Rôle et Affiliation
Nombre d'enfants
Gwenaëlle Boulet
Rédactrice en chef (Popie, Pomme d'Api), autrice de la BD "Ma vie de parent"
Trois
Julien Bisson
Directeur des rédactions (Le 1 hebdo), chroniqueur "Ma vie de parent"
Un
Marie Pernaud
Chroniqueuse (La maison des maternels), animatrice du podcast "Very Important Parents"
Quatre
Sonia de Viller
Journaliste et parente intervenant au cours du débat
Deux (au moins)
Le débat a également été enrichi par les témoignages d'auditeurs, offrant des perspectives vécues sur les défis abordés.
La discussion s'ouvre sur un exercice d'auto-notation, demandant aux invités de s'évaluer sur une échelle de 1 (parent exécrable) à 10 (parent parfait).
Les réponses révèlent immédiatement la complexité et la variabilité de la perception de soi en tant que parent.
• Gwenaëlle Boulet se donne un 8/10, justifiant cette note élevée par le fait que ses enfants n'ont pas été maltraités et vont globalement bien, tout en admettant leur laisser "suffisamment de quoi aller chez le psy plus tard".
• Julien Bisson souligne la fluctuation de sa performance : il s'évalue à 9/10 la veille au soir après un jeu de société, mais à 2/10 le matin même après avoir "hurlé sur son fils". Sa moyenne se situe donc autour de 5,5/10.
• Marie Pernaud abonde dans ce sens, affirmant que la qualité de sa parentalité varie selon les moments de la journée, notant que "le matin, c'est compliqué quand même".
• Florence, une auditrice de Haute-Savoie, se donne une moyenne de 7,5/10, reconnaissant que sa performance dépend des "circonstances de la vie".
Cette variabilité démontre que la parentalité n'est pas une compétence statique, mais un effort constant et situationnel.
Un thème majeur émerge rapidement : la tension entre l'enfant que les parents désirent et l'enfant qu'ils ont réellement.
• Florence, l'auditrice, définit le bon parent comme celui qui, dès la naissance, considère son enfant "comme un être à part entière" et non "comme sa possession".
L'objectif est de l'aider à se réaliser "selon ce qu'il est lui et non pas ce que je voulais moi, ce qui soit".
• Gwenaëlle Boulet confesse que c'est le "combat de sa vie".
Elle illustre cette lutte avec son désir que ses enfants aiment la littérature, un désir qui s'est heurté à leur indifférence et s'est avéré "contreproductif à souhait".
Elle trouve "hyper dur" d'accepter que son enfant puise "dans d'autres sources que les tiennes pour grandir".
• Julien Bisson conclut que pour s'approcher du "parent idéal", il faut d'abord "éviter de vouloir un enfant idéal".
Cet enfant idéal est celui sur lequel on projette ses propres attentes psychologiques et d'accomplissement.
• Marie Pernaud résume : être un bon parent, "c'est vraiment faire le deuil de l'enfant qu'on aurait voulu avoir".
Face à un conflit, la question à se poser est : "quel est l'enfant qu'on a en fait et comment on doit réagir par rapport à l'enfant qu'on a".
Sonia de Viller ajoute une nuance importante : on n'est pas le même parent pour chaque enfant.
"Je suis pas la même mère avec mon fils aîné et mon cadet et d'ailleurs il me le reproche".
Marie Pernaud confirme que chaque enfant révèle des facettes différentes, positives comme négatives, chez le parent.
La discussion met en évidence que la parentalité contemporaine est soumise à une série de pressions externes et internes qui complexifient la tâche.
L'accès à une masse d'informations sur le développement de l'enfant est perçu comme une arme à double tranchant.
• Gwenaëlle Boulet utilise l'analogie de l'effet Dunning-Kruger :
1. La "montagne de la stupidité" : Fin 19e/début 20e, les exigences se limitaient à s'assurer que l'enfant ne meure pas.
2. La "vallée de l'humilité" : L'arrivée de la psychanalyse et des neurosciences a fait chuter la confiance des parents, écrasés par les connaissances sur ce qu'il "faut surtout pas faire".
3. Le "plateau de la consolidation" : L'objectif est de remonter en faisant correspondre sa confiance et ses compétences, en utilisant ces connaissances tout en se faisant confiance.
• Julien Bisson qualifie les sciences de l'éducation de "bénédiction et malédiction".
Une bénédiction pour les savoirs apportés, une malédiction car elles "ont creusé énormément la distance entre le parent qu'on a l'impression d'être et le parent qu'on pense devoir être", créant un "mal-être parental énorme".
La société moderne impose une dynamique de comparaison et d'individualisme qui affecte directement les parents.
• La Compétition Parentale : Gwenaëlle Boulet décrit une "compète" ressentie dès la maternité (choisir la "super maternité") et qui se poursuit avec la scolarité (l'âge d'apprentissage de la lecture).
• L'Isolement : Julien Bisson lie cette compétition à une société avec "plus d'individualisme, plus d'isolement", ce qui renforce le sentiment d'être "seul" et "désarmé".
• Témoignage de Charlotte : Une auditrice d'Aix-en-Provence exprime sa difficulté à "créer une communauté de parents".
Elle se sent comme une "extraterrestre" lorsqu'elle propose des initiatives collectives ou parle de l'éducation au "vivre ensemble".
La recherche de la perfection parentale a un coût direct sur le bien-être des parents.
• Marie Pernaud alerte sur le risque d'épuisement face aux "injonctions". Les parents reçoivent une multitude d'informations et pensent devoir "absolument tout faire".
Elle rappelle le propos d'une Danoise : tant qu'il n'y a ni maltraitance et qu'il y a de l'amour, il ne peut y avoir de mauvaise éducation.
• Julien Bisson cite des chiffres issus d'un numéro du 1 hebdo sur la santé mentale des parents :
◦ Le mal-être parental touche 1 parent sur 5 (20%).
◦ Le burnout parental affecte 6 à 8 % des parents.
◦ Les femmes sont plus touchées, non par fragilité, mais parce qu'elles "portent encore aujourd'hui une charge parentale beaucoup plus importante que les hommes".
Face à l'idéal inaccessible, la discussion propose une approche plus réaliste et bienveillante, inspirée du concept du psychanalyste Donald Winnicott.
• Définition : Un parent suffisamment bon répond aux besoins de l'enfant sans être parfait et sans "faire trop".
• Évolution :
1. Nourrisson : Le parent répond immédiatement et exactement aux besoins du bébé (faim, réconfort).
2. Enfant : Le parent instaure progressivement "de la frustration gérable".
Il apprend à l'enfant à différer ses désirs, ce qui l'aide à grandir et à "vivre en société".
• Risque de l'anticipation : Anticiper systématiquement les besoins de l'enfant peut freiner son autonomie et son développement émotionnel.
L'erreur n'est pas seulement inévitable, elle est une composante de la relation.
• Reconnaître ses erreurs : Gwenaëlle Boulet insiste sur l'importance de pouvoir revenir vers son enfant et dire :
"Je suis désolé, je me suis emballée [...] j'avais pas envie de réagir comme ça". Cela permet de "réparer beaucoup de choses".
• Déculpabiliser l'enfant : Julien Bisson ajoute que cela aide l'enfant à comprendre que ce n'est "pas toujours de sa faute", car son objectif principal est de satisfaire ses parents.
• Le "Faux Choix" : Gwenaëlle Boulet partage une technique concrète : au lieu de demander "Tu veux prendre ta douche ?", poser la question "Tu veux prendre ta douche maintenant ou dans 5 minutes ?".
Cela offre à l'enfant un "terrain d'expérimentation du choix" tout en atteignant l'objectif du parent.
• L'Influence Partagée : Julien Bisson utilise la métaphore du "buffet" : le parent offre un buffet, mais ne contrôle pas ce que l'enfant va choisir.
De plus, il n'est "pas le seul à le nourrir" (grands-parents, amis, etc.). Il ne faut pas surestimer sa propre influence.
• Le Duo Parental : L'ajustement entre les deux parents, avec leurs bagages respectifs, est un défi mais aussi ce qui "sauve", permettant de prendre de la distance.
Intervenant/Source
Citation ou Idée Clé
Fiva (auditeur)
"Le parent parfait existe mais il n'a pas encore d'enfant."
Cécile Dancy (auditeur)
"Être un bon parent, c'est déjà être capable de travailler ses propres failles pour ne pas les faire peser sur nos enfants."
Peter Ustinov (cité)
"Les parents sont les os sur lesquelles les enfants se font les dents."
Russell Show (cité)
"Si nous accordons à nos enfants notre confiance, si nous les laissons suivre leur propre voix (...) nous allégerons notre vie tout en leur donnant les moyens de s'épanouir."
Ivan (auditeur)
Témoigne avec une grande émotion de sa souffrance en tant que père de deux adolescents.
Il reconnaît avoir projeté des attentes élevées sur son fils aîné, en réaction à sa propre relation difficile avec son père, ce qui a mené à une "cassure".
Il exprime son désarroi face à une situation complexe, concluant : "un bon parent, je ne sais pas ce que c'est [...] c'est simplement essayer de faire du mieux que je peux".
Le témoignage d'Ivan illustre de manière poignante le poids du passé, le risque de la surprotection et le sentiment de désarroi que peuvent ressentir les parents, même avec la volonté de bien faire.
Sa démarche de s'interroger, selon les intervenants, est déjà la preuve qu'il est "probablement un bon parent".
Warning in left_join(., secondary_data %>% select("PASSWORD", "VEGGIESCORE", : Detected an unexpected many-to-many relationship between `x` and `y`. ℹ Row 1 of `x` matches multiple rows in `y`. ℹ Row 86 of `y` matches multiple rows in `x`. ℹ If a many-to-many relationship is expected, set `relationship = "many-to-many"` to silence this warning.
remove warnings
Author response:
The following is the authors’ response to the original reviews
Public Reviews:
Reviewer #1 (Public Review):
Summary:
In this study, participants completed two different tasks. A perceptual choice task in which they compared the sizes of pairs of items and a value-different task in which they identified the higher value option among pairs of items with the two tasks involving the same stimuli. Based on previous fMRI research, the authors sought to determine whether the superior frontal sulcus (SFS) is involved in both perceptual and value-based decisions or just one or the other. Initial fMRI analyses were devised to isolate brain regions that were activated for both types of choices and also regions that were unique to each. Transcranial magnetic stimulation was applied to the SFS in between fMRI sessions and it was found to lead to a significant decrease in accuracy and RT on the perceptual choice task but only a decrease in RT on the value-different task. Hierarchical drift-diffusion modelling of the data indicated that the TMS had led to a lowering of decision boundaries in the perceptual task and a lower of non-decision times on the value-based task. Additional analyses show that SFS covaries with model-derived estimates of cumulative evidence and that this relationship is weakened by TMS.
Strengths:
The paper has many strengths including the rigorous multi-pronged approach of causal manipulation, fMRI and computational modelling which offers a fresh perspective on the neural drivers of decision making. Some additional strengths include the careful paradigm design which ensured that the two types of tasks were matched for their perceptual content while orthogonalizing trial-to-trial variations in choice difficulty. The paper also lays out a number of specific hypotheses at the outset regarding the behavioural outcomes that are tied to decision model parameters and are well justified.
Weaknesses:
(1.1) Unless I have missed it, the SFS does not actually appear in the list of brain areas significantly activated by the perceptual and value tasks in Supplementary Tables 1 and 2. Its presence or absence from the list of significant activations is not mentioned by the authors when outlining these results in the main text. What are we to make of the fact that it is not showing significant activation in these initial analyses?
You are right that the left SFS does not appear in our initial task-level contrasts. Those first analyses were deliberately agnostic to evidence accumulation (i.e., average BOLD by task, irrespective of trial-by-trial evidence). Consistent with prior work, SFS emerges only when we model the parametric variation in accumulated perceptual evidence.
Accordingly, we ran a second-level GLM that included trial-wise accumulated evidence (aE) as a parametric modulator. In that analysis, the left SFS shows significant aE-related activity specifically during perceptual decisions, but not during value-based decisions (SVC in a 10-mm sphere around x = −24, y = 24, z = 36).
To avoid confusion, we now:
(i) explicitly separate and label the two analysis levels in the Results; (ii) state up front that SFS is not expected to appear in the task-average contrast; and (iii) add a short pointer that SFS appears once aE is included as a parametric modulator. We also edited Methods to spell out precisely how aE is constructed and entered into GLM2. This should make the logic of the two-stage analysis clearer and aligns the manuscript with the literature where SFS typically emerges only in parametric evidence models.
(1.2) The value difference task also requires identification of the stimuli, and therefore perceptual decision-making. In light of this, the initial fMRI analyses do not seem terribly informative for the present purposes as areas that are activated for both types of tasks could conceivably be specifically supporting perceptual decision-making only. I would have thought brain areas that are playing a particular role in evidence accumulation would be best identified based on whether their BOLD response scaled with evidence strength in each condition which would make it more likely that areas particular to each type of choice can be identified. The rationale for the authors' approach could be better justified.
We agree that both tasks require early sensory identification of the items, but the decision-relevant evidence differs by design (size difference vs. value difference), and our modelling is targeted at the evidence integration stage rather than initial identification.
To address your concern empirically, we: (i) added session-wise plots of mean RTs showing a general speed-up across the experiment (now in the Supplement); (ii) fit a hierarchical DDM to jointly explain accuracy and RT. The DDM dissociates decision time (evidence integration) from non-decision time (encoding/response execution).
After cTBS, perceptual decisions show a selective reduction of the decision boundary (lower accuracy, faster RTs; no drift-rate change), whereas value-based decisions show no change to boundary/drift but a decrease in non-decision time, consistent with faster sensorimotor processing or task familiarity. Thus, the TMS effect in SFS is specific to the criterion for perceptual evidence accumulation, while the RT speed-up in the value task reflects decision-irrelevant processes. We now state this explicitly in the Results and add the RT-by-run figure for transparency.
(1.2.1) The value difference task also requires identification of the stimuli, and therefore perceptual decision-making. In light of this, the initial fMRI analyses do not seem terribly informative for the present purposes as areas that are activated for both types of tasks could conceivably be specifically supporting perceptual decision-making only.
Thank you for prompting this clarification.
The key point is what changes with cTBS. If SFS supported generic identification, we would expect parallel cTBS effects on drift rate (or boundary) in both tasks. Instead, we find: (a) boundary decreases selectively in perceptual decisions (consistent with SFS setting the amount of perceptual evidence required), and (b) non-decision time decreases selectively in the value task (consistent with speed-ups in encoding/response stages). Moreover, trial-by-trial SFS BOLD predicts perceptual accuracy (controlling for evidence), and neural-DDM model comparison shows SFS activity modulates boundary, not drift, during perceptual choices.
Together, these converging behavioral, computational, and neural results argue that SFS specifically supports the criterion for perceptual evidence accumulation rather than generic visual identification.
(1.2.2) I would have thought brain areas that are playing a particular role in evidence accumulation would be best identified based on whether their BOLD response scaled with evidence strength in each condition which would make it more likely that areas particular to each type of choice can be identified. The rationale for the authors' approach could be better justified.
We now more explicitly justify the two-level fMRI approach. The task-average contrast addresses which networks are generally more engaged by each domain (e.g., posterior parietal for PDM; vmPFC/PCC for VDM), given identical stimuli and motor outputs. This complements, but does not substitute for, the parametric evidence analysis, which is where one expects accumulation-related regions such as SFS to emerge. We added text clarifying that the first analysis establishes domain-specific recruitment at the task level, whereas the second isolates evidence-dependent signals (aE) and reveals that left SFS tracks accumulated evidence only for perceptual choices. We also added explicit references to the literature using similar two-step logic and noted that SFS typically appears only in parametric evidence models.
(1.3) TMS led to reductions in RT in the value-difference as well as the perceptual choice task. DDM modelling indicated that in the case of the value task, the effect was attributable to reduced non-decision time which the authors attribute to task learning. The reasoning here is a little unclear.
(1.3.1) Comment: If task learning is the cause, then why are similar non-decision time effects not observed in the perceptual choice task?
Great point. The DDM addresses exactly this: RT comprises decision time (DT) plus non-decision time (nDT). With cTBS, PDM shows reduced DT (via a lower boundary) but stable nDT; VDM shows reduced nDT with no change to boundary/drift. Hence, the superficially similar RT speed-ups in both tasks are explained by different latent processes: decision-relevant in PDM (lower criterion → faster decisions, lower accuracy) and decision-irrelevant in VDM (faster encoding/response). We added explicit language and a supplemental figure showing RT across runs, and we clarified in the text that only the PDM speed-up reflects a change to evidence integration.
(1.3.2) Given that the value-task actually requires perceptual decision-making, is it not possible that SFS disruption impacted the speed with which the items could be identified, hence delaying the onset of the value-comparison choice?
We agree there is a brief perceptual encoding phase at the start of both tasks. If cTBS impaired visual identification per se, we would expect longer nDT in both tasks or a decrease in drift rate. Instead, nDT decreases in the value task and is unchanged in the perceptual task; drift is unchanged in both. Thus, cTBS over SFS does not slow identification; rather, it lowers the criterion for perceptual accumulation (PDM) and, separately, we observe faster non-decision components in VDM (likely familiarity or motor preparation). We added a clarifying sentence noting that item identification was easy and highly overlearned (static, large food pictures), and we cite that nDT is the appropriate locus for identification effects in the DDM framework; our data do not show the pattern expected of impaired identification.
(1.4) The sample size is relatively small. The authors state that 20 subjects is 'in the acceptable range' but it is not clear what is meant by this.
We have clarified what we mean and provided citations. The sample (n = 20) matches or exceeds many prior causal TMS/fMRI studies targeting perceptual decision circuitry (e.g., Philiastides et al., 2011; Rahnev et al., 2016; Jackson et al., 2021; van der Plas et al., 2021; Murd et al., 2021). Importantly, we (i) use within-subject, pre/post cTBS differences-in-differences with matched tasks; (ii) estimate hierarchical models that borrow strength across participants; and (iii) converge across behavior, latent parameters, regional BOLD, and connectivity. We now replace the vague phrase with a concrete statement and references, and we report precision (HDIs/SEs) for all main effects.
Reviewer #2 (Public Review):
Summary:
The authors set out to test whether a TMS-induced reduction in excitability of the left Superior Frontal Sulcus influenced evidence integration in perceptual and value-based decisions. They directly compared behaviour - including fits to a computational decision process model - and fMRI pre and post-TMS in one of each type of decision-making task. Their goal was to test domain-specific theories of the prefrontal cortex by examining whether the proposed role of the SFS in evidence integration was selective for perceptual but not value-based evidence.
Strengths:
The paper presents multiple credible sources of evidence for the role of the left SFS in perceptual decision-making, finding similar mechanisms to prior literature and a nuanced discussion of where they diverge from prior findings. The value-based and perceptual decision-making tasks were carefully matched in terms of stimulus display and motor response, making their comparison credible.
Weaknesses:
(2.1) More information on the task and details of the behavioural modelling would be helpful for interpreting the results.
Thank you for this request for clarity. In the revision we explicitly state, up front, how the two tasks differ and how the modelling maps onto those differences.
(1) Task separability and “evidence.” We now define task-relevant evidence as size difference (SD) for perceptual decisions (PDM) and value difference (VD) for value-based decisions (VDM). Stimuli and motor mappings are identical across tasks; only the evidence to be integrated changes.
(2) Behavioural separability that mirrors task design. As reported, mixed-effects regressions show PDM accuracy increases with SD (β=0.560, p<0.001) but not VD (β=0.023, p=0.178), and PDM RTs shorten with SD (β=−0.057, p<0.001) but not VD (β=0.002, p=0.281). Conversely, VDM accuracy increases with VD (β=0.249, p<0.001) but not SD (β=0.005, p=0.826), and VDM RTs shorten with VD (β=−0.016, p=0.011) but not SD (β=−0.003, p=0.419).
(3 How the HDDM reflects this. The hierarchical DDM fits the joint accuracy–RT distributions with task-specific evidence (SD or VD) as the predictor of drift. The model separates decision time from non-decision time (nDT), which is essential for interpreting the different RT patterns across tasks without assuming differences in the accumulation process when accuracy is unchanged.
These clarifications are integrated in the Methods (Experimental paradigm; HDDM) and in Results (“Behaviour: validity of task-relevant pre-requisites” and “Modelling: faster RTs during value-based decisions is related to non-decision-related sensorimotor processes”).
(2.2) The evidence for a choice and 'accuracy' of that choice in both tasks was determined by a rating task that was done in advance of the main testing blocks (twice for each stimulus). For the perceptual decisions, this involved asking participants to quantify a size metric for the stimuli, but the veracity of these ratings was not reported, nor was the consistency of the value-based ones. It is my understanding that the size ratings were used to define the amount of perceptual evidence in a trial, rather than the true size differences, and without seeing more data the reliability of this approach is unclear. More concerning was the effect of 'evidence level' on behaviour in the value-based task (Figure 3a). While the 'proportion correct' increases monotonically with the evidence level for the perceptual decisions, for the value-based task it increases from the lowest evidence level and then appears to plateau at just above 80%. This difference in behaviour between the two tasks brings into question the validity of the DDM which is used to fit the data, which assumes that the drift rate increases linearly in proportion to the level of evidence.
We thank the reviewer for raising these concerns, and we address each of them point by point:
2.2.1. Comment: It is my understanding that the size ratings were used to define the amount of perceptual evidence in a trial, rather than the true size differences, and without seeing more data the reliability of this approach is unclear.
That is correct—we used participants’ area/size ratings to construct perceptual evidence (SD).
To validate this choice, we compared those ratings against an objective image-based size measure (proportion of non-black pixels within the bounding box). As shown in Author response image 3, perceptual size ratings are highly correlated with objective size across participants (Pearson r values predominantly ≈0.8 or higher; all p<0.001). Importantly, value ratings do not correlate with objective size (Author response image 2), confirming that the two rating scales capture distinct constructs. These checks support using participants’ size ratings as the participant-specific ground truth for defining SD in the PDM trials.
Author response image 1.
Objective size and value ratings are unrelated. Scatterplots show, for each participant, the correlation between objective image size (x-axis; proportion of non-black pixels within the item box) and value-based ratings (y-axis; 0–100 scale). Each dot is one food item (ratings averaged over the two value-rating repetitions). Across participants, value ratings do not track objective size, confirming that value and size are distinct constructs.
Author response image 2.
Perceptual size ratings closely track objective size. Scatterplots show, for each participant, the correlation between objective image size (x-axis) and perceptual area/size ratings (y-axis; 0–100 scale). Each dot is one food item (ratings averaged over the two perceptual ratings). Perceptual ratings are strongly correlated with objective size for nearly all participants (see main text), validating the use of these ratings to construct size-difference evidence (SD).
(2.2.2) More concerning was the effect of 'evidence level' on behaviour in the value-based task (Figure 3a). While the 'proportion correct' increases monotonically with the evidence level for the perceptual decisions, for the value-based task it increases from the lowest evidence level and then appears to plateau at just above 80%. This difference in behaviour between the two tasks brings into question the validity of the DDM which is used to fit the data, which assumes that the drift rate increases linearly in proportion to the level of evidence.
We agree that accuracy appears to asymptote in VDM, but the DDM fits indicate that the drift rate still increases monotonically with evidence in both tasks. In Supplementary figure 11, drift (δ) rises across the four evidence levels for PDM and for VDM (panels showing all data and pre/post-TMS). The apparent plateau in proportion correct during VDM reflects higher choice variability at stronger preference differences, not a failure of the drift–evidence mapping. Crucially, the model captures both the accuracy patterns and the RT distributions (see posterior predictive checks in Supplementary figures 11-16), indicating that a monotonic evidence–drift relation is sufficient to account for the data in each task.
Author response image 3.
HDDM parameters by evidence level. Group-level posterior means (± posterior SD) for drift (δ), boundary (α), and non-decision time (τ) across the four evidence levels, shown (a) collapsed across TMS sessions, (b) for PDM (blue) pre- vs post-TMS (light vs dark), and (c) for VDM (orange) pre- vs post-TMS. Crucially, drift increases monotonically with evidence in both tasks, while TMS selectively lowers α in PDM and reduces τ in VDM (see Supplementary Tables for numerical estimates).
(2.3) The paper provides very little information on the model fits (no parameter estimates, goodness of fit values or simulated behavioural predictions). The paper finds that TMS reduced the decision bound for perceptual decisions but only affected non-decision time for value-based decisions. It would aid the interpretation of this finding if the relative reliability of the fits for the two tasks was presented.
We appreciate the suggestion and have made the quantitative fit information explicit:
(1) Parameter estimates. Group-level means/SDs for drift (δ), boundary (α), and nDT (τ) are reported for PDM and VDM overall, by evidence level, pre- vs post-TMS, and per subject (see Supplementary Tables 8-11).
(2) Goodness of fit and predictive adequacy. DIC values accompany each fit in the tables. Posterior predictive checks demonstrate close correspondence between simulated and observed accuracy and RT distributions overall, by evidence level, and across subjects (Supplementary Figures 11-16).
Together, these materials document that the HDDM provides reliable fits in both tasks and accurately recovers the qualitative and quantitative patterns that underlie our inferences (reduced α for PDM only; selective τ reduction in VDM).
(2.4) Behaviourally, the perceptual task produced decreased response times and accuracy post-TMS, consistent with a reduced bound and consistent with some prior literature. Based on the results of the computational modelling, the authors conclude that RT differences in the value-based task are due to task-related learning, while those in the perceptual task are 'decision relevant'. It is not fully clear why there would be such significantly greater task-related learning in the value-based task relative to the perceptual one. And if such learning is occurring, could it potentially also tend to increase the consistency of choices, thereby counteracting any possible TMS-induced reduction of consistency?
Thank you for pointing out the need for a clearer framing. We have removed the speculative label “task-related learning” and now describe the pattern strictly in terms of the HDDM decomposition and neural results already reported:
(1) VDM: Post-TMS RTs are faster while accuracy is unchanged. The HDDM attributes this to a selective reduction in non-decision time (τ), with no change in decision-relevant parameters (α, δ) for VDM (see Supplementary Figure 11 and Supplementary Tables). Consistent with this, left SFS BOLD is not reduced for VDM, and trialwise SFS activity does not predict VDM accuracy—both observations argue against a change in VDM decision formation within left SFS.
(2) PDM: Post-TMS accuracy decreases and RTs shorten, which the HDDM captures as a lower decision boundary (α) with no change in drift (δ). Here, left SFS BOLD scales with accumulated evidence and decreases post-TMS, and trialwise SFS activity predicts PDM accuracy, all consistent with a decision-relevant effect in PDM.
Regarding the possibility that faster VDM RTs should increase choice consistency: empirically, consistency did not change in VDM, and the HDDM finds no decision-parameter shifts there. Thus, there is no hidden counteracting increase in VDM accuracy that could mask a TMS effect—the absence of a VDM accuracy change is itself informative and aligns with the modelling and fMRI.
Reviewer #3 (Public Review):
Summary:
Garcia et al., investigated whether the human left superior frontal sulcus (SFS) is involved in integrating evidence for decisions across either perceptual and/or value-based decision-making. Specifically, they had 20 participants perform two decision-making tasks (with matched stimuli and motor responses) in an fMRI scanner both before and after they received continuous theta burst transcranial magnetic stimulation (TMS) of the left SFS. The stimulation thought to decrease neural activity in the targeted region, led to reduced accuracy on the perceptual decision task only. The pattern of results across both model-free and model-based (Drift diffusion model) behavioural and fMRI analyses suggests that the left SLS plays a critical role in perceptual decisions only, with no equivalent effects found for value-based decisions. The DDM-based analyses revealed that the role of the left SLS in perceptual evidence accumulation is likely to be one of decision boundary setting. Hence the authors conclude that the left SFS plays a domain-specific causal role in the accumulation of evidence for perceptual decisions. These results are likely to add importance to the literature regarding the neural correlates of decision-making.
Strengths:
The use of TMS strengthens the evidence for the left SFS playing a causal role in the evidence accumulation process. By combining TMS with fMRI and advanced computational modelling of behaviour, the authors go beyond previous correlational studies in the field and provide converging behavioural, computational, and neural evidence of the specific role that the left SFS may play.
Sophisticated and rigorous analysis approaches are used throughout.
Weaknesses:
(3.1) Though the stimuli and motor responses were equalised between the perception and value-based decision tasks, reaction times (according to Figure 1) and potential difficulty (Figure 2) were not matched. Hence, differences in task difficulty might represent an alternative explanation for the effects being specific to the perception task rather than domain-specificity per se.
We agree that RTs cannot be matched a priori, and we did not intend them to be. Instead, we equated the inputs to the decision process and verified that each task relied exclusively on its task-relevant evidence. As reported in Results—Behaviour: validity of task-relevant pre-requisites (Fig. 1b–c), accuracy and RTs vary monotonically with the appropriate evidence regressor (SD for PDM; VD for VDM), with no effect of the task-irrelevant regressor. This separability check addresses differences in baseline RTs by showing that, for both tasks, behaviour tracks evidence as designed.
To rule out a generic difficulty account of the TMS effect, we relied on the within-subject differences-in-differences (DID) framework described in Methods (Differences-in-differences). The key Task × TMS interaction compares the pre→post change in PDM with the pre→post change in VDM while controlling for trialwise evidence and RT covariates. Any time-on-task or unspecific difficulty drift shared by both tasks is subtracted out by this contrast. Using this specification, TMS selectively reduced accuracy for PDM but not VDM (Fig. 3a; Supplementary Fig. 2a,c; Supplementary Tables 5–7).
Finally, the hierarchical DDM (already in the paper) dissociates latent mechanisms. The post-TMS boundary reduction appears only in PDM, whereas VDM shows a change in non-decision time without a decision-relevant parameter change (Fig. 3c; Supplementary Figs. 4–5). If unmatched difficulty were the sole driver, we would expect parallel effects across tasks, which we do not observe.
(3.2) No within- or between-participants sham/control TMS condition was employed. This would have strengthened the inference that the apparent TMS effects on behavioural and neural measures can truly be attributed to the left SFS stimulation and not to non-specific peripheral stimulation and/or time-on-task effects.
We agree that a sham/control condition would further strengthen causal attribution and note this as a limitation. In mitigation, our design incorporates several safeguards already reported in the manuscript:
· Within-subject pre/post with alternating task blocks and DID modelling (Methods) to difference out non-specific time-on-task effects.
· Task specificity across levels of analysis: behaviour (PDM accuracy reduction only), computational (boundary reduction only in PDM; no drift change), BOLD (reduced left-SFS accumulated-evidence signal for PDM but not VDM; Fig. 4a–c), and functional coupling (SFS–occipital PPI increase during PDM only; Fig. 5).
· Matched stimuli and motor outputs across tasks, so any peripheral sensations or general arousal effects should have influenced both tasks similarly; they did not.
Together, these converging task-selective effects reduce the likelihood that the results reflect non-specific stimulation or time-on-task. We will add an explicit statement in the Limitations noting the absence of sham/control and outlining it as a priority for future work.
(3.3) No a priori power analysis is presented.
We appreciate this point. Our sample size (n = 20) matched prior causal TMS and combined TMS–fMRI studies using similar paradigms and analyses (e.g., Philiastides et al., 2011; Rahnev et al., 2016; Jackson et al., 2021; van der Plas et al., 2021; Murd et al., 2021), and was chosen a priori on that basis and the practical constraints of cTBS + fMRI. The within-subject DID approach and hierarchical modelling further improve efficiency by leveraging all trials.
To address the reviewer’s request for transparency, we will (i) state this rationale in Methods—Participants, and (ii) ensure that all primary effects are reported with 95% CIs or posterior probabilities (already provided for the HDDM as pmcmcp_{\mathrm{mcmc}}pmcmc). We also note that the design was sensitive enough to detect RT changes in both tasks and a selective accuracy change in PDM, arguing against a blanket lack of power as an explanation for null VDM accuracy effects. We will nevertheless flag the absence of a formal prospective power analysis in the Limitations.
Recommendations for the Authors:
Reviewer #1 (Recommendations For The Authors):
Some important elements of the methods are missing. How was the site for targeting the SFS with TMS identified? The methods described how M1 was located but not SFS.
Thank you for catching this omission. In the revised Methods we explicitly describe how the left SFS target was localized. Briefly, we used each participant’s T1-weighted anatomical scan and frameless neuronavigation to place a 10-mm sphere at the a priori MNI coordinates (x = −24, y = 24, z = 36) derived from prior work (Heekeren et al., 2004; Philiastides et al., 2011). This sphere was transformed to native space for each participant. The coil was positioned tangentially with the handle pointing posterior-lateral, and coil placement was continuously monitored with neuronavigation throughout stimulation. (All of these procedures mirror what we already report for M1 and are now stated for SFS as well.)
Where to revise the manuscript:
Methods → Stimulation protocol. After the first sentence naming cTBS, insert:<br /> “The left SFS target was localized on each participant’s T1-weighted anatomical image using frameless neuronavigation. A 10-mm radius sphere was centered at the a priori MNI coordinates x = −24, y = 24, z = 36 (Heekeren et al., 2004; Philiastides et al., 2011), then transformed to native space. The MR-compatible figure-of-eight coil was positioned tangentially over the target with the handle oriented posterior-laterally, and its position was tracked and maintained with neuronavigation during stimulation.”
It is not clear how participants were instructed that they should perform the value-difference task. Were they told that they should choose based on their original item value ratings or was it left up to them?
We agree the instruction should be explicit. Participants were told_: “In value-based blocks, choose the item you would prefer to eat at the end of the experiment.”_ They were informed that one VDM trial would be randomly selected for actual consumption, ensuring incentive-compatibility. We did not ask them to recall or follow their earlier ratings; those ratings were used only to construct evidence (value difference) and to define choice consistency offline.
Where to revise the manuscript:
Methods → Experimental paradigm.
Add a sentence to the VDM instruction paragraph:
“In value-based (LIKE) blocks, participants were instructed to choose the item they would prefer to consume at the end of the experiment; one VDM trial was randomly selected and implemented, making choices incentive-compatible. Prior ratings were used solely to construct value-difference evidence and to score choice consistency; participants were not asked to recall or match their earlier ratings.”
Line 86 Introduction, some previous studies were conducted on animals. Why it is problematic that the studies were conducted in animals is not stated. I assume the authors mean that we do not know if their findings will translate to the human brain? I think in fairness to those working with animals it might be worth an extra sentence to briefly expand on this point.
We appreciate this and will clarify that animal work is invaluable for circuit-level causality, but species differences and putative non-homologous areas (e.g., human SFS vs. rodent FOF) limit direct translation. Our point is not that animal studies are problematic, but that establishing causal roles in humans remains necessary.
Revision:
Introduction (paragraph discussing prior animal work). Replace the current sentence beginning “However, prior studies were largely correlational”
“Animal studies provide critical causal insights, yet direct translation to humans can be limited by species-specific anatomy and potential non-homologies (e.g., human SFS vs. frontal orienting fields in rodents). Therefore, establishing causal contributions in the human brain remains essential.”
Line 100-101: "or whether its involvement is peripheral and merely functionally supporting a larger system" - it is not clear what you mean by 'supporting a larger system'
We meant that observed SFS activity might reflect upstream/downstream support processes (e.g., attentional control or working-memory maintenance) rather than the computation of evidence accumulation itself. We have rephrased to avoid ambiguity.
Revision:
Introduction. Replace the phrase with:
“or whether its observed activity reflects upstream or downstream support processes (e.g., attention or working-memory maintenance) rather than the accumulation computation per se.”
The authors do have to make certain assumptions about the BOLD patterns that would be expected of an evidence accumulation region. These assumptions are reasonable and have been adopted in several previous neuroimaging studies. Nevertheless, it should be acknowledged that alternative possibilities exist and this is an inevitable limitation of using fMRI to study decision making. For example, if it turns out that participants collapse their boundaries as time elapses, then the assumption that trials with weaker evidence should have larger BOLD responses may not hold - the effect of more prolonged activity could be cancelled out by the lower boundaries. Again, I think this is just a limitation that could be acknowledged in the Discussion, my opinion is that this is the best effort yet to identify choice-relevant regions with fMRI and the authors deserve much credit for their rigorous approach.
Agreed. We already ground our BOLD regressors in the DDM literature, but acknowledge that alternative mechanisms (e.g., time-dependent boundaries) can alter expected BOLD–evidence relations. We now add a short limitation paragraph stating this explicitly.
Revision:
Discussion (limitations paragraph). Add:
“Our fMRI inferences rest on model-based assumptions linking accumulated evidence to BOLD amplitude. Alternative mechanisms—such as time-dependent (collapsing) boundaries—could attenuate the prediction that weaker-evidence trials yield longer accumulation and larger BOLD signals. While our behavioural and neural results converge under the DDM framework, we acknowledge this as a general limitation of model-based fMRI.”
Reviewer #2 (Recommendations For The Authors):
Minor points
I suggest the proportion of missed trials should be reported.
Thank you for the suggestion. In our preprocessing we excluded trials with no response within the task’s response window and any trials failing a priori validity checks. Because non-response trials contain neither a choice nor an RT, they are not entered into the DDM fits or the fMRI GLMs and, by design, carry no weight in the reported results. To keep the focus on the data that informed all analyses, we now (i) state the trial-inclusion criteria explicitly and (ii) report the number of analysed (valid) trials per task and run. This conveys the effective sample size contributing to each condition without altering the analysis set.
Revision:
Methods → (at the end of “Experimental paradigm”): “Analyses were conducted on valid trials only, defined as trials with a registered response within the task’s response window and passing pre-specified validity checks; trials without a response were excluded and not analysed.”
Results → “Behaviour: validity of task-relevant pre-requisites” (add one sentence at the end of the first paragraph): “All behavioural and fMRI analyses were performed on valid trials only (see Methods for inclusion criteria).”
Figure 4 c is very confusing. Is the legend or caption backwards?
Thanks for flagging. We corrected the Figure 4c caption to match the colouring and contrasts used in the panel (perceptual = blue/green overlays; value-based = orange/red; ‘post–pre’ contrasts explicitly labeled). No data or analyses were changed, just the wording to remove ambiguity.
Revision:
Figure 4 caption (panel c sentence). Replace with:
“(c) Post–pre contrasts for the trialwise accumulated-evidence regressor show reduced left-SFS BOLD during perceptual decisions (green overlay), with a significantly stronger reduction for perceptual vs value-based decisions (blue overlay). No reduction is observed for value-based decisions.”
Even if not statistically significant it may be of interest to add the results for Value-based decision making on SFS in Supplementary Table 3.
Done. We now include the SFS small-volume results for VDM (trialwise accumulated-evidence regressor) alongside the PDM values in the same table, with exact peak, cluster size, and statistics.
Revision:
Supplementary Table 3 (title):
“Regions encoding trialwise accumulated evidence (parametric modulation) during perceptual and value-based decisions, including SFS SVC results for both tasks.”
Model comparisons: please explain how model complexity is accounted for.
We clarify that model evidence was compared using the Deviance Information Criterion (DIC), which penalizes model fit by an effective number of parameters (pD). Lower DIC indicates better out-of-sample predictive performance after accounting for model complexity.
Revision:
Methods → Hierarchical Bayesian neural-DDM (last paragraph). Add:
“Model comparison used the Deviance Information Criterion (DIC = D̄ + pD), where pD is the effective number of parameters; thus DIC penalizes model complexity. Lower DIC denotes better predictive accuracy after accounting for complexity.”
Reviewer #3 (Recommendations For The Authors):
The following issues would benefit from clarification in the manuscript:
- It is stated that "Our sample size is well within acceptable range, similar to that of previous TMS studies." The sample size being similar to previous studies does not mean it is within an acceptable range. Whether the sample size is acceptable or not depends on the expected effect size. It is perfectly possible that the previous studies cited were all underpowered. What implications might the lack of an a priori power analysis have for the interpretation of the results?
We agree and have revised our wording. We did not conduct an a priori power analysis. Instead, we relied on a within-participant design that typically yields higher sensitivity in TMS–fMRI settings and on convergence across behavioural, computational, and neural measures. We now acknowledge that the absence of formal power calculations limits claims about small effects (particularly for null findings in VDM), and we frame those null results cautiously.
Revision:
Discussion (limitations). Add:
“The within-participant design enhances statistical sensitivity, yet the absence of an a priori power analysis constrains our ability to rule out small effects, particularly for null results in VDM.”
- I was confused when trying to match the results described in the 'Behaviour: validity of task-relevant pre-requisites' section on page 6 to what is presented in Figure 1. Specifically, Figure 1C is cited 4 times but I believe two of these should be citing Figure 1B?
Thank you—this was a citation mix-up. The two places that referenced “Fig. 1C” but described accuracy should in fact point to Fig. 1B. We corrected both citations.
Revision:
Results → Behaviour: validity… Change the two incorrect “Fig. 1C” references (when describing accuracy) to “Fig. 1B”.
- Also, where is the 'SD' coefficient of -0.254 (p-value = 0.123) coming from in line 211? I can't match this to the figure.
This was a typographical error in an earlier draft. The correct coefficients are those shown in the figure and reported elsewhere in the text (evidence-specific effects: for PDM RTs, SD β = −0.057, p < 0.001; for VDM RTs, VD β = −0.016, p = 0.011; non-relevant evidence terms are n.s.). We removed the erroneous value.
Revision:
Results → Behaviour: validity… (sentence with −0.254). Delete the incorrect value and retain the evidence-specific coefficients consistent with Fig. 1B–C.
- It is reported that reaction times were significantly faster for the perceptual relative to the value-based decision task. Was overall accuracy also significantly different between the two tasks? It appears from Figure 3 that it might be, But I couldn't find this reported in the text.
To avoid conflating task with evidence composition, we did not emphasize between-task accuracy averages. Our primary tests examine evidence-specific effects and TMS-induced changes within task. For completeness, we now report descriptive mean accuracies by task and point readers to the figure panels that display accuracy as a function of evidence (which is the meaningful comparison in our matched-evidence design). We refrain from additional hypothesis testing here to keep the analyses aligned with our preregistered focus.
Revision:
Results → Behaviour: validity… Add:
“For completeness, group-mean accuracies by task are provided descriptively in Fig. 3a; inferential tests in the manuscript focus on evidence-specific effects and TMS-induced changes within task.”
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public review):
Lack of Sensitivity Analyses for some Key Methodological Decisions: Certain methodological choices in this manuscript diverge from approaches used in previous works. In these cases, I recommend the following: (i) The authors could provide a clear and detailed justification for these deviations from established methods, and (ii) supplementary sensitivity analyses could be included to ensure the robustness of the findings, demonstrating that the results are not driven primarily by these methodological changes. Below, I outline the main areas where such evaluations are needed:
This detailed guidance is incredibly valuable, and we are grateful. Work of this kind is in its relative infancy, and there are so many design choices depending on the data available, questions being addressed, and so on. Help us navigate that has been extremely useful. In our revised manuscript we are very happy to add additional justification for design choices made, and wherever possible test the impact of those choices. It is certainly the case that different approaches have been used across the handful of papers published in this space, and, unlike in other areas of systems neuroscience, we have yet to reach the point where any of these approaches are established. We agree with the reviewer that wherever possible these design choices should be tested.
Use of Communicability Matrices for Structural Connectivity Gradients: The authors chose to construct structural connectivity gradients using communicability matrices, arguing that diffusion map embedding "requires a smooth, fully connected matrix." However, by definition, the creation of the affinity matrix already involves smoothing and ensures full connectedness. I recommend that the authors include an analysis of what happens when the communicability matrix step is omitted. This sensitivity test is crucial, as it would help determine whether the main findings hold under a simpler construction of the affinity matrix. If the results significantly change, it could indicate that the observations are sensitive to this design choice, thereby raising concerns about the robustness of the conclusions. Additionally, if the concern is related to the large range of weights in the raw structural connectivity (SC) matrix, a more conventional approach is to apply a log-transformation to the SC weights (e.g., log(1+𝑆𝐶<sub>𝑖𝑗</sub>)), which may yield a more reliable affinity matrix without the need for communicability measures.
The reason we used communicability is indeed partly because we wanted to guarantee a smooth fully connected matrix, but also because our end goal for this project was to explore structure-function coupling in these low-dimensional manifolds. Structural communicability – like standard metrics of functional connectivity – includes both direct and indirect pathways, whereas streamline counts only capture direct communication. In essence we wanted to capture not only how information might be routed from one location to another, but also the more likely situation in which information propagates through the system.
In the revised manuscript we have given a clearer justification for why we wanted to use communicability as our structural measure (Page 4, Line 179):
“To capture both direct and indirect paths of connectivity and communication, we generated weighted communicability matrices using SIFT2-weighted fibre bundle capacity (FBC). These communicability matrices reflect a graph theory measure of information transfer previously shown to maximally predict functional connectivity (Esfahlani et al., 2022; Seguin et al., 2022). This also foreshadowed our structure-function coupling analyses, whereby network communication models have been shown to increase coupling strength relative to streamline counts (Seguin et al., 2020)”.
We have also referred the reader to a new section of the Results that includes the structural gradients based on the streamline counts (Page 7, line 316):
“Finally, as a sensitivity analysis, to determine the effect of communicability on the gradients, we derived affinity matrices for both datasets using a simpler measure: the log of raw streamline counts. The first 3 components derived from streamline counts compared to communicability were highly consistent across both NKI (r<sub>s</sub> = 0.791, r<sub>s</sub> = 0.866, r<sub>s</sub> = 0.761) and the referred subset of CALM (r<sub>s</sub> = 0.951, r<sub>s</sub> = 0.809, r<sub>s</sub> = 0.861), suggesting that in practice the organisational gradients are highly similar regardless of the SC metric used to construct the affinity matrices”.
Methodological ambiguity/lack of clarity in the description of certain evaluation steps: Some aspects of the manuscript’s methodological description are ambiguous, making it challenging for future readers to fully reproduce the analyses based on the information provided. I believe the following sections would benefit from additional detail and clarification:
Computation of Manifold Eccentricity: The description of how eccentricity was computed (both in the results and methods sections) is unclear and may be problematic. The main ambiguity lies in how the group manifold origin was defined or computed. (1) In the results section, it appears that separate manifold origins were calculated for the NKI and CALM groups, suggesting a dataset-specific approach. (2) Conversely, the methods section implies that a single manifold origin was obtained by somehow combining the group origins across the three datasets, which seems contradictory. Moreover, including neurodivergent individuals in defining the central group manifold origin in conceptually problematic. Given that neurodivergent participants might exhibit atypical brain organization, as suggested by Figure 1, this inclusion could skew the definition of what should represent a typical or normative brain manifold. A more appropriate approach might involve constructing the group manifold origin using only the neurotypical participants from both the NKI and CALM datasets. Given the reported similarity between group-level manifolds of neurotypical individuals in CALM and NKI, it would be reasonable to expect that this combined origin should be close to the origin computed within neurotypical samples of either NKI or CALM. As a sanity check, I recommend reporting the distance of the combined neurotypical manifold origin to the centres of the neurotypical manifolds in each dataset. Moreover, if the manifold origin was constructed while utilizing all samples (including neurodivergent samples) I think this needs to be reconsidered.
This is a great point, and we are very happy to clarify. Separate manifolds were calculated for the NKI and CALM participants, hence a dataset-specific approach. Indeed, in the long-run our goal was to explore individual differences in these manifolds, relative to the respective group-level origins, and their intersection across modalities, so manifold eccentricity was calculated at an individual level for subsequent analyses. At the group level, for each modality, we computed 3 manifold origins: one for NKI, one for the referred subset of CALM, and another for the neurotypical portion of CALM. Crucially, because the manifolds are always normal, in each case the manifold origin point is near-zero (extremely near-zero, to the 6<sup>th</sup> or 7<sup>th</sup> decimal place). In other words, we do indeed calculate the origin separately each time we calculate the gradients, but the origin is zero in every case. As a result, differences in the origin point cannot be the source of any differences we observe in manifold eccentricity between groups or individuals. We have updated the Methods section with the manifold origin points for each dataset and clarified our rationale (Page 16, Line 1296):
“Note that we used a dataset-specific approach when we computed manifold eccentricity for each of the three groups relative to their group-level origin: neurotypical CALM (SC origin = -7.698 x 10<sup>-7</sup>, FC origin = 6.724 x 10<sup>-7</sup>), neurodivergent CALM (SC origin = -6.422 x 10 , FC origin = 1.363 x 10 ), and NKI (SC origin = -7.434 x 10 , FC origin = 4.308 x 10<sup>-6</sup>). Eccentricity is a relative measure and thus normalised relative to the origin. Because of this normalisation, each time gradients are constructed the manifold origin is necessarily near-zero, meaning that differences in manifold eccentricity of individual nodes, either between groups or individuals, are stem from the eccentricity of that node rather than a difference in origin point”.
We clarified the computation of the respective manifold origins within the Results section, and referred the reader to the relevant Methods section (Page 9, line 446):
“For each modality (2 levels: SC and FC) and dataset (3 levels: neurotypical CALM, neurodivergent CALM, and NKI), we computed the group manifold origin as the mean of their respective first three gradients. Because of the normal nature of the manifolds this necessarily means that these origin points will be very near-zero, but we include the exact values in the ‘Manifold Eccentricity’ methodology sub-section”.
Individual-Level Gradients vs. Group-Level Gradients: Unlike previous studies that examined alterations in principal gradients (e.g., Xia et al., 2022; Dong et al., 2021), this manuscript focuses on gradients derived directly from individual-level data. In contrast, earlier works have typically computed gradients based on grouped data, such as using a moving window of individuals based on age (Xia et al.) or evaluating two distinct age groups (Dong et al.). I believe it is essential to assess the sensitivity of the findings to this methodological choice. Such an evaluation could clarify whether the observed discrepancies with previous reports are due to true biological differences or simply a result of different analytical strategies.
This is a brilliant point. The central purpose of our project was to test how individual differences in these gradients, and their intersection across modalities, related to differences in phenotype (e.g. cognitive difficulties). These necessitated calculating gradients at the level of individuals and building a pipeline to do so, given that we could find no other examples. Nonetheless, despite this different goal and thus approach, we had expected to replicate a couple of other key findings, most prominently the ‘swapping’ of gradients shown by Dong et al. (2021). We were also surprised that we did not find this changing in order. The reviewer is right and there could be several design features that produce the difference, and in the revised manuscript we test several of them. We have added the following text to the manuscript as a sensitivity analysis for the Results sub-section titled “Stability of individual-level gradients across developmental time” (Page 7, Line 344 onwards):
“One possibility is that our observation of gradient stability – rather than a swapping of the order for the first two gradients (Dong et al., 2021) – is because we calculated them at an individual level. To test this, we created subgroups and contrasted the first two group-level structural and functional gradients derived from children (younger than 12 years old) versus those from adolescents (12 years old and above), using the same age groupings as prior work (Dong et al., 2021). If our use of individually calculated gradients produces the stability, then we should observe the swapping of gradients in this sensitivity analysis. Using baseline scans from NKI, the primary structural gradient in childhood (N = 99) as shown in Figure 1f, this was highly correlated (r<sub>s</sub> = 0.995) with those derived from adolescents (N = 123). Likewise, the secondary structural gradient in childhood was highly consistent in adolescence (r<sub>s</sub> = 0.988). In terms of functional connectivity, the principal gradient in childhood (N = 88) was highly consistent in adolescence (r<sub>s</sub> = 0.990, N = 125). The secondary gradient in childhood was again highly similar in adolescence (r<sub>s</sub> = 0.984). The same result occurred in the CALM dataset: In the baseline referred subset of CALM, the primary and secondary communicability gradients derived from children (N = 258) and adolescents (N = 53) were near-identical (r<sub>s</sub> = 0.991 and r<sub>s</sub> = 0.967, respectively). Alignment for the primary and secondary functional gradients derived from children (N = 130) and adolescents (N = 43) were also near-identical (r<sub>s</sub> = 0.972 and r<sub>s</sub> = 0.983, respectively). These consistencies across development suggest that gradients of communicability and functional connectivity established in childhood are the same as those in adolescence, irrespective of group-level or individual-level analysis. Put simply, our failure to replicate the swapping of gradient order in Dong et al. (2021) is not the result of calculating gradients at the level of individual participants.”
Procrustes Transformation: It is unclear why the authors opted to include a Procrustes transformation in this analysis, especially given that previous related studies (e.g., Dong et al.) did not apply this step. I believe it is crucial to evaluate whether this methodological choice influences the results, particularly in the context of developmental changes in organizational gradients. Specifically, the Procrustes transformation may maximize alignment to the group-level gradients, potentially masking individual-level differences. This could result in a reordering of the gradients (e.g., swapping the first and second gradients), which might obscure true developmental alterations. It would be informative to include an analysis showing the impact of performing vs. omitting the Procrustes transformation, as this could help clarify whether the observed effects are robust or an artifact of the alignment procedure. (Please also refer to my comment on adding a subplot to Figure 1). Additionally, clarifying how exactly the transformation was applied to align gradients across hemispheres, individuals, and/or datasets would help resolve ambiguity.
The current study investigated individual differences in connectome organisation, rather than group-level trends (Dong et al., 2021). This necessitates aligning individual gradients to the corresponding group-level template using a Procrustes rotation. Without a rotation, there is no way of knowing if you are comparing ‘like with like’: the manifold eccentricity of a given node may appear to change across individuals simply due to subtle differences in the arbitrary orientation of the underlying manifolds. We also note that prior work examining individual differences in principal alignment have used Procrustes (Xia et al., 2022), who demonstrated emergence of the principal gradient across development, albeit with much smaller effects than Dong and colleagues (2021). Nonetheless, we agree, the Procrustes rotation could be another source of the differences we observed with the previous paper (Dong et al. 2021). We explored the impact of the Procrustes rotation on individual gradients as our next sensitivity analysis. We recalculated everyone’s gradients without Procrustes rotation. We then tested the alignment of each participant with the group-level gradients using Spearman’s correlations, followed by a series of generalised linear models to predict principal gradient alignment using head motion, age, and sex. The expected swapping of the first and second functional gradient (Dong et al., 2021) would be represented by a decrease in the spatial similarity of each child’s principal functional gradient to the principal childhood group-level gradient, at the onset of adolescence (~age 12). However, there is no age effect on this unrotated alignment, suggesting that the lack of gradient swapping in our data does not appear to be the result of the Procrustes rotation. When you use unrotated individual gradients the alignment is remarkably consistent across childhood and adolescence. Alignment is, however, related to head motion, which is often related to age. To emphasise the importance of motion, particularly in relation to development, we conducted a mediation analysis between the relationship between age and principal alignment (without correcting for motion), with motion as a mediator, within the NKI dataset. Before accounting for motion, the relationship between age and principal alignment is significant, but this can be entirely accounted for by motion. In our revised manuscript we have included this additional analysis in the Results sub-section titled “Stability of individual-level gradients across developmental time”, following on from the above point about the effect of group-level versus individual-level analysis (Page 8, Line 400):
“A second possible discrepancy between our results and that of prior work examining developmental change in group-level functional gradients (Dong et al., 2021) was the use of Procrustes alignment. Such alignment of individual-level gradients to group-level templates is a necessary step to ensure valid comparisons between corresponding gradients across individuals, and has been implemented in sliding-window developmental work tracking functional gradient development (Xia et al., 2022). Nonetheless, we tested whether our observation of stable principal functional and communicability gradients may be an artefact of the Procrustes rotation. We did this by modelling how individual-level alignment without Procrustes rotation to the group-level templates varies with age, head motion, and sex, as a series of generalised linear models. We included head motion as the magnitude of the Procrustes rotation has been shown to be positively correlated with mean framewise displacement (Sasse et al., 2024), and prior group-level work (Dong et al., 2021) included an absolute motion threshold rather than continuous motion estimates. Using the baseline referred CALM sample, there was no significant relationship between alignment and age (β = -0.044, 95% CI = [-0.154, 0.066], p = 0.432) after accounting for head motion and sex. Interestingly, however head motion was significantly associated with alignment ( β = -0.318, 95% CI = [-0.428, -.207], p = 1.731 x 10<sup>-8</sup>), such that greater head motion was linked to weaker alignment. Note that older children tended to have exhibit less motion for their structural scans (r<sub>s</sub> = 0.335, p < 0.001). We observed similar trends in functional alignment, whereby tighter alignment was significantly predicted by lower head motion (β = -0.370, 95% CI = [-0.509, -0.231], p = 1.857 x 10<sup>-7</sup>), but not by age (β= 0.049, 95% CI = [-0.090, 0.187], p = 0.490). Note that age and head motion for functional scans were not significantly related (r<sub>s</sub> = -0.112, p = 0.137). When repeated for the baseline scans of NKI, alignment with the principal structural gradient was not significantly predicted by either scan age (β = 0.019, 95% CI = [-0.124, 0.163], p = 0.792) or head motion (β = -0.133, 95% CI = [-0.175, 0.009], p = 0.067) together in a single model, where age and motion were negatively correlated (r<sub>s</sub> = -0.355, p < 0.001). Alignment with the principal functional gradient was significantly predicted by head motion (β = -0.183, 95% CI = [-0.329, -0.036], p = 0.014) but not by age (β= 0.066, 95% CI = [-0.081, 0.213], p = 0.377), where age and motion were also negatively correlated (r<sub>s</sub> = -0.412, p < 0.001). Across modalities and datasets, alignment with the principal functional gradient in NKI was the only example in which there was a significant correlation between alignment and age (r<sub>s</sub> = 0.164, p = 0.017) before accounting for head motion and sex. This suggests that apparent developmental effects on alignment are minimal, and where they do exist they are removed after accounting for head motion. Put together this suggests that the lack of order swapping for the first two gradients is not the result of the Procrustes rotation – even without the rotation there is no evidence for swapping”.
“To emphasise the importance of head motion in the appearance of developmental change in alignment, we examined whether accounting for head motion removes any apparent developmental change within NKI. Specifically, we tested whether head motion mediates the relationship between age and alignment (Figure 1X), controlling for sex, given that higher motion is associated with younger children (β= -0.429, 95% CI = [0.552, -0.305], p = 7.957 x 10<sup>-11</sup>), and stronger alignment is associated with reduced motion (β = -0.211, 95% CI = [-0.344, -0.078], p = 2.017 x 10<sup>-3</sup>). Motion mediated the relationship between age and alignment (β = 0.078, 95% CI = [0.006, 0.146], p = 1.200 x 10<sup>-2</sup>), accounting for 38.5% variance in the age-alignment relationship, such that the link between age and alignment became non-significant after accounting for motion (β = 0.066, 95% CI = [-0.081, 0.214], p = 0.378). This firstly confirms our GLM analyses, where we control for motion and find no age associations. Moreover, this suggests that caution is required when associations between age and gradients are observed. In our analyses, because we calculate individual gradients, we can correct for individual differences in head motion in all our analyses. However, other than using an absolute motion threshold and motion-matched child and adolescent groups, individual differences in motion were not accounted for by prior work which demonstrated a flipping of the principal functional gradients with age (Dong et al., 2021)”.
We further clarify the use of Procrustes rotation as a separate sub-section within the Methods (Page 25, Line 1273):
“Procrustes Rotation
For group-level analysis, for each hemisphere we constructed an affinity matrix using a normalized angle kernel and applied diffusion-map embedding. The left hemisphere was then aligned to the right using a Procrustes rotation. For individual-level analysis, eigenvectors for the left hemisphere were aligned with the corresponding group-level rotated eigenvectors. No alignment was applied across datasets. The only exception to this was for structural gradients derived from the referred CALM cohort. Specifically, we aligned the principal gradient of the left hemisphere to the secondary gradient of the right hemisphere: this was due to the first and second gradients explaining a very similar amount of variance, and hence their order was switched”.
SC-FC Coupling Metric: The approach used to quantify nodal SC-FC coupling in this study appears to deviate from previously established methods in the field. The manuscript describes coupling as the "Spearman-rank correlation between Euclidean distances between each node and all others within structural and functional manifolds," but this description is unclear and lacks sufficient detail. Furthermore, this differs from what is typically referred to as SC-FC coupling in the literature. For instance, the cited study by Park et al. (2022) utilizes a multiple linear regression framework, where communicability, Euclidean distance, and shortest path length are independent variables predicting functional connectivity (FC), with the adjusted R-squared score serving as the coupling index for each node. On the other hand, the Baum et al. (2020) study, also cited, uses Spearman correlation, but between raw structural connectivity (SC) and FC values. If the authors opt to introduce a novel coupling metric, it is essential to demonstrate its similarity to these previous indices. I recommend providing an analysis (supplementary) showing the correlation between their chosen metric and those used in previous studies (e.g., the adjusted R-squared scores from Park et al. or the SC-FC correlation from Baum et al.). Furthermore, if the metrics are not similar and results are sensitive to this alternative metric, it raises concerns about the robustness of the findings. A sensitivity analysis would therefore be helpful (in case the novel coupling metric is not like previous ones) to determine whether the reported effects hold true across different coupling indices.
This is a great point, and we are happy to take the reviewer’s recommendation. There are multiple different ways of calculating structure-function coupling. For our set of questions, it was important that our metric incorporated information about the structural and functional manifolds, rather than being a separate approach that is unrelated to these low-dimensional embeddings. Put simply, we wanted our coupling measure to be about the manifolds and gradients outlined in the early sections of the results. We note that the multiple linear regression framework was developed by Vázquez-Rodríguez and colleagues (2019), whilst the structure-function coupling computed in manifold space by Park and colleagues (2022) was operationalised as a linear correlation between z-transformed functional connectomes and structural differentiation eigenvectors. To clarify how this coupling was calculated, and to justify why we developed a new coupling method based on manifolds rather than borrow an existing approach from the literature, we have revised the manuscript to make this far clearer for readers (Page 13, line 604):
“To examine the relationship between each node’s relative position in structural and functional manifold space, we turned our attention to structure-function coupling. Whilst prior work typically computed coupling using raw streamline counts and functional connectivity matrices, either as a correlation (Baum et al., 2020) or through a multiple linear regression framework (Vázquez-Rodríguez et al., 2019), we opted to directly incorporate low-dimensional embeddings within our coupling framework. Specifically, as opposed to correlating row-wise raw functional connectivity with structural connectivity eigenvectors (Park et al., 2022), our metric directly incorporates the relative position of each node in low-dimensional structural and functional manifold spaces. Each node was situated in a low-dimensional 3D space, the axes of which were each participant’s gradients, specific to each modality. For each participant and each node, we computed the Euclidean distance with all other nodes within structural and functional manifolds separately, producing a vector of size 200 x 1 per modality. The nodal coupling coefficient was the Spearman correlation between each node’s Euclidean distance to all other nodes in structural manifold space, and that in functional manifold space. Put simply, a strong nodal coupling coefficient suggests that that node occupies a similar location in structural space, relative to all other nodes, as it does in functional space”.
We also agree with the reviewer’s recommendation to compare this to some of the more standard ways of calculating coupling. We compare our metric with 3 others (Baum et al., 2020; Park et al., 2022; VázquezRodríguez et al., 2019), and find that all metrics capture the core developmental sensorimotor-to-association axis (Sydnor et al., 2021). Interestingly, manifold-based coupling measures captured this axis more strongly than non-manifold measures. We have updated the Results accordingly (Page 14, Line 638):
“To evaluate our novel coupling metric, we compared its cortical spatial distribution to three others (Baum et al., 2020; Park et al., 2022; Vázquez-Rodríguez et al., 2019), using the group-level thresholded structural and functional connectomes from the referred CALM cohort. As shown in Figure 4c, our novel metric was moderately positively correlated to that of a multi-linear regression framework (r<sub>s</sub> = 0.494, p<sub>spin</sub> = 0.004; Vázquez-Rodríguez et al., 2019) and nodal correlations of streamline counts and functional connectivity (r<sub>s</sub> = 0.470, p<sub>spin</sub> = 0.005; Baum et al., 2020). As expected, our novel metric was strongly positively correlated to the manifold-derived coupling measure (r<sub>s</sub> = 0.661, p<sub>spin</sub> < 0.001; Park et al., 2022), more so than the first (Z(198) = 3.669, p < 0.001) and second measure (Z(198) = 4.012, p < 0.001). Structure-function coupling is thought to be patterned along a sensorimotor-association axis (Sydnor et al., 2021): all four metrics displayed weak-tomoderate alignment (Figure 4c). Interestingly, the manifold-based measures appeared most strongly aligned with the sensorimotor-association axis: the novel metric was more strongly aligned than the multi-linear regression framework (Z(198) = -11.564, p < 0.001) and the raw connectomic nodal correlation approach (Z(198) = -10.724, p < 0.001), but the previously-implemented structural manifold approach was more strongly aligned than the novel metric (Z(198) = -12.242, p < 0.001). This suggests that our novel metric exhibits the expected spatial distribution of structure-function coupling, and the manifold approach more accurately recapitulates the sensorimotor-association axis than approaches based on raw connectomic measures”.
We also added the following to the legend of Figure 4 on page 15:
“d. The inset Spearman correlation plot of the 4 coupling measures shows moderate-to-strong correlations (p<sub>spin</sub> < 0.005 for all spatial correlations). The accompanying lollypop plot shows the alignment between the sensorimotor-to-association axis and each of the 4 coupling measures, with the novel measure coloured in light purple (p<sub>spin</sub> < 0.007 for all spatial correlations)”.
Prediction vs. Association Analysis: The term “prediction” is used throughout the manuscript to describe what appear to be in-sample association tests. This terminology may be misleading, as prediction generally implies an out-of-sample evaluation where models trained on a subset of data are tested on a separate, unseen dataset. If the goal of the analyses is to assess associations rather than make true predictions, I recommend refraining from the term “prediction” and instead clarifying the nature of the analysis. Alternatively, if prediction is indeed the intended aim (which would be more compelling), I suggest conducting the evaluations using a k-fold cross-validation framework. This would involve training the Generalized Additive Mixed Models (GAMMs) on a portion of the data and training their predictive accuracy on a held-out sample (i.e. different individuals). Additionally, the current design appears to focus on predicting SC-FC coupling using cognitive or pathological dimensions. This is contrary to the more conventional approach of predicting behavioural or pathological outcomes from brain markers like coupling. Could the authors clarify why this reverse direction of analysis was chosen? Understanding this choice is crucial, as it impacts the interpretation and potential implications of the findings.
We have replaced “prediction” with “association” across the manuscript. However, for analyses corresponding to Figure 5, which we believe to be the most compelling, we conducted a stratified 5-fold cross-validation procedure, outlined below, repeated 100 times to account for random variation in the train-test splits. To assess whether prediction accuracy in the test splits was significantly greater than chance, we compared our results to those derived from a null dataset in which cognitive factor 2 scores had been permuted across participants. To account for the time-series element and block design of our data, in that some participants had 2 or more observations, we permuted entire participant blocks of cognitive factor 2 scores, keeping all other variables, including covariates, the same. Included in our manuscript are methodological details and results pertaining to this procedure. Specifically, the following has been added to the Results (Page 16, Line 758):
“To examine the predictive value of the second cognitive factor for global and network-level structure-function coupling, operationalised as a Spearman rank correlation coefficient, we implemented a stratified 5-fold crossvalidation framework, and predictive accuracy compared with that of a null data frame with cognitive factor 2 scores permuted across participant blocks (see ‘GAMM cross-validation’ in the Methods). This procedure was repeated 100 times to account for randomness in the train-test splits, using the same model specification as above. Therefore, for each of the 5 network partitions in which an interaction between the second cognitive factor and age was a significant predictor of structure-function coupling (global, visual, somato-motor, dorsal attention, and default-mode), we conducted a Welch’s independent-sample t-test to compare 500 empirical prediction accuracies with 500 null prediction accuracies. Across all 5 network partitions, predictive accuracy of coupling was significantly higher than that of models trained on permuted cognitive factor 2 scores (all p < 0.001). We observed the largest difference between empirical (M = 0.029, SD = 0.076) and null (M = -0.052, SD = 0.087) prediction accuracy in the somato-motor network [t (980.791) = 15.748, p < 0.001, Cohen’s d = 0.996], and the smallest difference between empirical (M = 0.080, SD = 0.082) and null (M = 0.047, SD = 0.081) prediction accuracy in the dorsal attention network [t (997.720) = 6.378, p < 0.001, Cohen’s d = 0.403]. To compare relative prediction accuracies, we ordered networks by descending mean accuracy and conducted a series of Welch’s independent sample t-tests, followed by FDR correction (Figure 5X). Prediction accuracy was highest in the default-mode network (M = 0.265, SD = 0.085), two-fold that of global coupling (t(992.824) = 25.777, p<sub>FDR</sub> = 5.457 x 10<sup>-112</sup>, Cohen’s d = 1.630, M = 0.131, SD = 0.079). Global prediction accuracy was significantly higher than the visual network (t (992.644) = 9.273, p<sub>FDR</sub> = 1.462 x 10<sup>-19</sup>, Cohen’s d = 0.586, M = 0.083, SD = 0.085), but visual prediction accuracy was not significantly higher than within the dorsal attention network (t (997.064) = 0.554, p<sub>FDR</sub> = 0.580, Cohen’s d = 0.035, M = 0.080, SD = 0.082). Finally, prediction accuracy within the dorsal attention network was significantly stronger than that of the somato-motor network [t (991.566) = 10.158, p<sub>FDR</sub> = 7.879 x 10<sup>-23</sup>, Cohen’s d = 0.642 M = 0.029, SD = 0.076]. Together, this suggests that out-of-sample developmental predictive accuracy for structure-function coupling, using the second cognitive factor, is strongest in the higher-order default-mode network, and lowest in the lower-order somatosensory network”.
We have added a separate section for GAMM cross-validation in the Methods (Page 27, Line 1361):
GAMM cross-validation
“We implemented a 5-fold cross validation procedure, stratified by dataset (2 levels: CALM or NKI). All observations from any given participant were assigned to either the testing or training fold, to prevent data leakage, and the cross-validation procedure was repeated 100 times, to account for randomness in data splits. The outcome was predicted global or network-level structure-function coupling across all test splits, operationalised as the Spearman rank correlation coefficient. To assess whether prediction accuracy exceeded chance, we compared empirical prediction accuracy with that of GAMMs trained and tested on null data in which cognitive factor 2 scores were permuted across subjects. The number of observations formed 3 exchangeability blocks (N = 320 with one observation, N = 105 with two observations, and N = 33 with three observations), whereby scores from a participant with two observations were replaced by scores from another participant with two observations, with participant-level scores kept together, and so on for all numbers of observations. We compared empirical and null prediction accuracies using independent sample t-tests as, although the same participants were examined, the shuffling meant that the relative ordering of participants within both distributions was not preserved. For parallelisation and better stability when estimating models fit on permuted data, we used the bam function from the mgcv R package (Wood, 2017)”.
We also added a justification for why we predicted coupling using behaviour or psychopathology, rather than vice versa (Page 27, Line 1349):
“When using our GAMMs to test for the relationship between cognition and psychopathology and our coupling metrics, we opted to predict structure-function coupling using cognitive or psychopathological dimensions, rather than vice versa, to minimise multiple comparisons. In the current framework, we corrected for 8 multiple comparisons within each domain. This would have increased to 16 multiple comparison corrections for predicting two cognitive dimensions using network-level coupling, and 24 multiple comparison corrections for predicting three psychopathology dimensions. Incorporating multiple networks as predictors within the same regression framework introduces collinearity, whilst the behavioural dimensions were orthogonal: for example, coupling is strongly correlated between the somato-motor and ventral attention networks (r<sub>s</sub> = 0.721), between the default-mode and frontoparietal networks (r<sub>s</sub> = 0.670), and between the dorsal attention and fronto-parietal networks (r<sub>s</sub> = 0.650)”.
Finally, we noticed a rounding error in the ages of the data frame containing the structure-function coupling values and the cognitive/psychopathology dimensions. We rectified this and replaced the GAMM results, which largely remained the same.
In typical applications of diffusion map embedding, sparsification (e.g., retaining only the top 10 of the strongest connections) is often employed at the vertex-level resolution to ensure computational feasibility. However, since the present study performs the embedding at the level of 200 brain regions (a considerably coarser resolution), this step may not be necessary or justifiable. Specifically, for FC, it might be more appropriate to retain all positive connections rather than applying sparsification, which could inadvertently eliminate valuable information about lower-strength connections. Whereas for SC, as the values are strictly non-negative, retaining all connections should be feasible and would provide a more complete representation of the structural connectivity patterns. Given this, it would be helpful if the authors could clarify why they chose to include sparsification despite the coarser regional resolution, and whether they considered this alternative approach (using all available positive connections for FC and all non-zero values for SC). It would be interesting if the authors could provide their thoughts on whether the decision to run evaluations at the resolution of brain regions could itself impact the functional and structural manifolds, their alteration with age, and or their stability (in contrast to Dong et al. which tested alterations in highresolution gradients).
This is another great point. We could retain all connections, but we usually implement some form of sparsification to reduce noise, particularly in the case of functional connectivity. But we nonetheless agree with the reviewer’s point. We should check what impact this is having on the analysis. In brief, we found minimal effects of thresholding, suggesting that the strongest connections are driving the gradient (Page 7, Line 304):
“To assess the effect of sparsity on the derived gradients, we examined group-level structural (N = 222) and functional (N = 213) connectomes from the baseline session of NKI. The first three functional connectivity gradients derived using the full connectivity matrix (density = 92%) were highly consistent with those obtained from retaining the strongest 10% of connections in each row (r<sub>1</sub> = 0.999, r<sub>2</sub> = 0.998, r<sub>3</sub> < 0.999, all p < 0.001). Likewise, the first three communicability gradients derived from retaining all streamline counts (density = 83%) were almost identical to those obtained from 10% row-wise thresholding (r<sub>1</sub> = 0.994, r<sub>2</sub> = 0.963, r<sub>3</sub> = 0.955, all p < 0.001). This suggests that the reported gradients are driven by the strongest or most consistent connections within the connectomes, with minimal additional information provided by weaker connections. In terms of functional connectivity, such consistency reinforces past work demonstrating that the sensorimotor-toassociation axis, the major axis within the principal functional connectivity gradient, emerges across both the top- and bottom-ranked functional connections (Nenning et al., 2023)”.
Furthermore, we appreciate the nudge to share our thoughts on whether the difference between vertex versus nodal metrics could be important here, particularly regarding thresholds. To combine this point with R2’s recommendation to expand the Discussion, we have added the following paragraph (Page 19, Line 861):
“We consider the role of thresholding, cortical resolution, and head motion as avenues to reconcile the present results with select reports in the literature (Dong et al., 2021; Xia et al., 2022). We would suggest that thresholding has a greater effect on vertex-level data, rather than parcel-level. For example, a recent study revealed that the emergence of principal vertex-level functional connectivity gradients in childhood and adolescence are indeed threshold-dependent (Dong et al., 2024). Specifically, the characteristic unimodal organisation for children and transmodal organisation for adolescents only emerged at the 90% threshold: a 95% threshold produced a unimodal organisation in both groups, whilst an 85% threshold produced a transmodal organisation in both groups. Put simply, the ‘swapping’ of gradient orders only occurs at certain thresholds. Furthermore, our results are not necessarily contradictory to this prior report (Dong et al., 2021): developmental changes in high-resolution gradients may be supported by a stable low-dimensional coarse manifold. Indeed, our decision to use parcellated connectomes was partly driven by recent work which demonstrated that vertex-level functional gradients may be derived using biologically-plausible but random data with sufficient spatial smoothing, whilst this effect is minimal at coarser resolutions (Watson & Andrews, 2023). We observed a gradual increase in the variance of individual connectomes accounted for by the principal functional connectivity gradient in the referred subset of CALM, in line with prior vertex-level work demonstrating a gradual emergence of the sensorimotor-association axis as the principal axis of connectivity (Xia et al., 2022), as opposed to a sudden shift. It is also possible that vertex-level data is more prone to motion artefacts in the context of developmental work. Transitioning from vertex-level to parcel-level data involves smoothing over short-range connectivity, thus greater variability in short-range connectivity can be observed in vertex-level data. However, motion artefacts are known to increase short-range connectivity and decrease long-range connectivity, mimicking developmental changes (Satterthwaite et al., 2013). Thus, whilst vertexlevel data offers greater spatial resolution in representation of short-range connectivity relative to parcel-level data, it is possible that this may come at the cost of making our estimates of the gradients more prone to motion”.
Evaluating the consistency of gradients across development: the results shown in Figure 1e are used as evidence suggesting that gradients are consistent across ages. However, I believe additional analyses are required to identify potential sources of the observed inconsistency compared to previous works. The claim that the principal gradient explains a similar degree of variance across ages does not necessarily imply that the spatial structure remains the same. The observed variance explanation is hence not enough to ascertain inconsistency with findings from Dong et al., as the spatial configuration of gradients may still change over time. I suggest the following additional analyses to strengthen this claim. Alignment to group-level gradients: Assess how much of the variance in individual FC matrices is explained by each of the group-level gradients (G1, G2, and G3, for both FC and SC). This analysis could be visualized similarly to Figure 1e, with age on the x-axis and variance explained on the y-axis. If the explained variance varies as a function of age, it may indicate that the gradients are not as consistent as currently suggested.
This is another great suggestion. In the additional analyses above (new group-level analyses and unrotated gradient analyses) we rule-out a couple of the potential causes of the different developmental trends we observe in our data – namely the stability of the gradients over time. The suggested additional analysis is a great idea, and we have implemented it as follows (Page 8, Line 363):
“To evaluate the consistency of gradients across development, across baseline participants with functional connectomes from the referred CALM cohort (N = 177), we calculated the proportion of variance in individuallevel connectomes accounted for by group-level functional gradients. Specifically, we calculated the proportion of variance in an adjacency matrix A accounted for by the vector v<sub>i</sub> as the fraction of the square of the scalar projection of v<sub>i</sub> onto A, over the Frobenius norm of A. Using a generalised linear model, we then tested whether the proportion of variance explained varies systematically with age, controlling for sex and headmotion. The variance in individual-level functional connectomes accounted for by the group-level principal functional gradient gradually increased with development (β= 0.111, 95% CI = [0.022, 0.199], p = 1.452 x 10<sup>-2</sup>, Cohen’s d = 0.367), as shown in Figure 1g, and decreased with higher head motion ( β = -10.041, 95% CI = [12.379, -7.702], p = 3.900 x 10<sup>-17</sup>), with no effect of sex (β= 0.071, 95% CI = [-0.380, 0.523], p = 0.757). We observed no developmental effects on the variance explained by the second (r<sub>s</sub> = 0.112, p = 0.139) or third (r<sub>s</sub> = 0.053, p = 0.482) group-level functional gradient. When repeated with the baseline functional connectivity for NKI (N = 213), we observed no developmental effects (β = 0.097, 95% CI = [-0.035, 0.228], p = 0.150) on the variance explained by the principal functional gradient after accounting for motion (β= -3.376, 95% CI = [8.281, 1.528], p = 0.177) and sex (β = -0.368, 95% CI = [-1.078, 0.342], p = 0.309). However, we observed significant developmental correlations between age and variance (r<sub>s</sub> = 0.137, p = 0.046) explained before accounting for head motion and sex. We observed no developmental effects on the variance explained by the second functional gradient (r<sub>s</sub> = -0.066, p = 0.338), but a weak negative developmental effect on the variance explained by the third functional gradient (r<sub>s</sub> = -0.189, p = 0.006). Note, however, the magnitude of the variance accounted for by the third functional gradient was very small (all < 1%). When applied to communicability matrices in CALM, the proportion of variance accounted for by the group-level communicability gradient was negligible (all < 1%), precluding analysis of developmental change”.
“To further probe the consistency of gradients across development, we examined developmental changes in the standard deviation of gradient values, corresponding to heterogeneity, following prior work examining morphological (He et al., 2025) and functional connectivity gradients (Xia et al., 2022). Using a series of generalised linear models within the baseline referred subset of CALM, correcting for head motion and sex, we found that gradient variation for the principal functional gradient increased across development (= 0.219, 95% CI = [0.091, 0.347], p = 0.001, Cohen’s d = 0.504), indicating greater heterogeneity (Figure 1h), whilst gradient variation for the principal communicability gradient decreased across development (β = -0.154, 95% CI = [-0.267, -0.040], p = 0.008, Cohen’s d = -0.301), indicating greater homogeneity (Figure 1h). Note, a paired t-test on the 173 common participants demonstrated a significant effect of modality on gradient variability (t(172) = -56.639, p = 3.663 x 10<sup>-113</sup>), such that the mean variability of communicability gradients (M = 0.033, SD = 0.001) was less than half that of functional connectivity (M = 0.076, SD = 0.010). Together, this suggests that principal functional connectivity and communicability gradients are established early in childhood and display age-related refinement, but not replacement”.
The Issue of Abstraction and Benefits of the Gradient-Based View: The manuscript interprets the eccentricity findings as reflecting changes along the segregation-integration spectrum. Given this, it is unclear why a more straightforward analysis using established graph-theory metrics of segregationintegration was not pursued instead. Mapping gradients and computing eccentricity adds layers of abstraction and complexity. If similar interpretations can be derived directly from simpler graph metrics, what additional insights does the gradient-based framework offer? While the manuscript argues that this approach provides “a more unifying account of cortical reorganization”, it is not evident why this abstraction is necessary or advantageous over traditional graph metrics. Clarifying these benefits would strengthen the rationale for using this method.
This is a great point, and something we spent quite a bit of time considering when designing the analysis. The central goal of our project was to identify gradients of brain organisation across different datasets and modalities and then test how the organisational principles of those modalities align. In other words, how do structural and functional ‘spaces’ intersect, and does this vary across the cortex? That for us was the primary motivation for operationalising organisation as nodal location within a low-dimensional manifold space (Bethlehem et al., 2020; Gale et al., 2022; Park et al., 2021), using a simple composite measure to achieve compression, rather than as a series of graph metrics. The reason we subsequently calculated those graph metrics and tested for their association was simply to help us interpret what eccentricity within that lowdimensional space means. Manifold eccentricity was moderately positively correlated to graph-theory metrics of integration, leaving a substantial portion of variance unaccounted for, but that association we think is nonetheless helpful for readers trying to interpret eccentricity. However, since ME tells us about the relative position of a node in that low-dimensional space, it is also likely capturing elements of multiple graph theory measures. Following the Reviewer’s question, this is something we decided to test. Specifically, using 4 measures of segregation, including two new metrics requested by the Reviewer in a minor point (weighted clustering coefficient and normalized degree centrality), we conducted a dominance analysis (Budescu, 1993) with normalized manifold eccentricity of the group-level referred CALM structural connectome. We also detail the use of gradient measures in developmental contexts, and how they can be complementary to traditional graph theory metrics.
We have added the following to the Results section (Page 10, Lines 472 onwards):
“To further contextualise manifold eccentricity in terms of integration and segregation beyond simple correlations, we conducted a multivariate dominance analysis (Budescu, 1993) of four graph theory metrics of segregation as predictors of nodal normalized manifold eccentricity within the group-level referred CALM structural and functional connectomes (Figure 2c). A dominance analysis assesses the relative importance of each predictor in a multilinear regression framework by fitting 2<sup>n</sup> – 1 models (where n is the number of predictors) and calculating the relative increase in adjusted R2 caused by adding each predictor to the model across both main effects and interactions. A multilinear regression model including weighted clustering coefficient, within-module degree Z-score, participation coefficient and normalized degree centrality accounted for 59% of variance in nodal manifold eccentricity in the group-level CALM structural connectome. Withinmodule degree Z score was the most important predictor (40.31% dominance), almost twice that of the participation coefficient (24.03% dominance) and normalized degree centrality (24.05% dominance) which made roughly equal contributions. The least important predictor was the weighted clustering coefficient (11.62% dominance). When the same approach was applied for the group-level referred CALM functional connectome, the 4 predictors accounted for 52% variability. However, in contrast to the structural connectome, functional manifold eccentricity seemed to incorporate the same graph theory metrics in different proportions. Normalized degree centrality was the most important predictor (47.41% dominance), followed by withinmodule degree Z-score (24.27%), and then the participation coefficient (15.57%) and weighted clustering coefficient (12.76%) which made approximately equal contributions. Thus, whilst structural manifold eccentricity was dominated most by within-module degree Z-score and least by the weighted clustering coefficient, functional manifold eccentricity was dominated most by normalized degree centrality and least by the weighted clustering coefficient. This suggests that manifold mapping techniques incorporate different aspects of integration dependent on modality. Together, manifold eccentricity acts as a composite measure of segregation, being differentially sensitive to different aspects of segregation, without necessitating a priori specification of graph theory metrics. Further discussion of the value of gradient-based metrics in developmental contexts and as a supplement to traditional graph theory analyses is provided in the ‘Manifold Eccentricity’ methodology sub-section”.
We added further justification to the manifold eccentricity Methods subsection (Page 26, line 1283):
“Gradient-based measures hold value in developmental contexts, above and beyond traditional graph theory metrics: within a sample of over 600 cognitively-healthy adults aged between 18 and 88 years old, sensitivity of gradient-based within-network functional dispersion to age were stronger and more consistent across networks compared to segregation (Bethlehem et al., 2020). In the context of microstructural profile covariance, modules resolved by Louvain community detection occupied distinct positions across the principal two gradients, suggesting that gradients offer a way to meaningfully order discrete graph theory analyses (Paquola et al., 2019)”.
We added the following to the Introduction section outlining the application of gradients as cortex-wide coordinate systems (Page 3, Line 121):
“Using the gradient-based approach as a compression tool, thus forgoing the need to specify singular graph theory metrics a priori, we operationalised individual variability in low-dimensional manifolds as eccentricity (Gale et al., 2022; Park et al., 2021). Crucially, such gradients appear to be useful predictors of phenotypic variation, exceeding edge-level connectomics. For example, in the case of functional connectivity gradients, their predictive ability for externalizing symptoms and general cognition in neurotypical adults surpassed that of edge-level connectome-based predictive modelling (Hong et al., 2020), suggesting that capturing lowdimensional manifolds may be particularly powerful biomarkers of psychopathology and cognition”.
We also added the following to the Discussion section (Page 18, Line 839):
“By capitalising on manifold eccentricity as a composite measure of segregation across development, we build upon an emerging literature pioneering gradients as a method to establish underlying principles of structural (Paquola et al., 2020; Park et al., 2021) and functional (Dong et al., 2021; Margulies et al., 2016; Xia et al., 2022) brain development without a priori specification of specific graph theory metrics of interest”.
It is unclear whether the statistical tests finding significant dataset effects are capturing effects of neurotypical vs. Neurodivergent, or simply different scanners/sites. Could the neurotypical portion of CALM also be added to distinguish between these two sources of variability affecting dataset effects (i.e. ideally separating this to the effect of site vs. neurotypicality would better distinguish the effect of neurodivergence).
At a group-level, differences in the gradients between the two cohorts are very minor. Indeed, in the manuscript we describe these gradients as being seemingly ‘universal’. But we agree that we should test whether we can directly attribute any simple main effects of ‘dataset’ are resulting from the different site or the phenotype of the participants. The neurotypical portion of CALM (collected at the same site on the same scanner) helped us show that any minor differences in the gradient alignments is likely due to the site/scanner differences rather than the phenotype of the participants. We took the same approach for testing the simple main effects of dataset on manifold eccentricity. To better parse neurotypicality and site effects at an individual-level, we conducted a series of sensitivity analyses. First, in response to the reviewer’s earlier comment, we conducted a series of nodal generalized linear models for communicability and FC gradients derived from neurotypical and neurodivergent portions of CALM, alongside NKI, and tested for an effect of neurotypicality above and beyond scanner. As at the group level, having those additional scans on a ‘comparison’ sample for CALM is very helpful in teasing apart these effects. We find that neurotypicality affects communicability gradient expression to a greater degree than functional connectivity. We visualised these results and added them to Figure 1. Second, we used the same approach but for manifold eccentricity. Again, we demonstrate greater sensitivity of neurotypicality to communicability at a global-level, but we cannot pin these effects down to specific networks because the effects do not survive the necessary multiple comparison correction. We have added these analyses to the manuscript (Page 13, Line 583):
“Much as with the gradients themselves, we suspected that much of the simple main effect of dataset could reflect the scanner / site, rather than the difference in phenotype. Again, we drew upon the CALM comparison children to help us disentangle these two explanations. As a sensitivity analysis to parse effects of neurotypicality and dataset on manifold eccentricity, we conducted a series of generalized linear models predicting mean global and network-level manifold eccentricity, for each modality. We did this across all the baseline data (i.e. including the neurotypical comparison sample for CALM) using neurotypicality (2 levels: neurodivergent or neurotypical), site (2 levels: CALM or NKI), sex, head motion, and age at scan (Figure 3X). We restricted our analysis to baseline scans to create more equally-balanced groups. In terms of structural manifold eccentricity (N = 313 neurotypical, N = 311 neurodivergent), we observed higher manifold eccentricity in the neurodivergent participants at a global level (β = 0.090, p = 0.019, Cohen’s d = 0.188) but the individual network level effects did not survive the multiple comparison correction necessary for looking across all seven networks, with the default-mode network being the strongest (β = 0.135, p = 0.027, p<sub>FDR</sub> = 0.109, Cohen’s d = 0.177). There was no significant effect of neurodiversity on functional manifold eccentricity (N = 292 neurotypical and N = 177 neurodivergent). This suggests that neurodiversity is significantly associated with structural manifold eccentricity, over and above differences in site, but we cannot distinguish these effects reliably in the functional manifold data”.
Third, we removed the Scheirer-Ray-Hare test from the results for two reasons. First, its initial implementation did not account for repeated measures, and therefore non-independence between observations, as the same participants may have contributed both structural and functional data. Second, if we wanted to repeat this analysis in CALM using the referred and control portions, a significant difference in group size existed, which may affect the measures of variability. Specifically, for baseline CALM, 311 referred and 91 control participants contributed SC data, whilst 177 referred and 79 control participants contributed FC data. We believe that the ‘cleanest’ parsing of dataset and site for effects of eccentricity is achieved using the GLMs in Figure 3.
We observed no significant effect of neurodivergence on the magnitude of structure-function coupling across development, and have added the following text (Page 14, Line 632):
“To parse effects of neurotypicality and dataset on structure-function coupling, we conducted a series of generalized linear models predicting mean global and network-level coupling using neurotypicality, site, sex, head motion, and age at scan, at baseline (N = 77 CALM neurotypical, N = 173 CALM neurodivergent, and N = 170 NKI). However, we found no significant effects of neurotypicality on structure-function coupling across development”.
Since we demonstrated no significant effects of neurotypicality on structure-function coupling magnitude across development, but found differential dataset-specific effects of age on coupling development, we added the following sentence at the end of the coupling trajectory results sub-section (Page 14, line 664):
“Together, these effects demonstrate that whilst the magnitude of structure-function coupling appears not to be sensitive to neurodevelopmental phenotype, its development with age is, particularly in higher-order association networks, with developmental change being reduced in the neurodivergent sample”.
Figure 1.c: A non-parametric permutation test (e.g. Mann-Whitney U test) could quantitatively identify regions with significant group differences in nodal gradient values, providing additional support for the qualitative findings.
This is a great idea. To examine the effect of referral status on nodal gradient values, whilst controlling for covariates (head motion and sex), we conducted a series of generalised linear models. We opted for this instead of a Mann-Whitney U test, as the former tests for differences in distributions, whilst the direction of the t-statistic for referral status from the GLM would allow us to specify the magnitude and direction of differences in nodal gradient values between the two groups. Again, we conducted this in CALM (referred vs control), at an individual-level, as downstream analyses suggested a main effect of dataset (which is reflected in the highly-similar group-level referred and control CALM gradients). We have updated the Results section with the following text (Page 6, Line 283):
“To examine the effect of referral status on participant-level nodal gradient values in CALM, we conducted a series of generalized linear models controlling for head motion, sex and age at scan (Figure 1d). We restricted our analyses to baseline scans to reduce the difference in sample size for the referred (311 communicability and 177 functional gradients, respectively) and control participants (91 communicability and 79 functional gradients, respectively), and to the principal gradients. For communicability, 42 regions showed a significant effect (p < 0.05) of neurodivergence before FDR correction, with 9 post FDR correction. 8 of these 9 regions had negative t-statistics, suggesting a reduced nodal gradient value and representation in the neurodivergent children, encompassing both lower-order somatosensory cortices alongside higher-order fronto-parietal and default-mode networks. The largest reductions were observed within the prefrontal cortices of the defaultmode network (t = -3.992, p = 6.600 x 10<sup>-5</sup>, p<sub>FDR</sub> = 0.013, Cohen’s d = -0.476), the left orbitofrontal cortex of the limbic network (t = -3.710, p = 2.070 x 10<sup>-4</sup>, p<sub>FDR</sub> = 0.020, Cohen’s d = -0.442) and right somato-motor cortex (t = -3.612, p = 3.040 x 10<sup>-4</sup>, p<sub>FDR</sub> = 0.020, Cohen’s d = -0.431). The right visual cortex was the only exception, with stronger gradient representation within the neurotypical cohort (t = 3.071, p = 0.002, p<sub>FDR</sub> = 0.048, Cohen’s d = 0.366). For functional connectivity, comparatively fewer regions exhibited a significant effect (p < 0.05) of neurotypicality, with 34 regions prior to FDR correction and 1 post. Significantly stronger gradient representation was observed in neurotypical children within the right precentral ventral division of the defaultmode network (t = 3.930, p = 8.500 x 10<sup>-5</sup>, p<sub>FDR</sub> = 0.017, Cohen’s d = 0.532). Together, this suggests that the strongest and most robust effects of neurodivergence are observed within gradients of communicability, rather than functional connectivity, where alterations in both affect higher-order associative regions”.
In the harmonization methodology, it is mentioned that “if harmonisation was successful, we’d expect any significant effects of scanner type before harmonisation to be non-significant after harmonisation”. However, given that there were no significant effects before harmonization, the results reported do not help in evaluating the quality of harmonization.
We agree with the Reviewer, and have removed the post-harmonisation GLMs, and instead stating that there were no significant effects of scanner type before harmonization.
Figure 3: It would be helpful to include a plot showing the GAMM predictions versus real observations of eccentricity (x-axis: predictions, y-axis: actual values).
To plot the GAMM-predicted smooth effects of age, which we used for visualisation purposes only, we used the get_predictions function from the itsadug R package. This creates model predictions using the median value of nuisance covariates. Thus, whilst we specified the entire age range, the function automatically chooses the median of head motion, alongside controlling for sex (default level: male) and, for each dataset-specific trajectory. Since the gamm4 package separates the fitted model into a gam and linear mixed effects model (which accounts for participant ID as a random effect), and the get_predictions function only uses gam, random effects are not modelled in the predicted smooths. Therefore, any discrepancy between the observed and predicted manifold eccentricity values is likely due to sensitivity to default choices of covariates other than age, or random effects. To prevent Figure 3 being too over-crowded, we opted to not include the predictions: these were strongly correlated with real structural manifold data, but less for functional manifold data especially where significant developmental change was absent.
The 30mm threshold for filtering short streamlines in tractography is uncommon. What is the rationale for using such a large threshold, given the potential exclusion of many short-range association fibres?
A minimum length of 30mm was the default for the MRtrix3 reconstruction workflow, and something we have previously used. In a previous project, we systematically varied the minimum fibre length and found that this had minimal impact on network organisation (e.g. Mousley et al. 2025). However, we accept that short-range association fibres may have been excluded and have included this in the Discussion as a methodological limitation, alongside our predictions for how the gradients and structure-function coupling may’ve been altered had we included such fibres (Page 20, Line 955):
“A potential methodological limitation in the construction of structural connectomes was the 30mm tract length threshold which, despite being the QSIprep reconstruction default (Cieslak et al., 2021), may have potentially excluded short-range association fibres. This is pertinent as tracts of different lengths exhibit unique distributions across the cortex and functional roles (Bajada et al., 2019) : short-range connections occur throughout the cortex but peak within primary areas, including the primary visual, somato-motor, auditory, and para-hippocampal cortices, and are thought to dominate lower-order sensorimotor functional resting-state networks, whilst long-range connections are most abundant in tertiary association areas and are recruited alongside tracts of varying lengths within higher-order functional resting-state networks. Therefore, inclusion of short-range association fibres may have resulted in a relative increase in representation of lower-order primary areas and functional networks. On the other hand, we also note the potential misinterpretation of short-range fibres: they may be unreliably distinguished from null models in which tractography is restricted by cortical gyri only (Bajada et al., 2019). Further, prior (neonatal) work has demonstrated that the order of connectivity of regions and topological fingerprints are consistent across varying streamline thresholds (Mousley et al., 2025), suggesting minimal impact”.
Given the spatial smoothing of fMRI data (6mm FWHM), it would be beneficial to apply connectome spatial smoothing to structural connectivity measures for consistent spatial smoothness.
This is an interesting suggestion but given we are looking at structural communicability within a parcellated network, we are not sure that it would make any difference. The data structural data are already very smooth. Nonetheless we have added the following text to the Discussion (Page 20, Line 968):
“Given the spatial smoothing applied to the functional connectivity data, and examining its correspondence to streamline-count connectomes through structure-function coupling, applying the equivalent smoothing to structural connectomes may improve the reliability of inference, and subsequent sensitivity to cognition and psychopathology. Connectome spatial smoothing involves applying a smoothing kernel to the two streamline endpoints, whereby variations in smoothing kernels are selected to optimise the trade-off between subjectlevel reliability and identifiability, thus increasing the signal-to-noise ratio and the reliability of statistical inferences of brain-behaviour relationships (Mansour et al., 2022). However, we note that such smoothing is more effective for high-resolution connectomes, rather than parcel-level, and so have only made a modest improvement (Mansour et al., 2022)”.
Why was harmonization performed only within the CALM dataset and not across both CALM and NKI datasets? What was the rationale for this decision?
We thought about this very carefully. Harmonization aims to remove scanner or site effects, whilst retaining the crucial characteristics of interest. Our capacity to retain those characteristics is entirely dependent on them being *fully* captured by covariates, which are then incorporated into the harmonization process. Even with the best set of measures, the idea that we can fully capture ‘neurodivergence’ and thus preserve it in the harmonisation process is dubious. Indeed, across CALM and NKI there are limited number of common measures (i.e. not the best set of common measures), and thus we are limited in our ability to fully capture the neurodivergence with covariates. So, we worried that if we put these two very different datasets into the harmonisation process we would essentially eliminate the interesting differences between the datasets. We have added this text to the harmonization section of the Methods (Page 24, Line 1225):
“Harmonization aims to retain key characteristics of interest whilst removing scanner or site effects. However, the site effects in the current study are confounded with neurodivergence, and it is unlikely that neurodivergence may be captured fully using common covariates across CALM and NKI. Therefore, to preserve variation in neurodivergence, whilst reducing scanner effects, we harmonized within the CALM dataset only”.
The exclusion of subcortical areas from connectivity analyses is not justified.
This is a good point. We used the Schaefer atlas because we had previously used this to derive both functional and structural connectomes, but we agree that it would have been good to include subcortical areas (Page 20, Line 977).
“A potential limitation of our study was the exclusion of subcortical regions. However, prior work has shed light on the role of subcortical connectivity in structural and functional gradients, respectively, of neurotypical populations of children and adolescents (Park et al., 2021; Xia et al., 2022). For example, in the context of the primary-to-transmodal and sensorimotor-to-visual functional connectivity gradients, the mean gradient scores within subcortical networks were demonstrated to be relatively stable across childhood and adolescence (Xia et al., 2022). In the context of structural connectivity gradients derived from streamline counts, which we demonstrated were highly consistent with those derived from communicability, subcortical structural manifolds weighted by their cortical connectivity were anchored by the caudate and thalamus at one pole, and by the hippocampus and nucleus accumbens at the opposite pole, with significant age-related manifold expansion within the caudate and thalamus (Park et al., 2021)”.
In the KNN imputation method, were uniform weights used, or was an inverse distance weighting applied?
Uniform weights were used, and we have updated the manuscript appropriately.
The manuscript should clarify from the outset that the reported sample size (N) includes multiple longitudinal observations from the same individuals and does not reflect the number of unique participants.
We have rectified the Abstract (Page 2, Line 64) and Introduction (Page 3, Line 138):
“We charted the organisational variability of structural (610 participants, N = 390 with one observation, N = 163 with two observations, and N = 57 with three) and functional (512 participants, N = 340 with one observation, N = 128 with two observations, and N = 44 with three)”.
The term “structural gradients” is ambiguous in the introduction. Clarify that these gradients were computed from structural and functional connectivity matrices, not from other structural features (e.g. cortical thickness).
We have clarified this in the Introduction (Page 3, Line 134):
“Applying diffusion-map embedding as an unsupervised machine-learning technique onto matrices of communicability (from streamline SIFT2-weighted fibre bundle capacity) and functional connectivity, we derived gradients of structural and functional brain organisation in children and adolescents…”
Page 5: The sentence, “we calculated the normalized angle of each structural and functional connectome to derive symmetric affinity matrices” is unclear and needs clarification.
We have clarified this within the second paragraph of the Results section (Page 4, Line 185):
“To capture inter-nodal similarity in connectivity, using a normalised angle kernel, we derived individual symmetric affinity matrices from the left and right hemispheres of each communicability and functional connectivity matrix. Varying kernels capture different but highly-related aspects of inter-nodal similarity, such as correlation coefficients, Gaussian kernels, and cosine similarity. Diffusion-map embedding is then applied on the affinity matrices to derive gradients of cortical organisation”.
Figure 1.a: “Affine A” likely refers to the affinity matrix. The term “affine” may be confusing; consider using a clearer label. It would also help to add descriptive labels for rows and columns (e.g. region x region).
Thank you for this suggestion! We have replaced each of the labels with “pairwise similarity”. We also labelled the rows and columns as regions.
Figure 1.d: Are the cross-group differences statistically significant? If so, please indicate this in the figure.
We have added the results of a series of linear mixed effects models to the legend of Figure 1 (Page 6, line 252):
“indicates a significant effect of dataset (p < 0.05) on variance explained within a linear mixed effects model controlling for head motion, sex, and age at scan”.
The sentence “whose connectomes were successfully thresholded” in the methods is unclear. What does “successfully thresholded” mean? Additionally, this seems to be the first mention of the Schaefer 100 and Brainnetome atlas; clarify where these parcellations are used.
We have amended the Methodology section (Page 23, Line 1138):
“For each participant, we retained the strongest 10% of connections per row, thus creating fully connected networks required for building affinity matrices. We excluded any connectomes in which such thresholding was not possible due to insufficient non-zero row values. To further ensure accuracy in connectome reconstruction, we excluded any participants whose connectomes failed thresholding in two alternative parcellations: the 100node Schaefer 7-network (Schaefer et al., 2018) and Brainnetome 246-node (Fan et al., 2016) parcellations, respectively”.
We have also specified the use of the Schaefer 200-node parcellation in the first sentence on the second Results paragraph.
The use of “streamline counts” is misleading, as the method uses SIFT2-weighted fibre bundle capacity rather than raw streamline counts. It would be better to refer to this measure as “SIFT2-weighted fibre bundle capacity” or “FBC”.
We replaced all instances of “streamline counts” with “SIFT2-weighted fibre bundle capacity” as appropriate.
Figure 2.c: Consider adding plots showing changes in eccentricity against (1) degree centrality, and (2) weighted local clustering coefficient. Additionally, a plot showing the relationship between age and mean eccentricity (averaged across nodes) at the individual level would be informative.
We added the correlation between eccentricity and both degree centrality and the weighted local clustering coefficient and included them in our dominance analysis in Figure 2. In terms of the relationship between age and mean (global) eccentricity, these are plotted in Figure 3.
Figure 2.b: Considering the results of the following sections, it would be interesting to include additional KDE/violin plots to show group differences in the distribution of eccentricity within 7 different functional networks.
As part of our analysis to parse neurotypicality and dataset effects, we tested for group differences in the distribution of structural and functional manifold eccentricity within each of the 7 functional networks in the referred and control portions of CALM and have included instances of significant differences with a coloured arrow to represent the direction of the difference within Figure 3.
Figure 3: Several panels lack axis labels for x and y axes. Adding these would improve clarity.
To minimise the amount of text in Figure 3, we opted to include labels only for the global-level structural and functional results. However, to aid interpretation, we added a small schematic at the bottom of Figure 3 to represent all axis labels.
The statement that “differences between datasets only emerged when taking development into account” seems inaccurate. Differences in eccentricity are evident across datasets even before accounting for development (see Fig 2.b and the significance in the Scheirer-Ray-Hare test).
We agree – differences in eccentricity across development and datasets are evident in structural and functional manifold eccentricity, as well as within structure-function coupling. However, effects of neurotypicality were particularly strong for the maturation of structure-function coupling, rather than magnitude. Therefore, we have rephrased this sentence in the Discussion (page 18, line 832):
“Furthermore, group-level structural and functional gradients were highly consistent across datasets, whilst differences between datasets were emphasised when taking development into account, through differing rates of structural and functional manifold expansion, respectively, alongside maturation of structure-function coupling”.
The handling of longitudinal data by adding a random effect for individuals is not clear in the main text. Mentioning this earlier could be helpful.
We have included this detail in the second sentence of the “developmental trajectories of structural manifold contraction and functional manifold expansion” results sub-section (page 11, line 503):
“We included a random effect for each participant to account for longitudinal data”.
Figure 4.b: Why were ranks shown instead of actual coefficient of variation values? Consider including a cortical map visualization of the coefficients in the supplementary material.
We visualised the ranks, instead of the actual coefficient of variation (CV) values, due to considerable variability and skew in the magnitude of the CV, ranging from 28.54 (in the right visual network) to 12865.68 (in the parietal portion of the left default-mode network), with a mean of 306.15. If we had visualised the raw CV values, these larger values would’ve been over-represented. We’ve also noticed and rectified an error in the labelling of the colour bar for Figure 4b: the minimum should be most variable (i.e. a rank of 1). To aid contextualisation of the ranks, we have added the following to the Results (page 14, line 626):
“The distribution of cortical coefficients of variation (CV) varied considerably, with the largest CV (in the parietal division of the left default-mode network) being over 400 times that of the smallest (in the right visual network). The distribution of absolute CVs was positively skewed, with a Fisher skewness coefficient g<sub>1</sub> of 7.172, meaning relatively few regions had particularly high inter-individual variability, and highly peaked, with a kurtosis of 54.883, where a normal distribution has a skewness coefficient of 0 and a kurtosis of 3”.
Reviewer #2 (Public review):
Some differences in developmental trajectories between CALM and NKI (e.g. Figure 4d) are not explained. Are these differences expected, or do they suggest underlying factors that require further investigation?
This is a great point, and we appreciate the push to give a fuller explanation. It is very hard to know whether these effects are expected or not. We certainly don’t know of any other papers that have taken this approach. In response to the reviewer’s point, we decided to run some more analyses to better understand the differences. Having observed stronger age effects on structure-function coupling within the neurotypical NKI dataset, compared to the absent effects in the neurodivergent portion of CALM, we wanted to follow up and test that it really is that coupling is more sensitive to the neurodivergent versus neurotypical difference between CALM and NKI (rather than say, scanner or site effects). In short, we find stronger developmental effects of coupling within the neurotypical portion of CALM, rather than neurodivergent, and have added this to the Results (page 15, line 701):
“To further examine whether a closer correspondence of structure-function coupling with age is associated with neurotypicality, we conducted a follow-up analysis using the additional age-matched neurotypical portion of CALM (N = 77). Given the widespread developmental effects on coupling within the neurotypical NKI sample, compared to the absent effects in the neurodivergent portion of CALM, we would expect strong relationships between age and structure-function coupling with the neurotypical portion of CALM. This is indeed what we found: structure-function coupling showed a linear negative relationship with age globally (F = 16.76, p<sub>FDR</sub> < 0.001, adjusted R<sup>2</sup> = 26.44%), alongside fronto-parietal (F = 9.24, p<sub>FDR</sub> = 0.004, adjusted R<sup>2</sup> = 19.24%), dorsalattention (F = 13.162, p<sub>FDR</sub> = 0.001, adjusted R<sup>2</sup>= 18.14%), ventral attention (F = 11.47, p<sub>FDR</sub> = 0.002, adjusted R<sup>2</sup>= 22.78), somato-motor (F = 17.37, p<sub>FDR</sub> < 0.001, adjusted R<sup>2</sup>= 21.92%) and visual (F = 11.79, p<sub>FDR</sub> = 0.002, adjusted R<sup>2</sup>= 20.81%) networks. Together, this supports our hypothesis that within neurotypical children and adolescents, structure-function coupling decreases with age, showing a stronger effect compared to their neurodivergent counterparts, in tandem with the emergence of higher-order cognition. Thus, whilst the magnitude of structure-function coupling across development appeared insensitive to neurotypicality, its maturation is sensitive. Tentatively, this suggests that neurotypicality is linked to stronger and more consistent maturational development of structure-function coupling, whereby the tethering of functional connectivity to structure across development is adaptive”.
In conjunction with the Reviewer’s later request to deepen the Discussion, we have included an additional paragraph attempting to explain the differences in neurodevelopmental trajectories of structure-function coupling (Page 19, Line 924):
“Whilst the spatial patterning of structure-function coupling across the cortex has been extensively documented, as explained above, less is known about developmental trajectories of structure-function coupling, or how such trajectories may be altered in those with neurodevelopmental conditions. To our knowledge, only one prior study has examined differences in developmental trajectories of (non-manifold) structure-function coupling in typically-developing children and those with attention-deficit hyperactivity disorder (Soman et al., 2023), one of the most common conditions in the neurodivergent portion of CALM. Namely, using cross-sectional and longitudinal data from children aged between 9 and 14 years old, they demonstrated increased coupling across development in higher-order regions overlapping with the defaultmode, salience, and dorsal attention networks, in children with ADHD, with no significant developmental change in controls, thus encompassing an ectopic developmental trajectory (Di Martino et al., 2014; Soman et al., 2023). Whilst the current work does not focus on any condition, rather the broad mixed population of young people with neurodevelopmental symptoms (including those with and without diagnoses), there are meaningful individual and developmental differences in structure-coupling. Crucially, it is not the case that simply having stronger coupling is desirable. The current work reveals that there are important developmental trajectories in structure-function coupling, suggesting that it undergoes considerable refinement with age. Note that whilst the magnitude of structure-function coupling across development did not differ significantly as a function of neurodivergence, its relationship to age did. Our working hypothesis is that structural connections allow for the ordered integration of functional areas, and the gradual functional modularisation of the developing brain. For instance, those with higher cognitive ability show a stronger refinement of structurefunction coupling across development. Future work in this space needs to better understand not just how structural or functional organisation change with time, but rather how one supports the other”.
The use of COMBAT may have excluded extreme participants from both datasets, which could explain the lack of correlations found with psychopathology.
COMBAT does not exclude participants from datasets but simply adjusts connectivity estimates. So, the use of COMBAT will not be impacting the links with psychopathology by removing participants. But this did get us thinking. Excluding participants based on high motion may have systematically removed those with high psychopathology scores, meaning incomplete coverage. In other words, we may be under-representing those at the more extreme end of the range, simply because their head-motion levels are higher and thus are more likely to be excluded. We found that despite certain high-motion participants being removed, we still had good coverage of those with high scores and were therefore sensitive within this range. We have added the following to the revised Methods section (Page 26, Line 1338):
“As we removed participants with high motion, this may have overlapped with those with higher psychopathology scores, and thus incomplete coverage. To examine coverage and sensitivity to broad-range psychopathology following quality control, we calculated the Fisher-Pearson skewness statistic g<sub>1</sub> for each of the 6 Conners t-statistic measures and the proportion of youth with a t-statistic equal to or greater than 65, indicating an elevated or very elevated score. Measures of inattention (g<sub>1</sub> = 0.11, 44.20% elevated), hyperactivity/impulsivity (g<sub>1</sub> = 0.48, 36.41% elevated), learning problems (g<sub>1</sub> = 0.45, 37.36% elevated), executive functioning (g<sub>1</sub> = 0.27, 38.16% elevated), aggression (g<sub>1</sub> = 1.65, 15.58% elevated), and peer relations (g<sub>1</sub> = 0.49, 38% elevated) were positively skewed and comprised of at least 15% of children with elevated or very elevated scores, suggesting sufficient coverage of those with extreme scores”.
There is no discussion of whether the stable patterns of brain organization could result from preprocessing choices or summarizing data to the mean. This should be addressed to rule out methodological artifacts.
This is a brilliant point. We are necessarily using a very lengthy pipeline, with many design choices to explore structural and functional gradients and their intersection. In conjunction with the Reviewer’s later suggestion to deepen the Discussion, we have added the following paragraph which details the sensitivity analyses we carried out to confirm the observed stable patterns of brain organization (Page 18, Line 863):
“That is, whilst we observed developmental refinement of gradients, in terms of manifold eccentricity, standard deviation, and variance explained, we did not observe replacement. Note, as opposed to calculating gradients based on group data, such as a sliding window approach, which may artificially smooth developmental trends and summarise them to the mean, we used participant-level data throughout. Given the growing application of gradient-based analyses in modelling structural (He et al., 2025; Li et al., 2024) and functional (Dong et al., 2021; Xia et al., 2022) brain development, we hope to provide a blueprint of factors which may affect developmental conclusions drawn from gradient-based frameworks”.
Although imputing missing data was necessary, it would be useful to compare results without imputed data to assess the impact of imputation on findings.
It is very hard to know the impact of imputation without simply removing those participants with some imputed data. Using a simulation experiment, we expressed the imputation accuracy as the root mean squared error normalized by the range of observable data in each scale. This produced a percentage error margin. We demonstrate that imputation accuracy across all measures is at worst within approximately 11% of the observed data, and at best within approximately 4% of the observed data, and have included the following in the revised Methods section (Page 27, Line 1348):
“Missing data
To avoid a loss of statistical power, we imputed missing data. 27.50% of the sample had one or more missing psychopathology or cognitive measures (equal to 7% of all values), and the data was not missing at random: using a Welch’s t-test, we observed a significant effect of missingness on age [t (264.479) = 3.029, p = 0.003, Cohen’s d = 0.296], whereby children with missing data (M = 12.055 years, SD = 3.272) were younger than those with complete data (M = 12.902 years, SD = 2.685). Using a subset with complete data (N = 456), we randomly sampled 10% of the values in each column with replacement and assigned those as missing, thereby mimicking the proportion of missingness in the entire dataset. We conducted KNN imputation (uniform weights) on the subset with complete data and calculated the imputation accuracy as the root mean squared error normalized by the observed range of each measure. Thus, each measure was assigned a percentage which described the imputation margin of error. Across cognitive measures, imputation was within a 5.40% mean margin of error, with the lowest imputation error in the Trail motor speed task (4.43%) and highest in the Trails number-letter switching task (7.19%). Across psychopathology measures, imputation exhibited a mean 7.81% error margin, with the lowest imputation error in the Conners executive function scale (5.75%) and the highest in the Conners peer relations scale (11.04%). Together, this suggests that imputation was accurate”.
The results section is extensive, with many reports, while the discussion is relatively short and lacks indepth analysis of the findings. Moving some results into the discussion could help balance the sections and provide a deeper interpretation.
We agree with the Reviewer and appreciate the nudge to expand the Discussion section. We have added 4 sections to the Discussion. The first explores the importance of the default-mode network as a region whose coupling is most consistently predicted by working memory across development and phenotypes, in terms of its underlying anatomy (Paquola et al., 2025) (Page 20, Line 977):
“An emerging theme from our work is the importance of the default-mode network as a region in which structure-function coupling is reliably predicted by working memory across neurodevelopmental phenotypes and datasets during childhood and adolescence. Recent neurotypical adult investigations combining highresolution post-mortem histology, in vivo neuroimaging, and graph-theory analyses have revealed how the underlying neuroanatomy of the default-mode network may support diverse functions (Paquola et al., 2025), and thus exhibit lower structure-function coupling compared to unimodal regions. The default-mode network has distinct neuroanatomy compared to the remaining 6 intrinsic resting-state functional networks (Yeo et al., 2011), containing a distinctive combination of 5 of the 6 von Economo and Koskinas cell types (von Economo & Koskinas, 1925), with an over-representation of heteromodal cortex, and uniquely balancing output across all cortical types. A primary cytoarchitectural axis emerges, beyond which are mosaic-like spatial topographies. The duality of the default-mode network, in terms of its ability to both integrate and be insulated from sensory information, is facilitated by two microarchitecturally distinct subunits anchored at either end of the cytoarchitectural axis (Paquola et al., 2025). Whilst beyond the scope of the current work, structure-function coupling and their predictive value for cognition may also differ across divisions within the default-mode network, particularly given variability in the smoothness and compressibility of cytoarchitectural landscapes across subregions (Paquola et al., 2025)”.
The second provides a deeper interpretation and contextualisation of greater sensitivity of communicability, rather than functional connectivity, to neurodivergence (Page 19, Lines 907):
“We consider two possible factors to explain the greater sensitivity of neurodivergence to gradients of communicability, rather than functional connectivity. First, functional connectivity is likely more sensitive to head motion than structural-based communicability and suffers from reduced statistical power due to stricter head motion thresholds, alongside greater inter-individual variability. Second, whilst prior work contrasting functional connectivity gradients from neurotypical adults with those with confirmed ASD diagnoses demonstrated vertex-level reductions in the default-mode network in ASD and marginal increases in sensorymotor communities (Hong et al., 2019), indicating a sensitivity of functional connectivity to neurodivergence, important differences remain. Specifically, whilst the vertex-level group-level differences were modest, in line with our work, greater differences emerged when considering step-wise functional connectivity (SFC); in other words, when considering the dynamic transitions of or information flow through the functional hierarchy underlying the static functional connectomes, such that ASD was characterised by initial faster SFC within the unimodal cortices followed by a lack of convergence within the default-mode network (Hong et al., 2019). This emphasis on information flow and dynamic underlying states may point towards greater sensitivity of neurodivergence to structural communicability – a measure directly capturing information flow – than static functional connectivity”.
The third paragraph situates our work within a broader landscape of reliable brain-behaviour relationships, focusing on the strengths of combining clinical and normative samples to refine our interpretation of the relationship between gradients and cognition, as well as the importance of equifinality in developmental predictive work (Page 20, line 994):
“In an effort to establish more reliable brain-behaviour relationships despite not having the statistical power afforded by large-scale, typically normative, consortia (Rosenberg & Finn, 2022), we demonstrated the development-dependent link between default-mode structure-function coupling and working memory generalised across clinical (CALM) and normative (NKI) samples, across varying MRI acquisition parameters, and harnessing within- and across-participant variation. Such multivariate associations are likely more reliable than their univariate counterparts (Marek et al., 2022), but can be further optimised using task-related fMRI (Rosenberg & Finn, 2022). The consistency, or lack of, of developmental effects across datasets emphasises the importance of validating brain-behaviour relationships in highly diverse samples. Particularly evident in the case of structure-function coupling development, through our use of contrasting samples, is equifinality (Cicchetti & Rogosch, 1996), a key concept in developmental neuroscience: namely, similar ‘endpoints’ of structure-function coupling may be achieved through different initialisations dependent on working memory.
The fourth paragraph details methodological limitations in response to Reviewer 1’s suggestions to justify the exclusion of subcortical regions and consider the role of spatial smoothing in structural connectome construction as well as the threshold for filtering short streamlines”.
While the methods are thorough, it is not always clear whether the optimal approaches were chosen for each step, considering the available data.
In response to Reviewer 1’s concerns, we conducted several sensitivity analyses to evaluate the robustness of our results in terms of procedure. Specifically, we evaluated the impact of thresholding (full or sparse), level of analysis (individual or group gradients), construction of the structural connectome (communicability or fibre bundle capacity), Procrustes rotation (alignment to group-level gradients before Procrustes), tracking the variance explained in individual connectomes by group-level gradients, impact of head motion, and distinguishing between site and neurotypicality effects. All these analyses converged on the same conclusion: whilst we observe some developmental refinement in gradients, we do not observe replacement. We refer the reviewer to their third point, about whether stable patterns of brain organization were artefactual.
The introduction is overly long and includes numerous examples that can distract readers unfamiliar with the topic from the main research questions.
We have removed the following from the Introduction, reducing it to just under 900 words:
“At a molecular level, early developmental patterning of the cortex arises through interacting gradients of morphogens and transcription factors (see Cadwell et al., 2019). The resultant areal and progenitor specialisation produces a diverse pool of neurones, glia, and astrocytes (Hawrylycz et al., 2015). Across childhood, an initial burst in neuronal proliferation is met with later protracted synaptic pruning (Bethlehem et al., 2022), the dynamics of which are governed by an interplay between experience-dependent synaptic plasticity and genomic control (Gottlieb, 2007)”.
“The trends described above reflect group-level developmental trends, but how do we capture these broad anatomical and functional organisational principles at the level of an individual?”
We’ve also trimmed the second Introduction paragraph so that it includes fewer examples, such as removal of the wiring-cost optimisation that underlies structural brain development, as well as removing specific instances of network segregation and integration that occur throughout childhood.
Author response:
The following is the authors’ response to the original reviews
Reviewer #1 (Public Review):
Overall, it's a well-performed study, however, causality between Plscr1 and Ifnlr1 expression needs to be more firmly established. This is because two recent studies of PLSCR1 KO cells infected with different viruses found no major differences in gene expression levels compared with their WT controls (Xu et al. Nature, 2023; LePen et al. PLoS Biol, 2024). There were also defects in the expression of other cytokines (type I and II IFNs plus TNF-alpha) so a clear explanation of why Ifnlr1 was chosen should also be given.
We appreciate the reviewer’s reference to the two recently published research on PLSCR1’s role in SARS-CoV-2 infections. We have also discussed those studies in the Introduction and Discussion sections of this manuscript. Here, we would like to clarify ourselves for the rationale of investigating Ifn-λr1 signaling.
The reviewer mentioned “defects in the expression of other cytokines (type I and II IFNs plus TNF-alpha)” and requested a clearer explanation of why Ifnlr1 was chosen for study. In our investigation of IAV infection, we observed no defects in the expression of type I and II IFNs or TNF-α in Plscr1<sup>-/-</sup> mice; rather, these cytokines were expressed at even higher levels compared to WT controls (Figures 2D and 3A). This indicates that the type I and II IFN and TNF-α signaling pathways remain intact and are not negatively affected by the loss of Plscr1. Notably, Ifn-λr1 expression is the only one among all IFNs and their receptors that is significantly impaired in Plscr1<sup>-/-</sup> mice (Figure 3A), justifying our focused investigation of this receptor. To further clarify this point, we have expanded the explanation under the section titled “Plscr1 Binds to Ifn-λr1 Promoter and Activates Ifn-λr1 Transcription in IAV Infection” within the Results. The reviewer noted that previously published studies “found no major differences in gene expression levels compared with their WT controls”, but neither study examined Ifn-λr1 expression.
(1) The authors propose that Plscr1 restricts IAV infection by regulating the type III IFN signaling pathway. While the data show a positive correlation between Ifnlr1 and Plscr1 levels in both mouse and cell culture models, additional evidence is needed to establish causality between the impaired type III IFN pathway, and the increased susceptibility observed in Plscr1-KO mice. To strengthen this conclusion, the following experiments could be undertaken: (i) Measure IAV titers in WT, Plscr1-KO, Ifnlr1-KO, and Plscr1/ Ifnlr1-double KO cells. If the antiviral activity of Plscr1 is highly dependent on Ifnlr1, there should be no further increase in IAV titers in double KO cells compared to single KO cells; (ii) over-express Plscr1 in Ifnlr1-KO cells to determine if it still inhibits IAV infection. If Plscr1's main action is to upregulate Ifnlr1, then it should not be able to rescue susceptibility since Ifnlr1 cannot be expressed in the KO background. If Plscr1 over-expression rescues viral susceptibility, then there are Ifnlr1-independent mechanisms involved. These experiments should help clarify the relative contribution of the type III IFN pathway to Plscr1-mediated antiviral immunity.
We agree with the reviewer that additional evidence is necessary to establish causality between the impaired type III IFN pathway and the increased susceptibility observed in Plscr1-KO mice. As requested by the reviewer, and one step further, we have measured IAV titers in Wt, Plscr1<sup>-/-</sup>, Ifn-λr1<sup>-/-</sup>, and Plscr1<sup>-/-</sup>Ifn-λr1<sup>-/-</sup> mouse lungs, which provided us with more comprehensive information at the tissue and organismal level compared to cell culture models. Our results are detailed under “The Anti-Influenza Activity of Plscr1 Is Highly Dependent on Ifn-λr1” within “Results” section and in Supplemental Figure 5. Importantly, there was no further increase in weight loss (Supplemental Figure 5B), total BAL cell counts (Supplemental Figure 5C), neutrophil percentages (Supplemental Figure 5D), and IAV titers (Supplemental Figure 5E) in Plscr1<sup>-/-</sup>Ifn-λr1<sup>-/-</sup> mouse lungs compared to Ifn-λr1<sup>-/-</sup> mouse lungs. These findings indicate that the antiviral activity of Plscr1 is largely dependent on Ifn-λr1.
We agree that overexpression of Plscr1 on an Ifn-λr1<sup>-/-</sup> background would provide additional evidence to support our conclusion from the Plscr1<sup>-/-</sup>Ifn-λr1<sup>-/-</sup> mice. In future studies, we plan to specifically overexpress Plscr1 in ciliated epithelial cells on the Ifn-λr1<sup>-/-</sup> background by breeding Plscr1<sup>floxStop</sup>Foxj1-Cre<sup>+</sup>Ifn-λr1<sup>-/-</sup> mice. In addition, ciliated epithelial cells isolated from Ifn-λr1<sup>-/-</sup> murine airways could be transduced with a Plscr1 construct for overexpression. We hypothesize that overexpression of Plscr1 in ciliated epithelial cells will not rescue susceptibility in Ifn-λr1<sup>-/-</sup> mice or cells, since our Plscr1<sup>-/-</sup>Ifn-λr1<sup>-/-</sup> mouse model suggest that Ifn-λr1-independent anti-influenza functions of Plscr1 are likely minor compared to its role in upregulating Ifn-λr1. These future plans have been added to the “Discussion” section, and we look forward to presenting our results in a forthcoming publication.
(3) In Figure 4, the authors demonstrate the interaction between Plscr1 and Ifnlr1. They suggest that this interaction modulates IFN-λ signaling. However, Figures 5C-E show that the 5CA mutant, which lacks surface localization and the ability to bind Ifnlr1, exhibits similar anti-flu activity to WT Plscr1. Does this mean the interaction between Plscr1 and Ifnlr1 is dispensable for Plscr1-mediated antiviral function? Can the authors compare the activation of IFN-λ signaling pathway in Plscr1-KO cells expressing empty vector, WT Plscr1, and 5CA mutant? This could be done by measuring downstream ISG expression or using an ISRE-luciferase reporter assay upon IFN-λ treatment.
We agree with the reviewer that downstream activation of the IFN-λ signaling pathway is a critical component of the proposed regulatory role of PLSCR1. As suggested, we attempted to perform an ISRE-luciferase reporter assay following IFN-λ treatment in PLSCR1 rescue cell lines by transfecting the cells with hGAPDH-rLuc (Addgene #82479) and pGL4.45 [luc2P/ISRE/Hygro] (Promega #E4041).
Despite extensive efforts over several months, we were unable to achieve expression of pGL4.45 [luc2P/ISRE/Hygro] in PLSCR1 rescue cells using either Lipofectamine 3000 or electroporation, as no firefly luciferase activity was detected at baseline or following IFN-λ treatment. In contrast, hGAPDH-rLuc was robustly expressed in these cells.
The pGL4.45 [luc2P/ISRE/Hygro] plasmid was obtained directly from Promega as a purified product, and its sequence was confirmed via whole plasmid sequencing. Additionally, both hGAPDH-rLuc and pGL4.45 [luc2P/ISRE/Hygro] were successfully expressed in 293T cells, indicating that neither the plasmids nor the transfection protocols are inherently faulty.
We suspect that prior modifications to the PLSCR1 rescue cells—such as CRISPR-mediated knockout and lentiviral transduction—may interfere with successful transfection of pGL4.45 [luc2P/ISRE/Hygro] through an as-yet-unknown mechanism. Although these results are disappointing, we will continue troubleshooting and plan to communicate in a separate manuscript once the luciferase assay is successfully established.
Reviewer #1 (Recommendations):
(1) In the introduction, the linkage between the paragraph discussing type III IFN and PLSCR1 needs to be better established. The mention of PLSCR1 being an ISG at the outset may help connect these two paragraphs and make the text appear more logical.
We apologize for the lack of linkage and logic between type 3 IFN and PLSCR1. We have introduced PLSCR1 as an ISG at the beginning of its paragraph as recommended.
(2) The statement that, “Intriguingly, PLSCR1 is also an antiviral ISG, as its expression can be highly induced by type 1 and 2 interferons in various viral infections[15, 16]. However, whether its expression can be similarly induced by type 3 interferon has not been studied yet.” is incorrect. Xu et al. tested the role of PLSCR1 in type III IFN-induced control of SARS-CoV-2 (ref. 24). This needs to be revised.
We apologize for the incorrect information in the introduction and have revised the paragraph with the proper citation.
(3) In Figure 3B, can the authors provide a comprehensive heatmap that includes all ISGs above the threshold, rather than only a subset? This would offer a more complete overview of the changes in type I, II, and III IFN pathways in Plscr1-KO mice.
As suggested by the reviewer, we have provided a comprehensive heatmap that includes all ISGs above the threshold in Figure 3C (previously Figure 3B). We identified a total of 1,113 ISGs in our dataset with a fold change ≥2. Enlarged heatmaps with gene names are provided in Supplemental Figure 1. Among those ISGs, 584 are regulated exclusively by type 1 IFNs, and 488 are regulated by both type 1 and type 2 interferons. Unfortunately, the Interferome database does not include information on type 3 IFN-inducible genes in mice[1]. Although many ISGs were robustly upregulated in Plscr1<sup>-/-</sup> infected lungs, consistent with inflammation data, a large subset of ISGs failed to be transcribed when Ifn-λr1 function was impaired, especially at 7 dpi. We suspect that those non-transcribed ISGs in Plscr1<sup>-/-</sup> mice may be specifically regulated by type 3 IFN and represent interesting targets for future research. These results have been added to “Plscr1 Binds to Ifn-λr1 Promoter and Activates Ifn-λr1 Transcription in IAV Infection” within “Results” section.
(4) In Figure 3C, 5B and 7H, immunoblots should also be included to measure changes of Ifnlr1/IFNLR1 protein level.
As requested by the reviewer, we have provided western blots measuring Ifn-λr1/IFN-λR1 protein level in Figure 5B and 7I. The protein expressions were consistent with the PCR results.
(5) In Figure 3H, the amount of RPL30 is also low in the anti-PLSCR1-treated and IgG samples, making it difficult to estimate if ChIP binding is genuinely impacted.
RPL30 Exon 3 serves as a negative control in the ChIP experiment and is not expected to bind either the anti-PLSCR1-treated or the IgG control samples. Anti-Histone H3 treatment is a positive control, with the treated sample expected to show binding to RPL30 Exon 3. We hope this clarification has addressed any further potential confusion from the reviewer.
(6) In Figure 4A, can the authors show a larger slice of the gel with molecular weight markers for both Plscr1 and Ifnlr1. In the coIP, the binding may be indirect through intermediate partners. Proximity ligation assay is a more direct assay for interaction and can be stated as such.
As suggested by the reviewer, we have included whole gel images of Figure 4A with molecular weight markers for both Plscr1 and Ifnlr1 in Supplemental Figure 3. We appreciate the reviewer’s affirmation of proximity ligation assay and have stated it as a more direct assay for interaction under “Plscr1 Interacts with Ifn-λr1 on Pulmonary Epithelial Cell Membrane in IAV Infection” in “Results” section.
(7) In Figure 5A, how is the expression of PLSCR1 WT and mutants driven by an EF-1α promoter can be further upregulated by IAV infection? Can the authors also use immunoblots to examine the protein level of PLSCR1?
We apologize for the confusion and appreciate the reviewer’s careful observation. We were initially surprised by this finding as well, but upon further investigation, we found out that the human PLSCR1 primers used in our qRT-PCR assay can still detect the transcription from the undisturbed portion of the endogenous PLSCR1 mRNA, even in PLSCR1<sup>-/-</sup> cells. In the original Figure 5A, data for vector-transduced PLSCR1<sup>-/-</sup> were not included because PCR was not performed on those samples at the time. After conducting PCR for vector-transduced PLSCR1<sup>-/-</sup> cells, we detected transcription of PLSCR1, which confirms that the signaling originates from endogenous DNA, but not from the EF-1α promoter-driven PLSCR1 plasmid. Please see Author response image 1 below.
Author response image 1.
The forward human PLSCR1 primer we used matches 15-34 nt of Wt PLSCR1, and the reverse primer matches 224-244 nt of Wt PLSCR1. CRISPR-Cas9 KO of PLSCR1 was mediated by sgRNAs in A549 cells and was performed by Xu et al[2]. sgRNA #1 matches 227-246 nt, sgRNA #2 matches 209-228 nt, and sgRNA #3 matches 689-708 nt of Wt PLSCR1. The sgRNAs likely introduced a short deletion or insertion that does not affect transcription. However, those endogenous mRNA transcripts cannot be translated to functional and detectable PLSCR1 proteins, as validated by our western blot (below), as well as western blots performed by Xu et al[2]. Therefore, our primers could amplify endogenous PLSCR1 transcripts upregulated by IAV infection, if 15-244 nt was not disturbed by CRISPR-Cas9 KO. By western blot, we confirmed that only endogenous PLSCR1 expression is upregulated by IAV infection, and exogenous protein expression of PLSCR1 plasmids driven by an EF-1α promoter are not upregulated by IAV infection.
Author response image 2.
To avoid confusion, we have removed the original Figure 5A from the manuscript.
(8) In Figure 5C, the loss of anti-flu activity with the H262Y mutant is modest, suggesting the loss of ifnlr1 transcription is only partly responsible for the susceptibility of Plscr1 KO cells. The anti-flu activity being independent of scramblase activity resembles the earlier discovery of SARS-CoV-2 (Xu et al., 2024). This could be stated in the results since it is an important point that scramblase activity is dispensable for several major human viruses and shifts the emphasis regarding mechanism. It has been appropriately noted in the discussion.
We appreciated the comments and have acknowledged the consistency of our results with those of Xu et al. under “Both Cell Surface and Nuclear PLSCR1 Regulates IFN-λ Signaling and Limits IAV Infection Independent of Its Enzymatic Activity” in the “Results” section.
Reviewer #2 (Recommendations):
(1) The statement that type I interferons are expressed by “almost all cells” is inaccurate (line 61). Type I IFN production is also context-dependent and often restricted to specific cell types upon infection or stimulation.
We apologize for the inaccurate description of the expression pattern of type 1 IFNs and have corrected the restricted cellular sources of type 1 IFNs in the “Introduction”.
(2) The antiviral response is assessed solely through flu M gene expression. Incorporating infectious virus titers (e.g., TCID50 or plaque assay) would provide a more robust and direct measure of antiviral activity.
As requested by the reviewer, we have performed plaque assays on all experiments where flu M gene expression levels were measured (Figure 1G, 5E and 7F, and Supplemental Figure 6E). The plaque assay results are consistent with the flu M gene expressions.
(3) While mRNA expression of interferons is measured, protein levels (e.g., through ELISA) should also be quantified to establish the functional relevance of IFN expression changes.
As requested by the reviewer, we have quantified the protein level of IFN-λ in mouse BAL with ELISA (Figure 2E). The ELISA results are consistent with the mRNA expressions of IFN-λ.
(4) It is unclear whether reduced IFNLR1 expression translates to defective downstream signaling or antiviral responses after IFN-λ treatment in PLSCR1-deficient cells. This is particularly pertinent given the increase in IFN-λ ligand in vivo, which might compensate for receptor downregulation.
We agree with the reviewer that downstream activation of the IFN-λ signaling pathway is a critical aspect of PLSCR1’s proposed regulatory role. To investigate this, we attempted an ISRE-luciferase reporter assay to assess downstream signaling following IFN-λ treatment in PLSCR1 rescue cells. Unfortunately, the experiment encountered unforeseen technical issues. For additional context, please refer to our response to Reviewer #1’s public review #3.
(5) Detailed gating strategies for immune cell subsets are absent and should be included for clarity and reproducibility.
We would like to clarify that the immune cell subsets in BAL fluids were counted manually following cytospin preparation and Diff-Quik staining (Figure 2B and 7H, and Supplemental Figures 2C, 5D, and 8D), rather than by flow cytometry. We hope this resolves the reviewer’s confusion.
(6) The study does not definitively establish that reduced IFN-λ signaling causes the observed in vivo phenotype. Increased morbidity and mortality in PLSCR1-deficient mice could also stem from elevated TNF-α levels and lung damage, as proinflammatory cytokines and/or enhanced lung damage are known contributors to influenza morbidity and mortality. This point warrants detailed discussions.
We agreed with the reviewer that this study does not guarantee a definitive causality between reduced IFN-λ signaling and increased morbidity of Plscr1<sup>-/-</sup> mice and more experiments are needed to reach the conclusion. We have acknowledged this limitation of our study in the “Discussion”, as requested by the reviewer. We hope to fully eliminate the confounding elements and definitively establish the proposed causality in future studies.
Reviewer #3 (Public review):
Summary:
Yang et al. have investigated the role of PLSCR1, an antiviral interferon-stimulated gene (ISG), in host protection against IAV infection. Although some antiviral effects of PLSCR1 have been described, its full activity remains incompletely understood.
This study now shows that Plscr1 expression is induced by IAV infection in the respiratory epithelium, and Plscr1 acts to increase Ifn-λr1 expression and enhance IFN-λ signaling possibly through protein-protein interactions on the cell membrane.
Strengths:
The study sheds light on the way Ifnlr1 expression is regulated, an area of research where little is known. The study is extensive and well-performed with relevant genetically modified mouse models and tools.
Weaknesses:
There are some issues that need to be clarified/corrected in the results and figures as presented.
Also, the study does not provide much information about the role of PLSCR1 in the regulation of Ifn-λr1 expression and function in immune cells. This would have been a plus.
We would like to thank the reviewer for the positive feedback and insightful comment regarding the roles of PLSCR1 and IFN-λR1 in immune cells. It is important to note that IFN-λR1 expression is highly restricted in immune cells and is primarily limited to neutrophils and dendritic cells[3]. While dendritic cells were not the focus of this study, we did examine all immune cell subsets in our single cell RNA seq data and performed infection experiments in Plscr1<sup>floxStop</sup>/LysM-Cre<sup>+</sup> mice. We have not observed any significant findings in these populations. On the other hand, we do have some interesting preliminary data suggesting a role for PLSCR1 in regulating Ifn-λr1 expression and function in neutrophils. These findings are discussed in detail in our response to reviewer #3’s recommendation #12.
Reviewer #3 (Recommendations):
(1) In Figure 1B, the Plscr1 label should be moved to the y-axis so that readers don't confuse it with the Plscr1-/- mice used in the other figure panels. The fact that WT mice were used should be added in the figure legend.
We apologize for the confusion in the figures. We have moved Plscr1 label to the y-axis in Figure 1B and have mentioned Wt mice were used in the figure legend.
(2) In Figure 1C and D, the type of dose leading to the presented data should be added to help the reader. Also, shouldn't statistics be added?
We appreciate the suggestion and have added doses to Figure 1C and 1D. We are confused about the request of adding statistics by the reviewer, as two-way ANOVA tests were used to compare weight losses, and the significance was labeled on the figures.
(3) In Figures 1, F, and G, it is not indicated whether sublethal or lethal dose was used for the IAV infection. This should be very clear in the figure and figure legend.
We apologize for the confusion of infection doses used in the figures. We have added doses to Figure 1F, 1G and 1H.
(4) In Figure 1, the CTCF abbreviation should be explained in the Figure legend.
We have explained CTCF in the figure legend as requested.
(5) In Figure 2B, this is percentages of what?
Figure 2B shows the percentages of each immune cell type within total BAL cells.
(6) In Figures 3A and B, transcriptomes for each condition are from how many mice? Also, what do heatmaps show? Fold induction, differences, etc, and from what? What is compared with what? In addition, is there a discordance between the RNAseq data of Figure 3A and the qPCR data of Fig. 3C in terms of Ifnlr1 expression?
In Figure 3A and 3C (previously 3B), RNA from the whole lungs of 9 mice per PBS-treated group and 4 mice per IAV-infected group were pooled for transcriptomic analysis. Figure 3A represents a heatmap of differential gene expression, while Figure 3C (previously 3B) represents fold changes in gene expression relative to uninfected controls. In both heatmaps, gene expression values are color-coded from row minimum (blue) to row maximum (red), enabling comparison across groups within each gene (row). The major comparison of interest in these heatmaps is between Wt infected mice versus Plscr1<sup>-/-</sup> infected mice. We have added this information to the figure legend.
We also acknowledge the reviewer’s observation regarding the discordance between the RNA seq data of Figure 3A and the qPCR data of Figure 3B (previously 3C) for Ifnlr1 expression. To address this, we have repeated the qRT-PCR experiment with additional samples at 7 dpi. In the updated results, Wt mice consistently show significantly higher Ifn-λr1 expression than Plscr1<sup>-/-</sup> infected mice at both 3 dpi and 7 dpi, consistent with the RNA seq data. However, a time-dependent discrepancy between the RNA-seq and qRT-PCR datasets remains: Ifn-λr1 expression continues to increase at 7 dpi in the RNA-seq data (Figure 3A), whereas it declines in the qRT-PCR results (Figure 3B). The reason for this discrepancy remains unclear and has been addressed in the Discussion section.
(7) In Figure 3D, have the authors checked whether the Ifnlr1 antibody they use is indeed specific for Ifnlr1? Have they used any blocking peptide for the anti-mouse Ifn-λr1 polyclonal antibody they are using? Also, in Figure 3E, the marker used for staining should be indicated in the pictures of the lung section.
Unfortunately, a blocking peptide is not available for the anti-mouse Ifn-λr1 polyclonal antibody used in our study. To assess antibody specificity, we have performed immunofluorescence staining of Ifn-λr1 on lung tissues from Ifn-λr1<sup>-/-</sup> mice using the same antibody. No signal was detected (Supplemental Figure 5A), supporting the specificity of the antibody for Ifn-λr1.
As requested by the reviewer, we have added the marker (Ifn-λr1) to the pictures of the lung section in Figure 3E.
(8) In Figure 5, it's better to move each graph's label that stands to the top (e.g. PLSCR1, IFN-λR1 etc) to the y-axis label so that it doesn't get confused with the mouse -/- label.
We apologize for the confusion and have moved the top label to the y-axis in Figure 5.
(9) In Figure 6A, it is claimed that the 'two-dimensional UMAP demonstrated that these main lung cell populations (epithelial, endothelial, mesenchymal, and immune) were dynamic over the course of infection.'. This is not clear by the data. The percentage of cells per cluster should be calculated.
As requested by the reviewer, the proportion (Supplemental Figure 6A) and cell count (Supplemental Figure 6B) of each cluster have been calculated and included in “PLSCR1 Expression Is Upregulated in the Ciliated Airway Epithelial Compartment of Mice following Flu Infection” under “Results” section. Together with the two-dimensional UMAP (Figure 6A), these data demonstrate that the main lung cell populations (epithelial, endothelial, mesenchymal, and immune) were dynamic over the course of infection. Following infection, many populations emerged, particularly within the immune cell clusters. At the same time, some clusters were initially depleted and later restored, such as microvascular endothelial cells (cluster 2). Other populations, such as interferon-responsive fibroblasts (cluster 20), showed a dramatic yet transient expansion during acute infection and disappeared after infection resolved.
(10) In Figure 6 B and C, the legend should indicate that these are Violin plots. Also, if AT2 cells don't express Plscr1, does that indicate that in these cells Plscr1 is not needed for IFN-λR1 expression?
As requested, we have indicated in the legend of Figure 6B and 6C that these are violin plots. Plscr1 is expressed at low levels in AT2 cells. However, it is unclear whether Plscr1 is needed for Ifn-λr1 expression in AT2 cells, and it would be interesting to investigate further.
(11) In lines 302-304, it is stated that 'Among the various epithelial populations, ciliated epithelial cells not only had 303 the highest aggregated expression of Plscr1, but also were the only epithelial cell 304 population in which significantly more Plscr1 was induced in response to IAV infection.'. Which data/ figure support this statement?
Figure 6B shows that among the various epithelial populations, ciliated epithelial cells had the highest aggregated expression of Plscr1. To better illustrate this statement, we have rearranged the order of cell clusters from highest to lowest Plscr1 expression, and added red dots to indicate the mean expression levels for each cluster in Figure 6B.
Ciliated epithelial cells also had the most significant increase in Plscr1 expression (p < 2.22e-16 and p = 6.7e-05) in early IAV infection at 3 dpi (Figure 6C and Supplemental Figure 7A-7K). In comparison, AT1 cells were the only other epithelial cluster to show Plscr1 upregulation at 3dpi, but to a much less extent (p = 0.033, Supplemental Figure 7J). Supplemental Figure 7 was added to better support the statement and the explanation was added to “PLSCR1 Expression Is Upregulated in the Ciliated Airway Epithelial Compartment of Mice following Flu Infection” under “Results” section.
(12) As earlier, if Plscr1 is not expressed in neutrophils (Figure 6F), does that mean IFN-λR1 expression does not require Plscr1 in these cells?
Although Plscr1 is expressed at lower levels in neutrophils compared to epithelial cells, it is still detectable. In fact, our preliminary data suggest that IFN-λR1 expression in neutrophils is dependent on Plscr1. We have isolated neutrophils from peripheral blood and BAL of IAV-infected Wt and Plscr1<sup>-/-</sup> mice using a mouse neutrophil enrichment kit. Quantitative PCR results showed that Plscr1<sup>-/-</sup> neutrophils exhibit significantly lower expression of Ifn-λr1, alongside elevated levels of Il-1β, Il-6 and Tnf-α in IAV infection (see figures below). These findings suggest that Plscr1 may play an anti-inflammatory role in neutrophils by upregulating Ifn-λr1. These data were not included in the current manuscript because they are beyond the scope of current study, but we hope to address the role of PLSCR1 in regulating IFN-λR1 expression and function in neutrophils in a future study.
Author response image 3.
(13) The Figure 7A legend is not well stated. Something like ' Schematic representation of the experimental design of...' should be included. Also, Figure 7J is not referenced in the text.
We apologize for the unclear Figure 7A legend and have changed it to “Schematic representation of the experimental design of ciliated epithelial cell conditional Plscr1 KI mice.” Figure 8 (previously Figure 7J) has now been referenced in the text.
(14) In the Methods, more specific information in some parts should be provided. For example, the clones of the antibodies used should be included.
Apart from the 10x technology, the kits used and the type of the Illumina sequencing should be provided. Information on how the QC was performed (threshold for reads/cell, detected genes/per cells, and % of mitochondrial genes etc) should be added.
We apologize for the missing information in the “Methods”. We have now provided the clones of the antibodies used, the kit used to generate single-cell transcriptomic libraries, the type of the Illumina sequencing, and the QC performance data.
References
(1) Rusinova, I., et al., Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res, 2013. 41(Database issue): p. D1040-6.
(2) Xu, D., et al., PLSCR1 is a cell-autonomous defence factor against SARS-CoV-2 infection. Nature, 2023. 619(7971): p. 819-827.
(3) Donnelly, R.P., et al., The expanded family of class II cytokines that share the IL-10 receptor-2 (IL-10R2) chain. J Leukoc Biol, 2004. 76(2): p. 314-21.
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public Review):
Here the authors discuss mechanisms of ligand binding and conformational changes in GlnBP (a small E Coli periplasmic binding protein, which binds and carries L-glutamine to the inner membrane ATP-binding cassette (ABC) transporter). The authors have distinguished records in this area and have published seminal works. They include experimentalists and computational scientists. Accordingly, they provide comprehensive, high-quality, experimental and computational work. They observe that apo- and holo- GlnBP does not generate detectable exchange between open and (semi-) closed conformations on timescales between 100 ns and 10 ms. Especially, the ligand binding and conformational changes in GlnBP that they observe are highly correlated. Their analysis of the results indicates a dominant induced-fit mechanism, where the ligand binds GlnBP prior to conformational rearrangements. They then suggest that an approach resembling the one they undertook can be applied to other protein systems where the coupling mechanism of conformational changes and ligand binding. They argue that the intuitive model where ligand binding triggers a functionally relevant conformational change was challenged by structural experiments and MD simulations revealing the existence of unliganded closed or semi-closed states and their dynamic exchange with open unbound conformations, discuss alternative mechanisms that were proposed, their merits and difficulties, concluding that the findings were controversial, which, they suggest is due to insufficient availability of experimental evidence to distinguish them. As to further specific conclusions they draw from their results, they determine that a conformational selection mechanism is incompatible with their results, but induced fit is. They thus propose induced fit as the dominant pathway for GlnBP, further supported by the notion that the open conformation is much more likely to bind substrate than the closed one based on steric arguments. Considering the landscape of substrate-free states, in my view, the closed state is likely to be the most stable and, thus most highly populated. As the authors note and I agree that state can be sterically infeasible for a deep-pocketed substrate. As indeed they also underscore, there is likely to be a range of open states. If the populations of certain states are extremely low, they may not be detected by the experimental (or computational) methods. The free energy landscape of the protein can populate all possible states, with the populations determined by their relative energies. In principle, the protein can visit all states. Whether a particular state is observed depends on the time the protein spends in that state. The frequencies, or propensities, of the visits can determine the protein function. As to a specific order of events, in my view, there isn't any. It is a matter of probabilities which depend on the populations (energies) of the states. The open conformation that is likely to bind is the most favorable, permitting substrate access, followed by minor, induced fit conformational changes. However, a key factor is the ligand concentration. Ligand binding requires overcoming barriers to sustain the equilibrium of the unliganded ensemble, thus time. If the population of the state is low, and ligand concentration is high (often the case in in vitro experiments, and high drug dosage scenarios) binding is likely to take place across a range of available states. This is however a personal interpretation of the data. The paper here, which clearly embodies massive careful, and high-quality work, is extensive, making use of a range of experimental approaches, including isothermal titration calorimetry, single-molecule Förster resonance energy transfer, and surface-plasmon resonance spectroscopy. The problem the authors undertake is of fundamental importance.
Reviewer #2 (Public Review):
The manuscript by Han et al and Cordes is a tour-de-force effort to distinguish between induced fit and conformational selection in glutamine binding protein (GlnBP).
We thank the referee for the recognition of the work and effort that has gone into this manuscript.
It is important to say that I don't agree that a decision needs to be made between these two limiting possibilities in the sense that whether a minor population can be observed depends on the experiment and the energy difference between the states. That said, the authors make an important distinction which is that it is not sufficient to observe both states in the ligand-free solution because it is likely that the ligand will not bind to the already closed state. The ligand binds to the open state and the question then is whether the ligand sufficiently changes the energy of the open state to effectively cause it to close. The authors point out that this question requires both a kinetic and a thermodynamic answer. Their "method" combines isothermal titration calorimetry, single-molecule FRET including key results from multi-parameter photon-by-photon hidden Markov modelling (mpH2MM), and SPR. The authors present this "method" of combination of experiments as an approach to definitively differentiate between induced fit and conformational selection. I applaud the rigor with which they perform all of the experiments and agree that others who want to understand the exact mechanism of protein conformational changes connected to ligand binding need to do such a multitude of different experiments to fully characterize the process. However, the situation of GlnBP is somewhat unique in the high affinity of the Gln (slow offrate) as compared to many small molecule binding situations such as enzyme-substrate complexes. It is therefore not surprising that the kinetics result in an induced fit situation.
For us these comments are an essential part of the conceptual aspects of our work and the resulting research. From a descriptive viewpoint, it is essential for us (and we tried to further highlight and stress this in the updated version of our paper) that IF and CS are two kinetic mechanisms of ligand binding. They imply – if active in a biomolecular system – a temporal order and timescale separation of ligand binding and conformational changes. Since we found many conflicting results for the binding mechanism of GlnBP, but also other SPBs, we decided to assess the situation in GlnBP.
In the case of the E-S complexes I am familiar with, the dissociation is much more rapid because the substrate binding affinity is in the micromolar range and therefore the re-equilibration of the apo state is much faster. In this case, the rate of closing and opening doesn't change much whether ligand is present or not. Here, of course, once the ligand is bound the re-equilibration is slow. Therefore, I am not sure if the conclusions based on this single protein are transferrable to most other protein-small molecule systems.
We do not argue that our results and interpretations are valid for most other protein-ligand systems may those be enzymes or simple ligand binders. Yet, based on the conservation of ABC-related SBPs and the fact that quite a few of them show sub-µM Kds, we render it likely to find many analogous situations as for GlnBP also based on our previous results e.g., from de Boer et al., eLife (2019).
I am also not sure if they are transferrable to protein-protein systems where both molecules the ligand and the receptor are expected to have multiscale dynamics that change upon binding.
As we argue above the two mechanisms IF/CS imply a clear temporal order and separation of timescales for ligand binding and conformational changes. These mechanisms are simple and extreme cases that we tested before more complex kinetic schemes are inferred for the description of ligand binding and conformational changes (which might not be necessary).
Strengths:
The authors provide beautiful ITC data and smFRET data to explore the conformational changes that occur upon Gln binding. Figure 3D and Figure 4 (mpH2MM data) provide the really critical data. The multi-parameter photon-by-photon hidden Markov modelling (mpH2MM) data. In the presence of glutamine concentrations near the Kd, two FRET-active sub-populations are identified that appear to interconvert on timescales slower than 10 ms. They then do a whole bunch of control experiments to look for faster dynamics (Figure 5). They also do TIRF smFRET to try to compare their results to those of previous publications. Here, they find several artifacts are occurring including inactivation of ~50% of the proteins. They also perform SPR experiments to measure the association rate of Gln and obtain expectedly rapid association rates on the order of 10<sup>^</sup>8 M-1s-1.
Thank you.
Weaknesses:
Looking at the traces presented in the supplementary figures, one can see that several of the traces have more than one molecule present. The authors should make sure that they use only traces with a single photobleaching event for each fluorophore. One can see steps in some of the green traces that indicate two green fluorophors (likely from 2 different molecules) in the traces. This is one of the frequent problems with TIRF smFRET with proteins, that only some of the spots represent single molecules and the rest need to be filtered out of the analysis.
We have inspected all TIRF data provided with the manuscript and assume that the referee refers to data shown in current Appendix Figure 4/5. We agree that those traces in which no photo bleaching occurs could potentially be questioned, yet they would not change our interpretations and thus decided to leave the figure as is.
The NMR experiments that the authors cite are not in disagreement with the work presented here. NMR is capable of detecting "invisible states" that occur in 1-5% of the population. SmFRET is not capable of detecting these very minor states. I am quite sure that if NMR spectroscopists could add very high concentrations of Gln they would also see a conversion to the closed population.
We agree with the referee that NMR is capable of detecting invisible states that occur in 1-5% of the population (see e.g., the paper cited in our manuscript by Tang, C et al., Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR. Nature 2007, 449, 1078). Yet, we see a strong disagreement between our work and papers on GlnBP, where a combination of NMR, FRET and MD was used (Feng, Y. et al., Conformational Dynamics of apo‐GlnBP Revealed by Experimental and Computational Analysis. Angewandte Chemie 2016, 55, 13990; Zhang, L. et al., Ligand-bound glutamine binding protein assumes multiple metastable binding sites with different binding affinities. Communications biology 2020, 3, 1). These inconsistencies were also noted by others in the field (Kooshapur, H. et al., NMR Analysis of Apo Glutamine‐Binding Protein Exposes Challenges in the Study of Interdomain Dynamics. Angewandte Chemie 2019, 58, 16899) and we reemphasize that this latest NMR publication comes to similar conclusions as we present in our manuscript.
Reviewer #1 (Recommendations For The Authors):
The paper embodies massive careful and high-quality work, and is extensive, making use of a range of experimental approaches, including isothermal titration calorimetry, single-molecule Förster resonance energy transfer, and surface-plasmon resonance spectroscopy. Considering this extensiveness, I do not see what more the authors can do.
We very much appreciate the assessment and positive comments of the referee, but still tried to incorporate simulation data to support our interpretations.
Reviewer #2 (Recommendations For The Authors):
(1) Looking at the traces presented in the supplementary figures, one can see that several of the traces have more than one molecule present. The authors should make sure that they use only traces with a single photobleaching event for each fluorophore. One can see steps in some of the green traces that indicate two green fluorophors (likely from 2 different molecules) in the traces. This is one of the frequent problems with TIRF smFRET with proteins, that only some of the spots represent single molecules and the rest need to be filtered out of the analysis.
See response above for iteration of TIRF data selection and analysis.
(2) The NMR experiments that the authors cite are not in disagreement with the work presented here. NMR is capable of detecting "invisible states" that occur in 1-5% of the population. SmFRET is not capable of detecting these very minor states. I am quite sure that if NMR spectroscopists could add very high concentrations of Gln they would also see a conversion to the closed population.
See response above.
Minor point:
(1) It is difficult to see what is going on between apo and holo in Figure 1B. Could the authors make Figure 1a, 1b apo, and 1b holo in the same orientation (by aligning D2 or D1 to each other in all figures) so one can see which helices are in the same place and which have moved?
We respectfully disagree and decided to keep this figure as it is
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public review):
Summary:
This study focuses on the bacterial metabolite TMA, generated from dietary choline. These authors and others have previously generated foundational knowledge about the TMA metabolite TMAO, and its role in metabolic disease. This study extends those findings to test whether TMAO's precursor, TMA, and its receptor TAAR5 are also involved and necessary for some of these metabolic phenotypes. They find that mice lacking the host TMA receptor (Taar5-/-) have altered circadian rhythms in gene expression, metabolic hormones, gut microbiome composition, and olfactory and innate behavior. In parallel, mice lacking bacterial TMA production or host TMA oxidation have altered circadian rhythms.
Strengths:
These authors use state-of-the-art bacterial and murine genetics to dissect the roles of TMA, TMAO, and their receptor in various metabolic outcomes (primarily measuring plasma and tissue cytokine/gene expression). They also follow a unique and unexpected behavioral/olfactory phenotype. Statistics are impeccable.
Weaknesses:
Enthusiasm for the manuscript is dampened by some ambiguous writing and the presentation of ideas in the introduction, both of which could easily be improved upon revision.
We apologize for the abbreviated and ambiguous writing style in our original submission. Given Reviewer 2 also suggested reorganizing and rewriting certain parts, we have spent time to remove ambiguity by adding additional points of clarification and adding more historical context to justify studying TMA-TAAR5 signaling in regulating host circadian rhythms. We have also reorganized the presentation of data aligned with this.
Reviewer #2 (Public review):
Summary:
In the manuscript by Mahen et al., entitled "Gut Microbe-Derived Trimethylamine Shapes Circadian Rhythms Through the Host Receptor TAAR5," the authors investigate the interplay between a host G protein-coupled receptor (TAAR5), the gut microbiota-derived metabolite trimethylamine (TMA), and the host circadian system. Using a combination of genetically engineered mouse and bacterial models, the study demonstrates a link between microbial signaling and circadian regulation, particularly through effects observed in the olfactory system. Overall, this manuscript presents a novel and valuable contribution to our understanding of hostmicrobe interactions and circadian biology. However, several sections would benefit from improved clarity, organization, and mechanistic depth to fully support the authors' conclusions.
Strengths:
(1) The manuscript addresses an important and timely topic in host-microbe communication and circadian biology.
(2) The studies employ multiple complementary models, e.g., Taar5 knockout mice, microbial mutants, which enhance the depth of the investigation.
(3) The integration of behavioral, hormonal, microbial, and transcript-level data provides a multifaceted view of the observed phenotype.
(4) The identification of olfactory-linked circadian changes in the context of gut microbes adds a novel perspective to the field.
Weaknesses:
While the manuscript presents compelling data, several weaknesses limit the clarity and strength of the conclusions.
(1) The presentation of hormonal, cytokine, behavioral, and microbiome data would benefit from clearer organization, more detailed descriptions, and functional grouping to aid interpretation.
We appreciate this comment and have reorganized the data to improve functional grouping and readability. We have also added additional detail to descriptions of the data in the revised figure legends and results.
(2) Some transitions-particularly from behavioral to microbiome data-are abrupt and would benefit from better contextual framing.
We agree with this comment, and have added additional language to provide smoother transitions. This in many cases brings in historical context of why we focused on both behavioral and microbiome alterations in this body of work.
(3) The microbial rhythmicity analyses lack detail on methods and visualization, and the sequencing metadata (e.g., sample type, sex, method) are not clearly stated.
We apologize for this, and have now added more detail in our methods, figures, and figure legends to ensure the reader can easily understand sample type, sex, and the methods used.
(4) Several figures are difficult to interpret due to dense layouts or vague legends, and key metabolites and gene expression comparisons are either underexplained or not consistently assessed across models.
Aligned with the last comment we now added more detail in our methods, figures, and figure legends to provide clear information. We have now provided additional data showing the same key metabolites, hormones, and gene expression alterations in each model if the same endpoints were measured.
(5) Finally, while the authors suggest a causal role for TAAR5 and its ligand in circadian regulation, the current data remain correlative; mechanistic experiments or stronger disclaimers are needed to support these claims.
We agree with this comment, and as a result have removed any language causally linking TMA and TAAR5 together in circadian regulation. Instead, we only state finding in each model and refrain from overinterpreting.
Reviewer #3 (Public review):
Summary:
Deletion of the TMA-sensor TAAR5 results in circadian alterations in gene expression, particularly in the olfactory bulb, plasma hormones, and neurobehaviors.
Strengths:
Genetic background was rigorously controlled.
Comprehensive characterization.
Weaknesses:
The weaknesses identified by this reviewer are minor.
Overall, the studies are very nicely done. However, despite careful experimentation, I note that even the controls vary considerably in their gene expression, etc, across time (eg, compare control graphs for Cry 1 in IB, 4B). It makes me wonder how inherently noisy these measurements are. While I think that the overall point that the Taar5 KO shows circadian changes is robust, future studies to dissect which changes are reproducible over the noise would be helpful.
We thank the reviewer for this insightful comment. We completely agree that there are clear differences in the circadian data in experiments from Taar5<sup>-/-</sup> mice and those from gnotobiotic mice where we have genetically deleted CutC. Although the data from Taar5<sup>-/-</sup> mice show nice robust circadian rhythms, the data from mice where microbial CutC is altered have inherently more “noise”. We attribute some of this to the fact that the Taar5<sup>-/-</sup> mouse experiment have a fully intact and diverse gut microbiome . Whereas, the gnotobiotic study with CutC manipulation includes only a 6 member microbiome community that does not represent the normal microbiome diversity in the gut. This defined synthetic community was used as a rigorous reductionist approach, but likely affected the normal interactions between a complex intact gut microbiome and host circadian rhythms. We have added some additional discussion to indicate this in the limitations section of the manuscript.
Impact:
These data add to the growing literature pointing to a role for the TMA/TMAO pathway in olfaction and neurobehavioral.
Reviewer #1 (Recommendations for the authors):
I suggest a revision of the writing and organization. The potential impact of the study after reading the introduction is unclear. One example, in the intro, " TMAO levels are associated with many human diseases including diverse forms of CVD5-12, obesity13,14, type 2 diabetes15,16, chronic kidney disease (CKD)17,18, neurodegenerative conditions including Parkinson's and Alzheimer's disease19,20, and several cancers21,22" It would be helpful to explain how the previous literature has distinguished that the driver of these phenotypes is TMA/TMAO and not increased choline intake. Basically, for a TMA/O novice reader, a more detailed intro would be helpful.
We appreciate this insightful comment and have now provided a more expansive historical context for the reader regarding the effects of choline consumption (which impacts many things, including choline, acetylcholine, phosphatidylcholine, TMA, TMAO, etc) versus the primary effects of TMA and TMAO.
There were also many uses of vague language (regulation/impact/etc). Directionality would be super helpful.
We thank the reviewer for this recommendation and have improved language as suggested to show directionality of our findings. The terms regulation, impact, shape etc. are used only when we describe multiple variable changing at the same time over the time course of a 24-hour circadian period (some increased and some decreased).
Reviewer #2 (Recommendations for the authors):
In the manuscript by Mahen et al., entitled "Gut Microbe-Derived Trimethylamine Shapes Circadian Rhythms Through the Host Receptor TAAR5," the authors investigate the interplay between a host G protein-coupled receptor (TAAR5), the gut microbiota-derived metabolite trimethylamine (TMA), and the host circadian system. Using a combination of genetically engineered mouse and bacterial models, the study demonstrates a link between microbial signaling and circadian regulation, particularly through effects observed in the olfactory system. Overall, this manuscript presents a novel and valuable contribution to our understanding of hostmicrobe interactions and circadian biology. However, several sections would benefit from improved clarity, organization, and mechanistic depth to fully support the authors' conclusions. Below are specific major and minor suggestions intended to enhance the presentation and interpretation of the data.
Major suggestions:
(1) Consider adding a schematic/model figure as Panel A early in the manuscript to help readers understand the experimental conditions and major comparisons being made.
We thank the reviewer for this recommendation and have added a graphical abstract figure to help the reader understand the major comparisons being made.
(2) Could the authors present body weight and food intake characteristics in Taar5 KO vs. WT animals?
We have added body weight data as requested in Figure 1, Figure supplement 1. Although we have not stressed these mice with a high fat diet for these behavioral studies, under chow-fed conditions studied here we did not find any significant differences in body weight. Given no difference in body weight, we did not collect data on food consumption and have mentioned this as a limitation in the discussion.
(3) Several figures, especially Figures 3 and 4, and Supplemental Figures, would benefit from more structured organization and expanded legends. Grouping related data into thematic panels (e.g., satiety vs. appetite hormones, behavioral domains) may help improve readability.
We appreciate the reviewer’s thoughtful comments and agree that reorganization would improve clarity. We have reorganized figures to improve clarity and have expanded the figure legends to provide more detail on experimental methods.
(4) Clarify and expand the description of hormonal and cytokine changes. For instance, the phrase "altered rhythmic levels" is vague - do the authors mean dampened, phase-shifted, enhanced, etc., relative to WT controls?
Given a similar suggestion was made by Reviewer 1, we have provided more precise language focused on directionality and which specific endpoints we are referring to. For anything looking at circadian rhythms, the revised manuscript includes specific indications when we are discussing mesor, amplitude, and acrophase alterations. The terms regulation, impact, shape etc. are used only when we describe multiple complex variables changing at the same time over the time course of a 24-hour circadian period (some increased and some decreased).
(5) Consider grouping hormones and cytokines functionally (e.g., satiety vs. appetite-stimulating, pro- vs. antiinflammatory) to better interpret how these changes relate to the KO phenotype.
We thank the reviewer for this recommendation, and have re-organized figure panels to reflect this.
(6) Please provide a more detailed description of the behavioral results, particularly those in Supplemental Figure 2.
We have both expanded the methods description in the revised figure legends, but have also added a more detailed description of the behavioral results.
(7) As with hormonal data, behavioral outcomes would be easier to follow if organized thematically (e.g., locomotor activity, anxiety-like behavior, circadian-related behavior), especially for readers less familiar with behavioral assays.
We appreciate this reviewer’s comment and agree that we can better group our data to show how each test is associated with the type of behavior it assesses. As a result we have reorganized the behavioral data into broad categories such as olfactory-related, innate, cognitive, depressive/anxiety-like, or social behaviors. We have also new data in each of these behavioral categories to provide a more comprehensive understanding of behavioral alterations seen in Taar5<sup>-/-</sup> mice.
(8) The following statement needs clarification: "Also, it is important to note that many behavioral phenotypes examined, including tests not shown, were unaltered in Taar5-/- mice (Figures S2G, S2H, and S2I)." Consider rephrasing to explicitly state the intended message: are the authors emphasizing a lack of behavioral phenotype, or highlighting specific unaltered aspects?
We apologize for this confusing statement, and have changed the verbiage to improve readability. To expand the comprehensive nature of this study, we also now include the tests that were “not shown” in the original submission to provide a more comprehensive understanding of behavioral alterations seen in Taar5<sup>-/-</sup> mice. These new data are included as 6 different figure supplements to main Figure 2.
(9) The transition from behavior to microbiome data feels abrupt. Can the authors better explain whether the behavioral changes are thought to result from gut microbial function, independent of TMA-Taar5 signaling?
We apologize for the poor transitions in our writing style. We have spent time to explain the previous findings linking the TMA pathway to circadian reorganization of the gut microbiome (mostly coming from our original paper Schugar R, et al. 2022, eLife) and how this correlates with behavioral phenotypes. Although at this point it is difficult to know whether the microbiome changes are driving behavioral changes, or vice versa it could be central TAAR5 signaling is altering oscillations in gut microbiome, we present our findings here as a framework for follow up studies to more precisely get at these questions. It is important to note that our experiment using defined community gnotobiotic mice with or without the capacity to produce TMA (i.e. CutC-null community) shows that clearly microbial TMA production can impact host circadian rhythms in the olfactory bulb. Additional experiments beyond the scope of this work will be required to test which phenotypes originate from TMA-TAAR5 signaling versus more broad effects of the restructured gut microbiome.
(10) For Figure 3A, please expand the microbiome results with more granularity:
(a) Indicate in the Results section whether the sequencing method was 16S amplicon or metagenomic.
Sequencing was done using 16S rRNA amplicon sequencing using methods published by our group (PMID: 36417437, PMID: 35448550).
(b) State whether samples were from males, females, or a mix.
We have indicated that all mice from Figure 1 were male mice in the revised figure legend.
(c) Clarify whether beta diversity is based on phylogenetic or non-phylogenetic metrics. Consider using both types if not already done.
Beta diversity was analyzed using the Bray-Curtis dissimilarity index as the metric. Details have been included in the methods section.
(d) Make lines partially transparent in the Beta-diversity plot so that individual points are visible.
We have now updated the Beta-diversity plot with individual points visualized.
(e) Clarify what percentage of variation in the Beta-diversity plot is explained by CCA1, and whether this low percentage suggests minimal community-level differences.
We have updated the Beta-diversity plot to include the R<sup>2</sup> and p-values associated with these data.
(f) Confirm if the y-axis on the Beta-diversity plot should be labeled CCA2 rather than "CCAA 1".
We appreciate this comments, given it identified a typographical error in the plot. The revised figure now include the proper label of CCA2 instead of CCAA 1.
(11) For Figure 3B:
(a) Provide a description of the taxonomy plot in the results.
We have added a description of the taxonomy plot in the revised results section.
(b) Add phylum-level labels and enlarge the legend to improve the readability of genus-level data.
We agree this is a good suggestion so have enlarged the legend for the genus-level data and have also added phylum-level plots as well in the revised manuscript in Figure 3, figure supplement 1.
(12) Rhythmicity of the microbiome is central to the manuscript. The current approach of comparing relative abundance at discrete time points is limiting.
We thank the reviewer for this comment. We agree with this statement that discrete timepoint are not enough to describe circadian rhythmicity. In addition to comparing genotypes at discrete time points, we also used a rigorous cosinor analysis to plot the data over a 24-hour time period, and those differences are shown in the figure itself as well as Table 1.
(a) Please describe how rhythmicity was determined, e.g., what data or statistical method supports the statement: "Taar5-/- mice showed loss of the normal rhythmicity for Dubosiella and Odoribacter genera yet gained in amplitude of rhythmicity for Bacteroides genera (Figure 3 and S3)."
We appreciate this reviewer comment. Rhythmicity was determined using a cosinor analysis by use of an R program. Cosinor analysis is a statistical method used to model and analyze rhythmic patterns in time-series data, typically assuming a sinusoidal (cosine) shape. It estimates key parameters like mesor (mean level), amplitude (height of oscillation), and acrophase (timing of the peak), making it especially useful in fields like chronobiology and circadian rhythm research. We have used this in previous research to describe circadian rhythms. We do plan to improve language considering directionality of these circadian changes.
(b) Supplemental Figure S3 needs reorganization to highlight key findings. It's not currently clear how taxa are arranged or what trends are being shown.
The data in Figure S3 show the entire 24-hour time course of the cecal taxa that were significantly altered for at least one time point between Taar5<sup>+/+</sup> and Taar5<sup>-/-</sup> mice. Given we showed time pointspecific alterations in the Main Figure 3, we thought these more expansive plots would be important to show to depict how the circadian rhythms were altered.
(c) Supplemental Table 1, which includes 16S features, should be referenced and discussed in the microbiome section.
We have now referenced and discussed Supplemental Table 1 which includes all cosinor statistics for microbiome and other data presented in circadian time point studies.
(13) Did the authors quantify the 16S rRNA gene via RT-PCR to determine if this was similar between KO and WT over the 24-hour period?
We did not quantify 16S rRNA gene via RT-PCR, but do not think adding this will change our overall interpretations.
(14) Reorganize Figure 4 to align with the order of results discussed-starting with TMA and TMAO, followed by related metabolites like choline, L-carnitine, and gamma-butyrobetaine.
We thank the reviewer for this comment. We have chosen this organization because it is ordered from substrates (choline, L-carnitine, and betaine) to the microbe-associated products (TMA then TMAO). We will improve the writing associated with this figure to clearly explain this organization.
(a) Although the changes in the latter metabolites are more modest, they may still have physiological relevance. Could the authors comment on their significance?
We appreciate this reviewer comment and agree. We have expanded the results and discussion to address this.
(15) The authors note similarities in circadian gene expression between Taar5 KO mice and Clostridium sporogenes WT vs. ΔcutC mice, but the gene patterns are not consistent.
(a) Can the authors clarify what conclusions can reasonably be drawn from this comparison?
We hesitate to make definitive conclusions in the manuscript on why the gene patterns are not consistent, because it would be speculation. However, one major factor likely driving differences is the status of the diversity of the gut microbiome in the different studies. For instance, in the studies using Taar5<sup>+/+</sup> and Taar5<sup>-/-</sup> mice there is a very diverse microbiome in these conventionally housed mice. In contrast, by design the experiment using Clostridium sporogenes WT vs. ΔcutC communities is a reductionist approach that allows us to genetically define TMA production. In these gnotobiotic mice, the simplified community has very limited diversity and this likely alters the host circadian rhythms in gene expression quite dramatically. Although it is impossible to directly compare the results between these experiments given the difference microbiome diversity, there are clearly alterations in host gene expression when we manipulate TMA production (i.e. ΔcutC community) or TMA sensing (i.e. Taar5<sup>-/-</sup>).
(16) Were circadian and metabolic genes (e.g., Arntl, Cry1, Per2, Pemt, Pdk4) also analyzed in brown adipose tissue of Taar5 KO mice, and how do these results compare to the Clostridium models?
We thank the reviewer for this comment. Unfortunately, we did not collect brown adipose tissue in our original Taar5 study. We plan on doing this in future follow up studies studying cold-induced thermogenesis that are beyond the scope of this manuscript. However, we have decided to include data from our two timepoint Taar5 study which looks at ZT2 (9am) and ZT14 (9pm). There are clear differences in circadian genes between these timepoints.
(17) To allow a more direct comparison, please ensure the same cytokines (e.g., IL-1β, IL-2, TNF-α, IFN-γ, IL6, IL-33) are reported for both the Taar5 KO and microbial models.
We thank the reviewer for this comment and now include data from the same cytokines for each study.
(18) What was the defined microbial community used to colonize germ-free mice with C. sporogenes strains? Did this community exhibit oscillatory behavior?
To define TMA levels using a genetically-tractable model of a defined microbial community, we leveraged access to the community originally described by our collaborator Dr. Federico Rey (University of Wisconsin – Madison) (PMID: 25784704). We chose this community because it provide some functional metabolic diversity and is well known to allow for sufficient versus deficient TMA production. We are thankful for the reviewer comments about oscillatory behavior of this defined community, and to be responsive have performed sequencing to detect the species over time. These data are now included in the revised manuscript and show that there are clear differences in the oscillatory behavior of the defined community members. These data provide additional support that bacterial TMA production not only alters host circadian rhythms, but also the rhythmic behavior of gut bacteria themselves which has never been described before.
(19) Can the authors explain the rationale for measuring additional metabolites such as tryptophan, indole acetic acid, phenylacetic acid, and phenylacetylglycine? How are these linked to CutC gene function or Taar5 signaling?
We appreciate that this could be confusing, but have included other gut microbial metabolites to be as comprehensive as possible. This is important to include because we have found in other gnotobiotic studies where we have genetically altered metabolite production, if we alter one gut microbe-derived metabolite there can be unexpected alterations in other distinct classes of microbe-derived metabolites (PMID: 37352836). This is likely due to the fact that complex microbe-microbe and microbehost interactions work together to define systemic levels of circulating metabolites, influencing both the production and turnover of distinct and unrelated metabolites.
(20) The authors make several strong claims suggesting that loss of Taar5 or disruption of its ligand directly alters the circadian gene network. However, the current data are correlative. The authors should clarify that these findings demonstrate associations rather than direct causal effects, unless additional mechanistic evidence is provided. Approaches such as studies conducted in constant darkness, measurements of wheelrunning behavior, or analyses that control for potential confounding factors, e.g., inflammation or metabolic disruption, would help establish whether the observed changes in clock gene expression are primary or secondary effects. The authors are encouraged to either soften these causal claims or acknowledge this limitation explicitly in the discussion.
We thank the reviewer for this comment. We agree and have softened our language about direct effects of TMA via TAAR5 because we agree the data presented here are correlative only.
Minor suggestions:
(1) Avoid repetitive phrases such as "it is important to note..." for improved flow. Rephrasing these instances will enhance readability.
We thank the reviewer for this suggestion and have deleted such repetitive phrases.
(2) For Figure 2, remove interpretations above he graphs and use simple, descriptive panel labels, similar to those in Supplemental Figure 2.
We have removed these interpretations as suggested, but have retained descriptive panel labels to help the reader understand what type of data are being presented.
Reviewer #3 (Recommendations for the authors):
Minor:
In Figure 1D, UCP1 does not appear to be significantly changed.
We thank the reviewer for this comment and agree that UCP1 gene expression is not significantly altered . However, given the key role that UCP1 plays in white adipose tissue beiging, which is suppressed by the TMAO pathway, we think it is critical to show that this effect appears unaffected by perturbed TMA-TAAR5 signaling.
It would be helpful, in the discussion, to summarize any consistent changes across Taar5 KO, CutC deletion, and FMO3 deletion.
We have added this to the discussion, but as discussed above we hesitate to make strong interpretations about consistency between the models because the microbiome diversity is so different between the studies, and we did not measure all endpoints in both models.
For the Cosinor analysis, it may be helpful to remove the p-values that are >0.05 from the figures.
We have now removed any non-significant p-values that are associated with our figures.
For Figure 2, Supplement 1E, what are the two bars for each genotype?
We appreciate the reviewer pointing this out and will further explain this test in the figure with labels and in the legend.
Reviewer #3 (Public review):
The paper by Maggi et al. builds on earlier work by the team (Paatero et al., 2018) on oriented junction-based lamellipodia (JBL). They validate the role of JBLs in guiding endothelial cell rearrangements and utilise high-resolution time-lapse imaging of novel transgenic strains to visualise the formation of distal junctions and their subsequent fusion with proximal junctions. Through functional analyses of Arp2/3 and actomyosin contractility, the study identifies JBLs as localized mechanical hubs, where protrusive forces drive distal junction formation, and actomyosin contractility brings together the distal and proximal junctions. This forward movement provides a unique directionality which would contribute to proper lumen formation, EC orientation, and vessel stability during these early stages of vessel development.
Time-lapse live imaging of VEC, ZO-1, and actin reveals that VEC and ZO-1 are initially deposited at the distal junction, while actin primarily localizes to the region between the proximal and distal sites. Using a photoconvertible Cdh5-mClav2 transgenic line, the origin of the VEC aggregates was examined. This convincingly shows that VE-cadherin was derived from pools outside the proximal junctions. However, in addition to de novo VEC derived from within the photoconverted cell, could some VEC also be contributed by the neighbouring endothelial cell to which the JBL is connected?
As seen for JAILs in cultured ECs, the study reveals that Arp2/3 is enhanced when JBLs form by live imaging of Arpc1b-Venus in conjunction with ZO-1 and actin. Therefore Arp2/3 likely contributes to the initial formation of the distal junction in the lamellopodium.
Inhibiting Arp2/3 with CK666 prevents JBL formation, and filopodia form instead of lamellopodia. This loss of JBLs leads to impaired EC rearrangements.
Is the effect of CK666 treatment reversible? Since only a short (30 min) treatment is used, the overall effect on the embryo would be minimal, and thus washing out CK666 might lead to JBL formation and normalized rearrangements, which would further support the role of Arp2/3.
From the images in Figure 4d it appears that ZO-1 levels are increased in the ring after CK666 treatment. Has this been investigated, and could this overall stabilization of adhesion proteins further prevent elongation of the ring?
To explore how the distal and proximal junctions merge, imaging of spatiotemporal imaging of Myl9 and VEC is conducted. It indicates that Myl9 is localized at the interjunctional fusion site prior to fusion. This suggests pulling forces are at play to merge the junctions, and indeed Y 27632 treatment reduces or blocks the merging of these junctions.
For this experiment, a truncated version of VEC was use,d which lacks the cytoplasmic domain. Why have the authors chosen to image this line, since lacking the cytoplasmic domain could also impair the efficiency of tension on VEC at both junction sites? This is as described in the discussion (lines 328-332).
Since the time-lapse movies involve high-speed imaging of rather small structures, it is understandable that these are difficult to interpret. Adding labels to indicate certain structures or proteins at essential timepoints in the movies would help the readers understand these.
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public Review):
Summary:
Ravichandran et al investigate the regulatory panels that determine the polarization state of macrophages. They identify regulatory factors involved in M1 and M2 polarization states by using their network analysis pipeline. They demonstrate that a set of three regulatory factors (RFs) i.e., CEBPB, NFE2L2, and BCL3 can change macrophage polarization from the M1 state to the M2 state. They also show that siRNA-mediated knockdown of those 3-RF in THP1-derived M0 cells, in the presence of M1 stimulant increases the expression of M2 markers and showed decreased bactericidal effect. This study provides an elegant computational framework to explore the macrophage heterogeneity upon different external stimuli and adds an interesting approach to understanding the dynamics of macrophage phenotypes after pathogen challenge.
Strengths:
This study identified new regulatory factors involved in M1 to M2 macrophage polarization. The authors used their own network analysis pipeline to analyze the available datasets. The authors showed 13 different clusters of macrophages that encounter different external stimuli, which is interesting and could be translationally relevant as in physiological conditions after pathogen challenge, the body shows dynamic changes in different cytokines/chemokines that could lead to different polarization states of macrophages. The authors validated their primary computational findings with in vitro assays by knocking down the three regulatory factors-NCB.
We thank the reviewer for reading our manuscript and for the encouraging comments.
Weaknesses:
One weakness of the paper is the insufficient analysis performed on all the clusters. They used macrophages treated with 28 distinct stimuli, which included a very interesting combination of pro- and anti-inflammatory cytokines/factors that can be very important in the context of in vivo pathogen challenge, but they did not characterize the full spectrum of clusters.
We have performed a functional enrichment analysis of all the clusters and added a section describing the results (Fig 1B). We believe this work will provide a basis for future experiments to characterize other clusters.
We have also performed a Principal Component Analysis (PCA) using hall mark genes of inflammation and the NCB panel alone to show the relative position of all clusters with respect to each other
Although they mentioned that their identified regulatory panels could determine the precise polarization state, they restricted their analysis to only the two well-established macrophage polarization states, M1 and M2. Analyzing the other states beyond M1 and M2 could substantially advance the field. They mentioned the regulatory factors involved in individual clusters but did not study the potential pathway involving the target genes of these regulatory factors, which can show the importance of different macrophage polarization states. Importantly, these findings were not validated in primary cells or using in vivo models.
We agree it would be useful to demonstrate the polarization switch in other systems as well. However, it is currently infeasible for us to perform these experiments.
Reviewer #2 (Public Review):
Summary:
The authors of this manuscript address an important question regarding how macrophages respond to external stimuli to create different functional phenotypes, also known as macrophage polarization. Although this has been studied extensively, the authors argue that the transcription factors that mediate the change in state in response to a specific trigger remain unknown. They create a "master" human gene regulatory network and then analyze existing gene expression data consisting of PBMC-derived macrophage response to 28 stimuli, which they sort into thirteen different states defined by perturbed gene expression networks. They then identify the top transcription factors involved in each response that have the strongest predicted association with the perturbation patterns they identify. Finally, using S. aureus infection as one example of a stimulus that macrophages respond to, they infect THP-1 cells while perturbing regulatory factors that they have identified and show that these factors have a functional effect on the macrophage response.
Strengths:
The computational work done to create a "master" hGRN, response networks for each of the 28 stimuli studied, and the clustering of stimuli into 13 macrophage states is useful. The data generated will be a helpful resource for researchers who want to determine the regulatory factors involved in response to a particular stimulus and could serve as a hypothesis generator for future studies.
The streamlined system used here - macrophages in culture responding to a single stimulus - is useful for removing confounding factors and studying the elements involved in response to each stimulus.
The use of a functional study with S. aureus infection is helpful to provide proof of principle that the authors' computational analysis generates data that is testable and valid for in vitro analysis.
We thank the reviewer for reading our manuscript and for the encouraging comments
Weaknesses:
Although a streamlined system is helpful for interrogating responses to a stimulus without the confounding effects of other factors, the reality is that macrophages respond to these stimuli within a niche and while interacting with other cell types. The functional analysis shown is just the first step in testing a hypothesis generated from this data and should be followed with analysis in primary human cells or in an in vivo model system if possible.
It would be helpful for the authors to determine whether the effects they see in the THP-1 immortalized cell line are reproduced in another macrophage cell line, or ideally in PBMC-derived macrophages.
We agree; It would be useful in the future to demonstrate the polarization switch in other systems as well. We believe the results we provide here will inform future studies on other systems.
The paper would benefit from an expanded explanation of the network mining approach used, as well as the cluster stability analysis and the Epitracer analysis. Although these approaches may be published elsewhere, readers with a non-computational background would benefit from additional descriptions.
We have elaborated on the network mining approach and added a schematic diagram (Fig S13) to describe the EpiTracer algorithm.
Although the authors identify 13 different polarization states, they return to the iM0/M1/M2 paradigm for their validation and functional assays. It would be useful to comment on the broader applications of a 13-state model.
We have included a new figure panel describing the functional enrichment analysis of all the clusters (Fig 1B) and added a section describing the results. We have also performed a Principal Component Analysis (PCA) using hallmark gene of inflammation and the NCB panel alone to show the relative position of all clusters with respect to each other. The PCA plot shows that C11(M1) and C3(M2) are roughly at two extreme ends, with other clusters between them, forming something resembling a punctuated continuum of states.
The relative contributions of each "switching factor" to the phenotype remain unclear, especially as knocking out each individual factor changes different aspects of the model (Fig. S5).
Fig S5 shows the effect on phenotype upon individual knockdown of the switching factors, from which we deduce that CEBPB has the largest contribution in determining the phenotype. However, we maintain that all three genes are necessary as a panel for M1/M2 switching.
Reviewer #1 (Recommendations For The Authors):
The manuscript by Ravichandran et al describes the networks of genes that they named j"RF" associated with M1 to M2 polarization of macrophages by using their computational pipelines. They have shown 13 clusters of human macrophage polarization state by using an available database of different combinatorial treatments with cytokines, endotoxin, or growth factors, which is interesting and could be useful in the research field. However, there are a few comments which will help to understand the subject more precisely.
(1,2) The authors claimed to identify key regulatory factors involved in the human macrophage polarization from M1 to M2. However, recent advances suggest that macrophage polarization cannot be restricted to M1 and M2 only, which is also supported by the authors' data that shows 13 clusters of macrophages. However, they only focused on the difference between clusters 11 and 3 considering conventional M1 and M2. It will be more interesting to analyze the other clusters and how they relate to the established and simplistic M1 and M2 paradigms.
It will be interesting to know if they found any difference in the enriched pathways among these different clusters considering the exclusive regulatory factors and their targets.
We appreciate the point and have addressed it as follows. In the revised manuscript, we have discussed the clusters in detail and have provided the key regulatory factors (RF) combinations and target genes that define distinct macrophage population states (Please refer: Data file S2, S3). We have also discussed the associated immunological processes with each cluster, particularly in relation to the C11 and C3 clusters. We have added a new panel in Fig 1 to illustrate a heatmap indicating the enrichment of pathways relevant to inflammation in each of the clusters (Fig 1B). Indeed, there is a substantial difference in the enrichment terms between the extreme ends (M1, M2) and significant differences in some of the pathways between clusters.
(3) The authors have shown the involvement of NCB at 72h post LPS treatment. Are these RF involved in late response genes or act at the earlier time point of LPS treatment? Understanding the RF involvement in the dynamic response of macrophages to any stimulant will be important.
Using the data available for different time points (30 mins to 72 hours), we plotted the fold change (with respect to unstimulated cells) in M1 and M2 clusters for each of the NCB genes and observe clear divergence in the trend at 24 hours and have provided them as newly added (Supplementary Figure 9 A, B, C).
(4) The authors showed that the knockdown of RF- NCB can switch the M1 to M2. However, they showed a few conventional markers known to be M2 markers. What happens if NCB is overexpressed or knocked down in other treatment conditions/other clusters? Is the RF-NCB only involved in these two specific stimulations or their overexpression can promote M2 polarization in any given stimuli?
It is an interesting question but for practical reasons, experimental work was limited to M1 and M2 clusters as the aim was to establish proof of concept and could not be scaled up for all clusters, which would require a large amount of work and possibly a separate study. We believe the description of the clusters that we have provided will enable the design of future experiments that will throw light on the significance of the intermediate clusters.
(5) The authors have shown that knockdown of RF- NCB decreases pathogen clearance, but what are their altered functions? Are they more efficient in cellular debris clearance or resolution of inflammation? The authors can check the mRNA expression of markers/cytokines involved in those processes, in the NCB knockdown condition.
Indeed. Expression levels were measured for the following genes: CXCL2, IL1B, iNOS, SOCS3 (which are pro-inflammatory markers), as well as MRC1, ARG1, TGFB, IL10 (anti-inflammatory markers), as shown in Fig 4B.
Minor comments:
(1, 2). How the authors evaluate the performance of their knowledge-based gene network. The authors should write the methods in detail, how they generated the simulated network, and evaluated the simulated dataset.
Gene network construction and module detection have many tools available. The authors need to mention which one they used. The authors should show whether their findings are consistent with at least another two module-detection methods (eg; "RedeR") to strengthen their claim.
We have added a schematic figure (Supplementary Fig S11) and detailed description of network construction and mining in the Methods section, as follows: We have reconstructed a comprehensive knowledge-based human Gene Regulatory Network (hGRN), which consists of Regulatory Factors (RF) to Target Gene (TG) and RF to RF interactions. To achieve this, we curated experimentally determined regulatory interactions (RF-TG, RF-RF) associated with human regulatory factors (Wingender et al., 2013). These interactions were sourced from several resources, including: (a) literature-curated resources like the Human Transcriptional Regulation Interactions database (HTRIdb) (Bovolenta et al., 2012), Regulatory Network Repository (RegNetwork) (Liu et al., 2015), Transcriptional Regulatory Relationships Unraveled by Sentence-based Text-mining (TRRUST) (Han et al., 2015), and the TRANSFAC resource from Harmonizome (Rouillard et al., 2016); (b) ChEA3, which contains ChIP-seq determined interactions (Keenan et al., 2019); and (c) high-confidence protein-protein binding interactions (RF-RF) from the human protein-protein interaction network-2 (hPPiN2) (Ravichandran et al., 2021). As a result, our hGRN comprises 27,702 nodes and 890,991 interactions. It is important to note that none of the edges/interactions in the hGRN are data-driven. We utilized this extensive hGRN, which encompasses the experimentally determined interactions/edges, to infer stimulant-specific hGRNs and top paths using our in-house network mining algorithm, ResponseNet. We have previously demonstrated that ResponseNet, which utilizes a knowledge-based network and a sensitive interrogation algorithm, outperformed data-driven network inference methods in capturing biologically relevant processes and genes, whose validation is reported earlier (Ravichandran and Chandra, 2019; Sambaturu et al., 2021).
We utilized our in-house response network approach to identify the stimulant-specific top active and repressed perturbations (Ravichandran and Chandra, 2019; Sambaturu et al., 2021). This is clearly described in the revised manuscript. To summarize, we generated stimulant-specific Gene Regulatory Networks (GRNs) by applying weights to the master human Gene Regulatory Network (hGRN) based on differential transcriptomic responses to stimulants (i.e., comparing stimulant-treated conditions to baseline). We then produced individually weighted networks for each stimulant and implemented a refined network mining technique to extract the most significant pathways. Furthermore, we have previously conducted a systematic comparison of our network mining strategy with other data-driven module detection methods, including jActiveModules (Ideker et al, 2002), WGCNA (Langfelder et al, 2008), and ARACNE (Margolin et al, 2006). Our findings demonstrated that our approach outperformed conventional data-driven network inference methods in capturing the biologically pertinent processes and genes (Ravichandran and Chandra, 2019). Since we have experimentally validated what we predicted from the network analysis, we do not see a need for performing the computational analysis with another algorithm. Moreover, different network analyses are based on different aspects of identifying functionally relevant genes or subnetworks. While each of them output useful information, given the scale of the network and the number of different biologically significant subnetworks and genes that could be present in an unbiased network such as what we have used, the output from different methods need not agree with each other as they may capture different aspects all together and hence is not guaranteed to be informative.
(3) Representation of Fig 2B is difficult to understand the authors' interpretation of 'the 3-RF combination has 1293 targets, 359 covering about 53% of the top-perturbed network' for general readers. If the authors can simplify the interpretation will be helpful for the readers.
This is replaced with clearer figures in the revised manuscript (Figure 2A, 2B), and the associated text is also rephrased for clarity.
Reviewer #2 (Recommendations For The Authors):
Major comments:
(1) It would be helpful for the authors to determine whether the effects they see in the THP-1 immortalized cell line are reproduced in another macrophage cell line, or ideally in PBMC-derived macrophages if this is feasible. If using PBMC- or bone marrow-derived macrophages is beyond the scope of what the authors can reasonably perform, they could consider using another macrophage cell line such as RAW 264.7 cells, which would also provide orthogonal validation from a mouse model.
At this point of time, it is unfortunately infeasible for us to perform these experiments, due to resource limitation. Moreover, it would require a lot of time. We hope that our work provides pointers for anyone working on mouse models or other model systems to design their studies on regulatory controls and the aspect of generalizability of our findings in Thp-1 cell lines to other systems will eventually emerge.
(2) It would be helpful for the authors to provide an expanded explanation of the network mining approach used, as well as the cluster stability analysis and the Epitracer analysis. Although these approaches may be published elsewhere, readers with a non-computational background would benefit from additional descriptions. A schematic figure would also be helpful to clarify their approach.
We have added a new schematic diagram in Supplementary figures (S13) and a detailed text in the Methods section describing the network mining analysis and epitracer identification in the revised manuscript.
(3) It would be helpful for the authors to comment on whether the thirteen polarization states that they identify align with other analyses that have been performed using data collected from stimulated macrophages, or whether this is a novel finding, especially as the original paper from which the primary data are derived identified 9 clusters. More broadly, since the authors eventually return to the M1-M2 paradigm, it is unclear whether there is any functional support for a 13-state model - it is also possible that macrophages exist along a continuum of stimulation states rather than in discrete clusters. This at least merits further discussion, which could focus on different axes of polarization as discussed and shown in the original paper.
As described in the manuscript, Clustering based on the differential transcriptome profile of RF-set1, which contains 265 transcription factors (TFs), in response to 28 stimulants, resulted in 13 distinct clusters. The cluster member associations inferred from RF-set1 were similar in number and pattern to those inferred from the entire differential transcriptome (n=12,164; Fig. S2, cophenetic coefficient = 0.68; p-value = 1.25e−51). Furthermore, the inferred cluster pattern largely matched the clustering pattern previously described for the same dataset (Xue et al., 2014). Our contribution: The pattern we observed from the top-ranked epicenters in each cluster suggests that a subset of differentially expressed genes (DEGs) present in our top networks is sufficient for achieving differentiation. Our gene-regulatory models suggest that saturated (SA and PA) and unsaturated (LA, LiA, and OA) fatty acids, which were previously grouped together, mediate distinct modes of resolution and are now separated into two sub-branches. Similarly, the effects of IFNγ and sLPS, previously combined, are now distinctly resolved, aligning with known regulatory differences (Hoeksema et al., 2015; Kang et al., 2019).
The principal takeaway from this analysis is not the exact number of clusters but rather the molecular basis it provides for the differentiation of functional states, with M1 and M2 representing two ends of the spectrum. Several other states are dispersed within the polarization spectrum, which we describe as a punctuated continuum. For our switching studies, we focused on clusters C11 (M1-like) and C2 (M2-like) due to their established functional relevance. However, future studies are required to explore the functional relevance of other clusters. We have added a discussion on this aspect as suggested.
(4) It would be helpful to define the contribution of each component of the NCB group to M1 polarization.
We assessed the impact of CEBPB, NFE2L2, and BCL3 on C2 (M1-like) polarization states by quantifying the expression levels of M1 and M2 markers. Our findings indicate that knocking down CEBPB led to a significant downregulation in the expression of M1 markers and an increase in M2 marker expression. In contrast, NFE2L2 and BCL3 knockdown resulted in decreased expression of M1 markers without a corresponding significant increase in M2 markers. These results suggest that CEBPB is crucial for M1 to the M2 transition. We have added a note on pg 22 to emphasize this better.
(5) NRF2, CEBPb, and BCL3 all have well-described roles in macrophage polarization. To add clarity to their discussion, the authors should cite relevant literature (eg PMIDs 15465827, 27211851, and others) and discuss how their findings extend what is currently known about the contribution of these individual proteins to macrophage responses.
The role of NFE2L2, CEBPB and BCL3 in macrophage polarization and state transition are described in the discussion section. The PMIDs mentioned by the reviewer are added as well.
(6) The effect size of NCB knockdown in the in vitro Staph aureus model shown in 4C is fairly small - bacterial killing assays typically require at least a log of difference to demonstrate a convincing effect. It would be helpful for the authors to include a positive control for this experiment (for example, STAT4) to frame the magnitude of their effect.
We thank the reviewer for the comment, however, we would like to point out that the difference in CFU plotted in log<sub>10</sub> scale, as per common practice. The CFUs are therefore almost halved due to the knockdown in absolute scale and reproduced multiple times with statistically significant results (p-value <0.01). We feel it is sufficient to demonstrate that the NCB geneset by themselves bring out a change in polarization and hence the killing effect. We have used STAT4 as a control for marker measurements as shown in Fig 3C. While carrying out CFU with siSTAT4 may add additional information, we have proceeded to perform the infection experiments with and without the NCB knockdown as that remains the main focus of the study.
Minor recommendations:
(1) Is there a difference between the data represented in Figure 1A-B and Figure S1? If this is the same data, there is no need to repeat it, and Figure 1 could be composed only of the current panels C and D.
We have removed Figure1 A and B as it illustrates the same point as Figure S1. We have retained Figures C and D and renamed them as new Figure 1A and C. In addition, we have added a new panel Fig 1B (in response to earlier points).
(2) Could Figure 2B be represented in a different way? The circles do not contain any readable information about the genes, and it may be less visually overwhelming to represent this with just the large and small triangles. Perhaps the individual genes represented by the circles could be listed in a supplemental table or Excel file.
We have provided a new Figure 2 A and B panels for the M1 and M2 clusters respectively, which has only the barcode genes along with a functional annotation. The full network is already provided in supplementary data.
(3) When indicating the N for all experiments performed in the figure legends, the authors should indicate whether these were technical or biological replicates.
We appreciate the reviewers for the suggestion. We have indicated what N is for all figure legends.
(4) Fig 3B: the y-axis is confusing - it appears that normalization is actually to the untreated cells.
Yes indeed. The normalization is with respect to the untreated cells as per standard practice. We have indicated this clearly in the legend.
(5) The 72-hour time point in Fig S8 shows unexpected results. Could the authors explain or propose a hypothesis for why CXCL2 and IL1b abruptly decrease while iNOS and MRC1 abruptly increase?
The purpose of the mentioned experiment was to standardize the time point of M1 polarization post S. aureus infection. In this regard, we profiled the expression levels of markers at various time points. We chose to study the 24 hour time point for all the future experiments based on the significant upregulation of NCB seen in the macrophages. We believe that the 72 hour time point may show effects that are different since the initial immune response would have waned leading to differences in cytokine dynamics. However, as this is not the focus of our study, we are not discussing this aspect further.
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public review):
Summary:
In their previous publication (Dong et al. Cell Reports 2024), the authors showed that citalopram treatment resulted in reduced tumor size by binding to the E380 site of GLUT1 and inhibiting the glycolytic metabolism of HCC cells, instead of the classical citalopram receptor. Given that C5aR1 was also identified as the potential receptor of citalopram in the previous report, the authors focused on exploring the potential of the immune-dependent anti-tumor effect of citalopram via C5aR1. C5aR1 was found to be expressed on tumor-associated macrophages (TAMs) and citalopram administration showed potential to improve the stability of C5aR1 in vitro. Through macrophage depletion and adoptive transfer approaches in HCC mouse models, the data demonstrated the potential importance of C5aR1-expressing macrophage in the anti-tumor effect of citalopram in vivo. Mechanistically, their in vitro data suggested that citalopram may regulate the phagocytosis potential and polarization of macrophages through C5aR1. Next, they tried to investigate the direct link between citalopram and CD8+T cells by including an additional MASH-associated HCC mouse model. Their data suggest that citalopram may upregulate the glycolytic metabolism of CD8+T cells, probability via GLUT3 but not GLUT1-mediated glucose uptake. Lastly, as the systemic 5-HT level is down-regulated by citalopram, the authors analyzed the association between a low 5-HT and a superior CD8+T cell function against a tumor. Although the data is informative, the rationale for working on additional mechanisms and logical links among different parts is not clear. In addition, some of the conclusion is also not fully supported by the current data.
We thank the reviewer for their comprehensive summary of our study and appreciate the valuable feedback. We have made improvements based on these comments, and a detailed response addressing each point is presented below.
Strengths:
The idea of repurposing clinical-in-used drugs showed great potential for immediate clinical translation. The data here suggested that the anti-depression drug, citalopram displayed an immune regulatory role on TAM via a new target C5aR1 in HCC.
We thank the reviewer for recognizing the strengths of our study.
Weaknesses:
(1) The authors concluded that citalopram had a 'potential immune-dependent effect' based on the tumor weight difference between Rag-/- and C57 mice in Figure 1. However, tumor weight differences may also be attributed to a non-immune regulatory pathway. In addition, how do the authors calculate relative tumor weight? What is the rationale for using relative one but not absolute tumor weight to reflect the anti-tumor effect?
We appreciate your insights into the potential contributions of non-immune regulatory pathways to the observed tumor weight differences between Rag1<sup>-/- </sup>and wild type C57BL/6 mice. Indeed, the anti-tumor effects of citalopram involve non-immune mechanisms. Previously, we have demonstrated the direct effects of citalopram on cancer cell proliferation, apoptosis, and metabolic processes (PMID: 39388353). In this study, we focused on immune-dependent mechanisms, utilizing Rag1<sup>-/- </sup> mice to investigate a potential immune-mediated effect. The relative tumor weight was calculated by assigning an arbitrary value of 1 to the Rag1<sup>-/- </sup> mice in the DMSO treatment group, with all other tumor weights expressed relative to this baseline. As suggested, we have included absolute tumor weight data in the revised Figure 1B, 1E, 1F, and 3B.
(2) The authors used shSlc6a4 tumor cell lines to demonstrate that citalopram's effects are independent of the conventional SERT receptor (Figure 1C-F). However, this does not entirely exclude the possibility that SERT may still play a role in this context, as it can be expressed in other cells within the tumor microenvironment. What is the expression profiling of Slc6a4 in the HCC tumor microenvironment? In addition, in Figure 1F, the tumor growth of shSlc6a4 in C57 mice displayed a decreased trend, suggesting a possible role of Slc6a4.
As suggested, we probed the expression pattern of SERT in HCC and its tumor microenvironment. Using a single cell sequencing dataset of HCC (GSE125449), we revealed that SERT is also expressed by T cells, tumor-associated endothelial cells, and cancer-associated fibroblasts (see revised Figure S2G). Therefore, we cannot fully rule out the possibility that citalopram may influence these cellular components within the TME and contribute to its therapeutic effects. In the revised manuscript, we have included and discussed this result. In Figure 1F, SERT knockdown led to a 9% reduction in tumor growth, however, this difference was not statistically significant (0.619 ± 0.099 g vs. 0.594 ± 0.129 g; p = 0.75).
(3) Why did the authors choose to study phagocytosis in Figures 3G-H? As an important player, TAM regulates tumor growth via various mechanisms.
We choose to investigate phagocytosis because citalopram targets C5aR1-expressing TAM. C5aR1 is a receptor for the complement component C5a, which plays a crucial role in mediating the phagocytosis process in macrophages. In the revised manuscript, we have highlighted this rationale.
(4) The information on unchanged deposition of C5a has been mentioned in this manuscript (Figures 3D and 3F), the authors should explain further in the manuscript, for example, C5a could bind to receptors other than C5aR1 and/or C5a bind to C5aR1 by different docking anchors compared with citalopram.
Thank you for your insightful comment. In Figure 3D, tumor growth was attenuated in C5ar1<sup>-/-</sup> recipients compared with C5ar1<sup>-/-</sup> recipients, whereas C5a deposition remained unchanged. This suggests that while C5a is still present, its interaction with C5aR1 is critical for influencing tumor growth dynamics. In Figure 3F, C5a deposition was not affected by citalopram treatment. Indeed, docking analysis and DARTS assay revealed that citalopram binds to the D282 site of C5aR1. Previous report has shown that mutations on E199 and D282 reduce C5a binding affinity to C5aR1 (PMID: 37169960). Therefore, the impact of citalopram is primarily on C5a/C5aR1 interactions and downstream signaling pathways, rather than on altering C5a levels. In the revised manuscript, we have included this interpretation.
(5) Figure 3I-M - the flow cytometry data suggested that citalopram treatment altered the proportions of total TAM, M1 and M2 subsets, CD4<sup>+</sup> and CD8<sup>+</sup>T cells, DCs, and B cells. Why does the author conclude that the enhanced phagocytosis of TAM was one of the major mechanisms of citalopram? As the overall TAM number was regulated, the contribution of phagocytosis to tumor growth may be limited.
We thank the reviewer’s valuable input. Indeed, recent studies have demonstrated that targeting C5aR1<sup>+</sup> TAMs can induce many anti-tumor effects, such as macrophage polarization and CD8<sup>+</sup> T cell infiltration (PMID: 30300579, PMID: 38331868, and PMID: 38098230). In the revised manuscript, we have clarified our conclusion to better articulate the relationship between citalopram treatment, TAM populations, and their phagocytic activity, with particular emphasis on the role of CD8<sup>+</sup> T cells. For macrophage phagocytosis, one possible explanation is that citalopram targets C5aR1 to enhance macrophage phagocytosis and subsequent antigen presentation and/or cytokine production, which promotes T cell recruitment and activity as well as modulate other aspects of tumor immunity. Given that the anti-tumor effects of citalopram are largely dependent on CD8<sup>+</sup> T cells, we conclude that CD8<sup>+</sup> T cells are essential for the effector mechanisms of citalopram.
(6) Figure 4 - what is the rationale for using the MASH-associated HCC mouse model to study metabolic regulation in CD8<sup>+</sup> T cells? The tumor microenvironment and tumor growth would be quite different. In addition, how does this part link up with the mechanisms related to C5aR1 and TAM? The authors also brought GLUT1 back in the last part and focused on CD8<sup>+</sup> T cell metabolism, which was totally separated from previous data.
We chose the MASH-associated HCC mouse model because it closely mimics the etiology of metabolic-associated fatty liver disease (MAFLD), which is a significant contributor to the development of cirrhosis and HCC. In addition to the MASH-associated HCC mouse model, the study also incorporated the orthotopic Hepa1-6 tumor model. In our previous publication (Dong et al., Cell Reports 2024), we employed both of these HCC models. Therefore, we utilized the same two mouse models in this study. The inclusion of CD8<sup>+</sup> T cells in our study is based on the understanding that citalopram targets GLUT1, which plays a crucial role in glucose uptake (PMID: 39388353). CD8<sup>+</sup>T cell function is heavily reliant on glycolytic metabolism, making it essential to investigate how citalopram’s effects on GLUT1 influence the metabolic pathways and functionality of these immune cells. In this study, we identified that the primary glucose transporter in CD8<sup>+</sup> T cells is GLUT3, rather than GLUT1. The data presented in Figure 4 aim to illustrate the additional effect of citalopram on peripheral 5-HT levels, which, in turn, influences CD8<sup>+</sup> T cell functionality. By linking these findings, we clarify how citalopram impacts both TAMs and CD8<sup>+</sup> T cells. CD8<sup>+</sup> T cells can be influenced by citalopram through various mechanisms, including TAM-dependent mechanisms, reduced systemic serum 5-HT concentrations, and unidentified direct effects. In the revised manuscript, we have enhanced the background information to avoid any gaps.
(7) Figure 5, the authors illustrated their mechanism that citalopram regulates CD8<sup>+</sup> T cell anti-tumor immunity through proinflammatory TAM with no experimental evidence. Using only CD206 and MHCII to represent TAM subsets obviously is not sufficient.
Thank you for your valuable comments. As noted by the reviewer, TAMs can influence CD8<sup>+</sup> T cell anti-tumor immunity through various mechanisms. In this study, we focused on elucidating the impact of citalopram on pro-inflammatory TAMs, which in turn affect CD8<sup>+</sup> T cell anti-tumor immunity and ultimately influence tumor outcomes. Therefore, in the mechanistic diagram, we highlighted the effect of citalopram on pro-inflammatory TAMs, while the causal relationship between TAMs and CD8<sup>+</sup> T cell anti-tumor immunity was indicated with a dotted line due to the limited evidence presented in this study. Additionally, we have expanded our discussion on how citalopram regulates CD8<sup>+</sup> T cell anti-tumor immunity through pro-inflammatory TAMs.
For the analysis of TAMs, we initially sorted CD45<sup>+</sup>F4/80<sup>+</sup>CD11b<sup>+</sup> cells and assessed M1/M2 polarization by measuring CD206 and MHCII expression. As an added strength, we isolated TAMs from the orthotopic GLUT1<sup>KD</sup> Hepa1-6 model using CD11b microbeads and conducted real-time qPCR analysis of M1-oriented (Il6, Ifnb1, and Nos2) and M2-oriented (Mrc1, Il10, and Arg1) markers. Consistent with our flow cytometry data, the qPCR results confirmed that citalopram induces a pro-inflammatory TAM phenotype (revised Figure S9A).
Reviewer #2 (Public review): Summary:
Dong et al. present a thorough investigation into the potential of repurposing citalopram, an SSRI, for hepatocellular carcinoma (HCC) therapy. The study highlights the dual mechanisms by which citalopram exerts anti-tumor effects: reprogramming tumor-associated macrophages (TAMs) toward an anti-tumor phenotype via C5aR1 modulation and suppressing cancer cell metabolism through GLUT1 inhibition while enhancing CD8+ T cell activation. The findings emphasize the potential of drug repurposing strategies and position C5aR1 as a promising immunotherapeutic target. However, certain aspects of experimental design and clinical relevance could be further developed to strengthen the study's impact.
We thank the reviewer’s thoughtful review and constructive feedback. As suggested, we have made improvements based on the feedback provided.
Strength:
It provides detailed evidence of citalopram's non-canonical action on C5aR1, demonstrating its ability to modulate macrophage behavior and enhance CD8+ T cell cytotoxicity. The use of DARTS assays, in silico docking, and gene signature network analyses offers robust validation of drug-target interactions. Additionally, the dual focus on immune cell reprogramming and metabolic suppression presents a thorough strategy for HCC therapy. By emphasizing the potential for existing drugs like citalopram to be repurposed, the study also underscores the feasibility of translational applications.
We sincerely appreciate the reviewer’s recognition of the detailed evidence supporting citalopram’s non-canonical action on C5aR1, along with the innovative methodologies employed and the promising potential for repurposing existing drugs in HCC therapy.
Major weaknesses/suggestions:
The dataset and signature database used for GSEA analyses are not clearly specified, limiting reproducibility. The manuscript does not fully explore the potential promiscuity of citalopram's interactions across GLUT1, C5aR1, and SERT1, which could provide a deeper understanding of binding selectivity. The absence of GLUT1 knockdown or knockout experiments in macrophages prevents a complete assessment of GLUT1's role in macrophage versus tumor cell metabolism. Furthermore, there is minimal discussion of clinical data on SSRI use in HCC patients. Incorporating survival outcomes based on SSRI treatment could strengthen the study's translational relevance.
By addressing these limitations, the manuscript could make an even stronger contribution to the fields of cancer immunotherapy and drug repurposing.
We appreciate the reviewer’s valuable suggestions. As suggested, we have included the following revisions:
(a) GSEA analyses: For GSEA analyses, we conducted RNA sequencing (RNA-seq) analysis on HCC-LM3 cells treated with citalopram or fluvoxamine, which led to the identification of 114 differentially expressed genes (DEGs; 80 co-upregulated and 34 co-downregulated), as reported previously (PMID: 39388353). These DEGs were then utilized to create an SSRI-related gene signature. Subsequently, we analyzed RNA-seq data from liver HCC (LIHC) samples in The Cancer Genome Atlas (TCGA) cohort, comprising 371 samples, categorizing them into high and low expression groups based on the median expression levels of each candidate target gene (such as C5AR1). Finally, we performed GSEA on the grouped samples (C5AR1-high versus C5AR1-low) using the SSRI-related gene signature. In the revised manuscript, we have included this information in the “Materials and Methods” section.
(b) Exploration of binding selectivity: We acknowledge the importance of exploring the potential promiscuity of citalopram’s interactions across GLUT1, C5aR1, and SERT1. While we cannot provide further experimental data to support this aspect, we have included the following points in the revised manuscript: 1) We emphasize the significance of exploring the relative binding affinities of citalopram to GLUT1, C5aR1, and SERT, as varying affinities could influence the drug’s overall efficacy. As highlighted in the current manuscript and our previous publication (PMID: 39388353), citalopram interacts with C5aR1 and GLUT1 through distinct binding sites and mechanisms, whereas its interaction with SERT is characterized by a more direct inhibition of serotonin binding (PMID: 27049939). To gain deeper insights into these interactions, employing techniques such as surface plasmon resonance or biolayer interferometry could provide valuable quantitative data on binding kinetics and affinities for each target. 2) We discuss how citalopram’s interactions with multiple targets may contribute to its therapeutic effects, particularly in the context of immune modulation and tumor progression. The potential for citalopram to exhibit diverse mechanisms of action through its interactions with these proteins warrants further investigation. A comprehensive understanding of these pathways could lead to the development of improved therapeutic strategies.
(c) GLUT1 knockdown in macrophages: In the revised manuscript, we revealed that TAMs predominantly express GLUT3 but not GLUT1 (Figures S8B and S8C). GLUT1 knockdown in THP-1 cells did not significantly impact their glycolytic metabolism (Figure S8D), whereas GLUT3 knockdown led to a marked reduction in glycolysis in THP-1 cells.
(d) Clinical data on SSRI use in HCC patients: Previously, we have reported that SSRIs use is associated with reduced disease progression in HCC patients (PMID: 39388353) (Cell Rep. 2024 Oct 22;43(10):114818.). As detailed below:
“We determined whether SSRIs for alleviating HCC are supported by real-world data. A total of 3061 patients with liver cancer were extracted from the Swedish Cancer Register. Among them, 695 patients had been administrated with post-diagnostic SSRIs. The Kaplan-Meier survival analysis suggested that patients who utilized SSRIs exhibited a significantly improved metastasis-free survival compared to those who did not use SSRIs, with a P value of log-rank test at 0.0002. Cox regression analysis showed that SSRI use was associated with a lower risk of metastasis (HR = 0.78; 95% CI, 0.62-0.99)”.
Reviewer #1 (Recommendations for the authors):
(1) Add experiments to address the questions listed in the weaknesses.
As suggested, related experiments are performed to strengthen the conclusions.
(2) It would be appreciated to show the expression profile of SERT or employ KO mouse models to eliminate the effect of SERT.
As suggested, analysis of a single-cell sequencing dataset of HCC (GSE125449) revealed that SERT is expressed not only in HCC cells but also in T cells, tumor-associated endothelial cells, and cancer-associated fibroblasts (Figure S2G). Consistently, SERT has been reported as an immune checkpoint restricting CD8 T cell antitumor immunity (PMID: 40403728). Furthermore, SERT KO mice (Cyagen Biosciences, S-KO-02549) was employed to investigate the effects of citalopram. However, the Slc6a4 gene knockout in mice resulted in a significant decrease in 5-HT levels in the brain and a lack of cortical columnar structures. Importantly, the mice exhibited an intolerance to citalopram treatment. Therefore, we did not pursue further investigation into the effects of citalopram in SERT KO mice.
(3) Due to the concern of specificity and animal health, it would be more direct if the authors could use, for example, C5ar1-fl/fl x Adgre1-Cre mouse models.
Thank you for your valuable suggestion. We fully agree with your comment regarding the value of introducing C5ar1-fl/fl and Adgre1-Cre mouse models, along with the necessary experimental setups, to substantiate this point. However, in our study, the C5ar1 KO mice exhibited normal overall appearance and viability, indicating that the model is generally healthy. Furthermore, we have validated the specific role of C5aR1 in macrophages through bone marrow reconstitution experiments, reinforcing the importance of C5aR1 in these cells. Therefore, we chose the current model to balance experimental effectiveness with considerations for animal health.
(4) For example, a GSEA or GO analysis of comparison of macrophages from C5ar1-/- or C5ar1+/- mice may point to the enriched pathway of phagocytosis in macrophages derived from C5ar1-/- rather than C5ar1+/- mice, and this information is helpful for the integrity of this work. Besides, it would be more reliable if a nucleus staining is included in Figures 3G and 3H.
As suggested, macrophages were isolated from tumor-bearing C5ar1<sup>-/-</sup> and C5ar1<sup>+/-</sup> mice and subsequently analyzed using RNA sequencing. The Gene Set Enrichment Analysis (GSEA) revealed a significant enrichment of the phagocytosis pathway in macrophages derived from C5ar1<sup>-/-</sup> mice compared to those from C5ar1<sup>+/-</sup> mice (see revised Figure S6A). While we acknowledge that the addition of a nucleus staining would enhance reliability, we would like to point out that this style of presentation is also commonly found in articles related to phagocytosis. Furthermore, this experiment involved a significant number of experimental mice, and in accordance with the 3Rs principle for animal experiments, we did not obtain additional sorted TAMs to perform the phagocytosis assay. Thank you for your understanding.
(5) In line 122, there is a typo, and it should be 'analysis'.
Thank you for pointing out the typo. It has been corrected to "analysis" in the revised manuscript.
(6) In line 217, there is no causal relationship between the contexts, and using 'as a result' may lead to misunderstanding.
As suggested, ‘as a result’ has been removed to avoid any misunderstanding.
(7) In line 322, please make sure if it should be HBS or PBS.
It is PBS, and revisions have been made.
(8) Figure S7, the calculation of cell proportions needs to use a consistent denominator.
As suggested, we calculated cell proportions using a consistent denominator (CD45<sup>+</sup> cells).
(9) Figure 4C, label error.
Thanks for your careful review. It has been corrected to "MASH".
Reviewer #2 (Recommendations for the authors):
Dong et al. present compelling evidence for repurposing citalopram, a selective serotonin reuptake inhibitor (SSRI), as a potential therapeutic for hepatocellular carcinoma (HCC). While the concept of SSRI repurposing is not novel, this manuscript provides valuable insights into the drug's dual mechanisms: targeting tumor-associated macrophages (TAMs) via C5aR1 modulation and enhancing CD8+ T cell activity, alongside inhibiting cancer cell metabolism through GLUT1 suppression. The findings underscore the promise of drug repurposing strategies and identify C5aR1 as a noteworthy immunotherapeutic target. Addressing the following points will enhance the manuscript's impact and relevance to cancer immunotherapy.
Specific Comments:
(1) The authors identify C5aR1 on TAMs as a direct target of citalopram, independent of its classical SERT target, using drug-induced gene signature network analysis and co-immunofluorescence of CD163+ macrophages with C5aR1. The DARTS assay further supports the binding of C5aR1 to citalopram, complemented by in silico docking analysis adapted from their previous GLUT1 study. Since GLUT1 and SERT1 are transporter proteins while C5aR1 is a GPCR, these heterogeneous binding interactions suggest potential promiscuity in SSRI-target engagement.
(a) Figure 2A: The authors identify C5aR1 as a target using GSEA but do not specify the dataset used (e.g., cancer or immune cells) or the signature database consulted. Providing this context would enhance reproducibility.
For GSEA, we performed RNA sequencing (RNA-seq) on HCC-LM3 cells treated with citalopram or fluvoxamine and identified 114 differentially expressed genes (DEGs), which included 80 genes that were co-upregulated and 34 that were co-downregulated, as previously documented (PMID: 39388353). These DEGs were subsequently used to develop an SSRI-related gene signature. We then employed the RNA-seq data from liver hepatocellular carcinoma (LIHC) samples within The Cancer Genome Atlas (TCGA) cohort, which included 371 samples. HCC samples in the TCGA cohort were categorized into high and low expression groups based on the median expression levels of each candidate target gene, such as C5AR1. Finally, we conducted GSEA on the grouped samples (such as C5AR1-high versus C5AR1-low) using the SSRI-related gene signature. For reproducibility, detailed information has been added to the “Materials and Methods” section of the revised manuscript.
(b) Figure 2F: Given citalopram's reported role in inhibiting GLUT1, a comparative discussion on the relative contributions of GLUT1 inhibition versus C5aR1 modulation in tumor suppression is warranted. Performing a DARTS assay for GLUT1 in THP-1 cells, which express high GLUT1 levels and exhibit upregulation in M1 macrophages (https://doi.org/10.1038/s41467-022-33526-z), would clarify SSRI interactions with macrophage metabolism.
As suggested, we first investigated citalopram treatment in THP-1 cells. The result showed the glycolytic metabolism of THP-1 cells remained largely unaffected following citalopram treatment, as evidenced by glucose uptake, lactate release, and extracellular acidification rate (ECAR) (Figure S8A). Next, we mined a single cell sequencing datasets of HCC and revealed that TAMs predominantly express GLUT3 but not GLUT1 (Figure S8B). Consistently, Western blotting analysis showed a higher expression of GLUT3 and minimal levels of GLUT1 in THP-1 cells (Figure S8C). Consistently, it has been well documented that GLUT1 expression increased after M1 polarization stimuli an GLUT3 expression increased after M2 stimulation in macrophages (PMID: 37721853, PMID: 36216803). GLUT1 knockdown in THP-1 cells did not significantly impact their glycolytic metabolism (Figure S8D), whereas GLUT3 knockdown led to a marked reduction in glycolysis in THP-1 cells. Based on these findings, we conclude that the effects of citalopram on macrophages are primarily mediated through targeting C5aR1 rather than GLUT1.
(c) Figures 2H-I: A comparison of drug-protein interactions across GLUT1, C5aR1, and SERT1 would be valuable to identify potential shared or distinct binding features.
Citalopram exhibits distinct binding characteristics across its various targets, including GLUT1, C5aR1, and its classical target, SERT. In the case of C5aR1, our in silico docking analysis identified two key binding conformations at the orthosteric site. The interactions involved significant electrostatic contacts between citalopram’s amino group and negatively charged residues like E199 and D282. Notably, D282’s accessibility and orientation towards the binding cavity suggest it plays a crucial role in citalopram binding, highlighting the importance of specific amino acid interactions at this site. For GLUT1 (PMID: 39388353), citalopram’s interaction also demonstrated notable hydrophobic contacts, particularly through the fluorophenyl group with residues V328, P385, and L325. The cyanophtalane group penetrated the substrate-binding cavity, indicating that citalopram could occupy a similar binding site as glucose, which is distinct from the binding mechanism observed in C5aR1. The involvement of E380 in both poses for GLUT1 further emphasizes the role of electrostatic interactions in mediating citalopram’s binding to this transporter. In contrast, for SERT (PMID: 27049939), citalopram locks the transporter in an outward-open conformation by occupying the central binding site, which is located between transmembrane helices 1, 3, 6, 8 and 10. This binding directly obstructs serotonin from accessing its binding site, illustrating a more definitive blockade mechanism. Additionally, the allosteric site at SERT, positioned between extracellular loops 4 and 6 and transmembrane helices 1, 6, 10, and 11, enhances this blockade by sterically hindering ligand unbinding, thus providing a clear explanation for the allosteric modulation of serotonin transport. In summary, while citalopram interacts with C5aR1 and GLUT1 through distinct binding sites and mechanisms, its interaction with SERT is characterized by a more straightforward blockade of serotonin binding. The unique structural and functional attributes of each target highlight the versatility of citalopram and suggest that its pharmacological effects may vary significantly depending on the specific protein being targeted. In the revised manuscript, we have included detailed information in the revised manuscript.
(2) The manuscript presents evidence that citalopram reprograms TAMs to an anti-tumor phenotype, enhancing their phagocytic capacity.
(a) Bone Marrow Reconstitution Experiments (Figure 3): The use of donor (dC5aR1) and recipient (rC5aR1) mice is significant but requires clarification. Explicitly defining donor and recipient terminology and including a schematic of the experimental design would improve reader comprehension.
We appreciate your valuable feedback. As suggested, the terminology for donor (dC5aR1) and recipient (rC5aR1) mice was defined: “we injected GLUT1<sup>KD</sup> Hepa1-6 cells into syngeneic recipient C5ar1<sup>-/-</sup> (rC5ar1<sup>-/-</sup> ) mice that had been reconstituted with donor C5ar1<sup>+/-</sup> (dC5ar1<sup>+/-</sup>) or C5ar1<sup>-/-</sup> (dC5ar1<sup>-/-</sup>) bone marrow (BM) cells to analyze the therapeutic effect of citalopram”. Additionally, we have included a schematic of the experimental design to enhance reader comprehension (see revised Figure 3E).
(b) GLUT1 Knockdown (KD) Tumor Cells: While GLUT1 KD tumor cells are utilized, the authors do not assess GLUT1 KD or knockout (KO) in macrophages. Testing the effect of citalopram on macrophages with GLUT1 KO/KD would help determine the relative importance of C5aR1 versus GLUT1 in mediating SSRI effects.
As responded above, GLUT1 knockdown in THP-1 cells did not significantly alter their glycolytic metabolism (Figure S8D). This observation can be explained by the predominant expression of GLUT3 in TAMs rather than GLUT1 (Figures S8B and S8C). Indeed, knockdown of GLUT3 led to a significant reduction in glycolysis in THP-1 cells (Figure S8C).
(c) C5aR1's Pro-Tumoral Role: The authors state that C5aR1 fosters an immunosuppressive microenvironment but omit a discussion of current literature on C5aR1's pro-tumoral role (e.g., https://doi.org/10.1038/s41467-024-48637-y, https://www.nature.com/articles/s41419-024-06500-4, https://doi.org/10.1016/j.ymthe.2023.12.010). Including this background in both the introduction and discussion would contextualize their findings.
Thanks for your valuable feedback. As suggested, we have revised the manuscript to include discussions on C5aR1’s pro-tumoral role, referencing the suggested studies in both the introduction and discussion sections for better context. As detailed below:
(1) Targeting C5aR1<sup>+</sup> TAMs effectively reverses tumor progression and enhances anti-tumor response;
(2) Targeting C5aR1 reprograms TAMs from a protumor state to an antitumor state, promoting the secretion of CXCL9 and CXCL10 while facilitating the recruitment of cytotoxic CD8<sup>+</sup> T cells;
(3) Moreover, citalopram induces TAM phenotypic polarization towards to a M1 proinflammatory state, which supports anti-tumor immune response within the TME.
(d) C5aR1 Expression in TAMs: Is C5aR1 expression constitutive in TAMs? Further details on C5aR1 expression dynamics in TAMs under different conditions could strengthen the discussion. Public datasets on TAMs in various states (e.g., https://www.nature.com/articles/s41586-023-06682-5, https://www.cell.com/cell/abstract/S0092-8674(19)31119-5, https://pubmed.ncbi.nlm.nih.gov/36657444/) may offer useful insights.
Thank you for your valuable suggestions. As suggested, we investigated the expression patterns of C5aR1 in TAMs using a HCC cohort (http://cancer-pku.cn:3838/HCC/). In the study conducted by Qiming Zhang et al. (PMID: 31675496), six distinct macrophage subclusters were identified, with M4-c1-THBS1 and M4-c2-C1QA showing significant enrichment in tumor tissues. M4-c1-THBS1 was enriched with signatures indicative of myeloid-derived suppressor cells (MDSCs), while M4-c2-C1QA exhibited characteristics that resembled those of TAMs as well as M1 and M2 macrophages. Our subsequent analysis revealed that C5aR1 is highly expressed in these two clusters, while expression levels in the other macrophage clusters were notably lower (see revised Figure S3).
(3) The manuscript shows that citalopram-induced reductions in systemic serotonin levels enhance CD8+ T cell activation and cytotoxicity, as evidenced by increased glycolytic metabolism and elevated IFN-γ, TNF-α, and GZMB expression.
(a) How CD8+ T cell activation is done in serotonin-deficient environments?
As reported (PMID: 34524861), one possible explanation is that serotonin may enhance PD-L1 expression on cancer cells, thereby impairing CD8<sup>+</sup> T cell function. A deficiency of serotonin in the tumor microenvironment can delay tumor growth by promoting the accumulation and effector functions of CD8<sup>+</sup> T cells while reducing PD-L1 expression. In addition to the SERT-mediated transport and 5-HT receptor signaling, CD8<sup>+</sup> T cells can express TPH1 (PMID: 38215751, PMID: 40403728), enabling them to synthesize endogenous 5-HT, which activates their activity through serotonylation-dependent mechanisms (PMID: 38215751). In the revised manuscript, we have incorporated these interpretations.
(4) Suggestions for the model figure revision-C5aR1 in TAMs without Citalopram (Figure 5).
(a) Including a control scenario depicting receptor status and function in TAMs without citalopram treatment would provide a clearer baseline for understanding citalopram's effects.
Thank you for your valuable input regarding the model figure revision. We have included a revised mechanism model that depicts the receptor status and function of C5aR1 in TAMs without citalopram treatment, as you suggested.
(5) Suggestions for addressing clinical relevance.
The study predominantly uses preclinical mouse models, although some human HCC data is analyzed (Figures 2B and 3O). However, there is no discussion of clinical data on SSRI use in HCC patients.
Incorporating an analysis of patient survival outcomes based on SSRI treatment (e.g., https://pmc.ncbi.nlm.nih.gov/articles/PMC5444756/, https://pmc.ncbi.nlm.nih.gov/articles/PMC10483320/) would enhance the translational relevance of the findings.
Previously, we reported that the use of SSRIs is associated with reduced disease progression in HCC patients, based on real-world data from the Swedish Cancer Register (PMID: 39388353). As suggested, we have further discussed the clinical relevance of SSRIs in the revised manuscript. As detailed below:
“In a study involving 308,938 participants with HCC, findings indicated that the use of antidepressants following an HCC diagnosis was linked to a decreased risk of both overall mortality and cancer-specific mortality (PMID: 37672269). These associations were consistently observed across various subgroups, including different classes of antidepressants and patients with comorbidities such as hepatitis B or C infections, liver cirrhosis, and alcohol use disorders. Similarly, our analysis of real-world data from the Swedish Cancer Register demonstrated that SSRIs are correlated with slower disease progression in HCC patients (PMID: 39388353). Given these insights, antidepressants, especially SSRIs, show significant potential as anticancer therapies for individuals diagnosed with HCC”.
Santé Mentale : Fausses Promesses et Solutions Collectives – Synthèse du Briefing
Ce document synthétise les analyses et propositions issues d'une table ronde sur la santé mentale, organisée par Psycom au ministère de la Santé.
Le constat central est la nécessité urgente de dépasser une vision individualiste de la santé mentale, où le fardeau repose sur l'individu et la psychiatrie, pour adopter une approche collective et systémique.
Les discussions ont mis en lumière plusieurs problématiques majeures : * l'expansion d'un marché du "bien-être" non réglementé, proposant des solutions pseudoscientifiques dangereuses qui engendrent une "perte de chance" pour les personnes en souffrance ; * la montée des dérives sectaires qui exploitent les vulnérabilités psychiques à des fins financières et d'emprise ; et * l'impact prépondérant sur la santé psychique (estimé à 50 %) des déterminants socio-économiques tels que * la précarité, * les discriminations ou * le logement
Face à ces défis, les experts proposent des solutions multi-niveaux.
Celles-ci incluent un renforcement de la régulation des pratiques non conventionnelles et des titres de "thérapeutes", le développement de l'esprit critique et de la métacognition au sein de la population, et une transformation profonde du soin psychiatrique vers des modèles plus humains, participatifs et moins coercitifs, à l'image de l'approche "Open Dialogue".
Enfin, le rôle crucial des collectivités locales est souligné, celles-ci pouvant agir concrètement sur l'environnement social et urbain pour promouvoir le bien-être et recréer du lien, incarnant ainsi le passage d'une "société du soin" à une "société du prendre soin" attentive aux inégalités et aux vulnérabilités.
--------------------------------------------------------------------------------
La présente analyse se fonde sur les échanges d'une table ronde filmée en septembre 2025 au ministère de la Santé, lors de la journée "Full Santé Mentale :
de l'intime au collectif" organisée par Psycom, un organisme public de lutte contre la stigmatisation en santé mentale.
Question centrale :
Comment sortir d’une vision trop individualiste de la santé mentale pour aller vers une réflexion plus collective ?
Comment passer d’une société du soin à une société du "prendre soin", attentive aux vulnérabilités et aux inégalités ?
Participants :
Nom
Fonction
Organisation
Sophia Feuillère
Responsable de l'innovation pédagogique
Psychom
Elisabeth Fetti
Documentariste, créatrice du podcast sur la métacognition
Méta de Choc
Samir Calfa
Conseiller santé
Miviludes (Mission interministérielle de vigilance)
Maeva Musso
Psychiatre, présidente de l'association des jeunes psychiatres
Hôpitaux Paris Est Val-de-Marne / AJPJA
Marie-Christine Sanier Coavran
Adjointe à la santé et à la lutte contre les exclusions, vice-présidente du réseau Ville Santé
Ville de Lille
Sophia Feuillère identifie trois idées reçues persistantes qui freinent une approche collective :
1. La frontière rigide entre santé mentale et psychiatrie : Le public perçoit souvent la psychiatrie comme un état figé réservé aux "malades", et la santé mentale comme un état tout aussi figé pour les "bien-portants".
Pour contrer cela, Psychom promeut une notion de mouvement et de rétablissement, notamment via son outil de la "boussole de la santé mentale".
2. La seule responsabilité de l'individu : Une croyance répandue veut qu'il suffirait d'outiller les individus (cohérence cardiaque, compétences psychosociales) pour qu'ils prennent soin d'eux. Cette vision omet les déterminants extérieurs.
L'approche systémique, illustrée par l'outil du "cosmos mental", est donc essentielle pour réintégrer le contexte collectif.
3. L'exclusivité de l'expertise médicale : L'idée que seuls les soignants peuvent parler de santé mentale reste forte.
Il est crucial de légitimer la posture du "prendre soin", que chaque citoyen peut adopter, distincte de celle du "soin", qui relève des professionnels qualifiés.
Elisabeth Fetti observe une explosion des offres de "bien-être" sur les médias sociaux, portées par des influenceurs souvent sans expertise.
• Narratif dominant : Le discours s'appuie sur l'expérience personnelle ("J'ai touché le fond et j'ai rebondi, donc faites comme moi"), mêlant développement personnel (sans fondement scientifique) et spiritualité.
• instrumentalisation de la science : Des termes comme "neurosciences" ou "physique quantique" sont utilisés pour conférer une fausse légitimité aux discours.
• Mécanismes de persuasion : L'"effet Barnum" est massivement utilisé.
Il s'agit de formuler des généralités vagues dans lesquelles chacun peut se reconnaître ("Tu veux réussir mais parfois tu te sens empêché"), créant un sentiment de confiance et de compréhension.
• Risques avérés :
◦ Perte de chance : Le risque le plus grave est le retard de diagnostic et de prise en charge adéquate pour des pathologies réelles (dépression, endométriose, addictions).
◦ Escalade de l'engagement : Les clients sont entraînés dans un cycle d'engagement financier et émotionnel croissant (séance gratuite, puis livre, puis stage, etc.), rendant difficile la remise en question et la réorientation.
◦ Culpabilisation : En cas d'échec, la responsabilité est retournée contre l'individu :
"Si ça ne marche pas, c'est que tu n'as pas assez travaillé sur toi".
◦ Effets paradoxaux : Certaines pratiques, comme la "pensée positive", peuvent aggraver l'anxiété chez les personnes les plus vulnérables, comme le montrent des études scientifiques.
Samir Calfa alerte sur l'émergence d'un "système de santé parallèle" où les dérives sectaires prolifèrent, notamment dans le champ de la santé mentale qui représente 40 % des signalements à la Miviludes.
• Mécanisme central : Il ne peut y avoir de dérive sectaire sans emprise mentale, une relation singulière entre le gourou et sa victime.
• Vide juridique : N'importe qui peut aujourd'hui inventer et proposer une méthode de prise en charge psychologique sans réglementation.
• Profil des victimes et motivations des gourous : Neuf victimes sur dix sont des femmes.
Les gourous recherchent systématiquement trois choses : l'argent, les faveurs sexuelles et le travail dissimulé (les victimes devenant des "sergents recruteurs").
• Double impact psychologique : La vulnérabilité psychique est une porte d'entrée vers ces dérives, et la sortie de l'emprise laisse des séquelles psychologiques profondes et durables ("l'organisation sectaire ne sort jamais de votre tête").
Une augmentation des suicides liés à ces phénomènes est constatée.
Maeva Musso insiste sur le poids des facteurs environnementaux et sociaux.
Elle prend l'exemple des enfants placés, qui agit comme une "loupe" sur ces phénomènes :
• Statistiques alarmantes : Cette population présente 8 fois plus de handicaps, 5 fois plus de troubles psychiques graves, compose un quart de la population SDF à 25 ans et a une espérance de vie inférieure de 20 ans à la moyenne générale.
• Répartition des facteurs de troubles psychiques :
◦ 50 % : Déterminants socio-économiques (précarité, logement, discriminations).
◦ 25 % : Résilience du système de santé.
◦ 25 % : Facteurs individuels (génétique, biologie), eux-mêmes influencés par l'environnement via l'épigénétique.
• Nécessité d'une approche interministérielle : Pour agir sur ces déterminants, une collaboration entre les ministères de la Santé, de l'Éducation, de la Justice, etc., est indispensable, via un délégué interministériel dédié.
Marie-Christine Sanier Coavran démontre comment les politiques locales peuvent directement influencer la santé mentale de la population, en s'appuyant sur l'exemple de la ville de Lille.
• Urbanisme et logement : La conception des habitations (éviter les grandes tours, intégrer balcons et jardins) et des espaces publics (créer des îlots de verdure avec bancs et jeux) est pensée pour favoriser les interactions sociales et réduire le stress environnemental (bruit, pollution).
• Mobilité : Des mesures comme la limitation de vitesse à 30 km/h et le développement des pistes cyclables réduisent le bruit et la pollution tout en encourageant l'activité physique, bénéfique pour la santé mentale.
• Inclusion sociale : L'accompagnement vers l'emploi est complété par la valorisation d'autres formes d'engagement, comme le bénévolat, qui permettent aux individus de retrouver une place et une reconnaissance dans la société.
Face à la prolifération des offres dangereuses, une réponse ferme de la puissance publique est nécessaire.
• Actions de la Miviludes (Samir Calfa) : La mission mène des actions de sensibilisation auprès des élus et des professionnels de santé, publie des guides, et travaille en partenariat avec les ordres professionnels. 19,6 % des signalements concernent des professionnels de santé déviants.
• Cadre légal (Samir Calfa) : La loi du 10 mai 2024 constitue une avancée majeure, punissant d'un an de prison et 30 000 € d'amende la promotion de pratiques non éprouvées ou l'incitation à l'abandon de soins.
• Appel à la réglementation (Samir Calfa) : Un encadrement strict des appellations comme "psychopraticien", "psy-conseil" ou "coach" est indispensable, tout comme un contrôle des structures d'accueil qui échappent actuellement à la supervision des Agences Régionales de Santé (ARS).
Maeva Musso plaide pour une réforme des pratiques psychiatriques, en s'inspirant de modèles innovants.
• L'approche "Open Dialogue" :
◦ Principes : Intervention systématique en binôme de professionnels, implication du réseau social du patient (famille, amis), transparence totale des discussions et décisions, et réactivité (prise en charge sous 24-48h). ◦
Résultats observés : Réduction du recours à la coercition (isolement, contention) et aux prescriptions médicamenteuses à long terme.
Forte déstigmatisation au niveau communautaire, car une large part de la population finit par participer à ces réunions.
• Revendications de l'AJPJA :
◦ Faire des usagers des acteurs : Les intégrer à tous les niveaux (politique, formation des internes, recherche participative).
◦ Abolir les pratiques coercitives : Mettre fin à l'isolement et à la contention.
◦ Reconnaître la responsabilité collective : Le véritable tabou actuel est la responsabilité collective dans l'augmentation des troubles psychiques.
Le développement d'une culture partagée de la santé mentale passe par l'éducation et l'outillage de la population.
• Pédagogie et intelligence collective (Sophia Feuillère) : Les solutions doivent être co-construites ("tous ensemble"), en écoutant les singularités et les "points de vue situés" de chacun.
Les méthodes d'intelligence collective sont un levier puissant pour y parvenir.
• Métacognition et esprit critique (Elisabeth Fetti) : Il est crucial de développer la capacité à appliquer l'esprit critique à ses propres pensées.
Cela passe par la connaissance des mécanismes cognitifs et par l'étude de parcours de vie où des personnes ont radicalement changé de croyances, afin de "rendre désirable le questionnement sur soi".
Marie-Christine Sanier Coavran souligne le potentiel immense des municipalités et des réseaux de villes.
• Rôle de catalyseur : Les villes ont la capacité d'écouter les besoins, de mobiliser tous les acteurs (associations, professionnels, habitants) et de coordonner l'action.
• Actions concrètes : Le réseau Ville Santé recense de nombreuses initiatives, comme la gratuité des transports (Dunkerque), le maintien au logement (Metz), ou l'accès à la culture et au sport comme outils de bien-être (Lille, Poitiers).
• Formation citoyenne : Les villes peuvent financer des formations comme les "Premiers Secours en Santé Mentale" ou la création d'"ambassadeurs santé" pour doter la population de réflexes de base.
• Rôle d'interpellation : Face à la pénurie de soignants (18 mois d'attente dans certains CMP), les élus locaux ont le devoir d'interpeller l'État pour obtenir plus de psychiatres et une meilleure reconnaissance des psychologues cliniciens.
La table ronde conclut unanimement que la santé mentale est une question éminemment politique.
Le véritable tabou n'est plus la souffrance psychique elle-même, mais le refus de reconnaître la responsabilité collective dans l'augmentation des troubles.
La sortie de la crise passe par un engagement politique fort, une action interministérielle coordonnée et une implication de toutes les strates de la société.
Le passage d'une logique de soin individuel à une culture partagée du "prendre soin" collectif est la condition sine qua non pour construire une société plus résiliente et attentive à la santé psychique de toutes et tous.
CGC
DOI: 10.1371/journal.pgen.1011461
Resource: Caenorhabditis Genetics Center (RRID:SCR_007341)
Curator: @Apiekniewska
SciCrunch record: RRID:SCR_007341
hs-FLP,tubP-GAL80,FRT19A/Y
DOI: 10.1186/s13041-021-00782-x
Resource: Bloomington Drosophila Stock Center (RRID:SCR_006457)
Curator: @bdscstockkeepers
SciCrunch record: RRID:SCR_006457
42230
DOI: 10.1038/s42003-025-08984-y
Resource: RRID:Addgene_42230
Curator: @olekpark
SciCrunch record: RRID:Addgene_42230
Addgene_61591
DOI: 10.1038/s42003-025-08984-y
Resource: RRID:Addgene_61591
Curator: @scibot
SciCrunch record: RRID:Addgene_61591
Addgene_47869
DOI: 10.1038/s42003-025-08984-y
Resource: None
Curator: @scibot
SciCrunch record: RRID:Addgene_47869
Addgene_84745
DOI: 10.1038/s42003-025-08984-y
Resource: RRID:Addgene_84745
Curator: @scibot
SciCrunch record: RRID:Addgene_84745
PX549
DOI: 10.1038/s41419-025-08166-y
Resource: RRID:Addgene_62988
Curator: @olekpark
SciCrunch record: RRID:Addgene_62988
Addgene_121192
DOI: 10.1038/s44318-025-00594-y
Resource: None
Curator: @scibot
SciCrunch record: RRID:Addgene_121192
Addgene_114699
DOI: 10.1038/s44318-025-00594-y
Resource: None
Curator: @scibot
SciCrunch record: RRID:Addgene_114699
Addgene_72833
DOI: 10.1038/s44318-025-00594-y
Resource: RRID:Addgene_72833
Curator: @scibot
SciCrunch record: RRID:Addgene_72833
Addgene_105927
DOI: 10.1038/s44318-025-00594-y
Resource: RRID:Addgene_105927
Curator: @scibot
SciCrunch record: RRID:Addgene_105927
RRID:Addgene_61580
DOI: 10.1038/s41467-025-65029-y
Resource: RRID:Addgene_61580
Curator: @scibot
SciCrunch record: RRID:Addgene_61580
No compras un peluche. Creas uno que es solo tuyo: eligiendo desde su forma hasta su ropa y accesorios, con cientos de combinaciones posibles. Cada Soulmate es único.
Quitarle las negritas a este texto
Author response:
The following is the authors’ response to the current reviews.
I thank the authors for their clarifications. The manuscript is much improved now, in my opinion. The new power spectral density plots and revised Figure 1 are much appreciated. However, there is one remaining point that I am unclear about. In the rebuttal, the authors state the following: "To directly address the question of whether the auditory signal was distracting, we conducted a follow-up MEG experiment. In this study, we observed a significant reduction in visual accuracy during the second block when the distractor was present (see Fig. 7B and Suppl. Fig. 1B), providing clear evidence of a distractor cost under conditions where performance was not saturated."
I am very confused by this statement, because both Fig. 7B and Suppl. Fig. 1B show that the visual- (i.e., visual target presented alone) has a lower accuracy and longer reaction time than visual+ (i.e., visual target presented with distractor). In fact, Suppl. Fig. 1B legend states the following: "accuracy: auditory- - auditory+: M = 7.2 %; SD = 7.5; p = .001; t(25) = 4.9; visual- - visual+: M = -7.6%; SD = 10.80; p < .01; t(25) = -3.59; Reaction time: auditory- - auditory +: M = -20.64 ms; SD = 57.6; n.s.: p = .08; t(25) = -1.83; visual- - visual+: M = 60.1 ms ; SD = 58.52; p < .001; t(25) = 5.23)."
These statements appear to directly contradict each other. I appreciate that the difficulty of auditory and visual trials in block 2 of MEG experiments are matched, but this does not address the question of whether the distractor was actually distracting (and thus needed to be inhibited by occipital alpha). Please clarify.
We apologize for mixing up the visual and auditory distractor cost in our rebuttal. The reviewer is right in that our two statements contradict each other.
To clarify: In the EEG experiment, we see significant distractor cost for auditory distractors in the accuracy (which can be seen in SUPPL Fig. 1A). We also see a faster reaction time with auditory distractors, which may speak to intersensory facilitation. As we used the same distractors for both experiments, it can be assumed that they were distracting in both experiments.
In our follow-up MEG-experiment, as the reviewer stated, performance in block 2 was higher than in block 1, even though there were distractors present. In this experiment, distractor cost and learning effects are difficult to disentangle. It is possible that participants improved over time for the visual discrimination task in Block 1, as performance at the beginning was quite low. To illustrate this, we divided the trials of each condition into bins of 10 and plotted the mean accuracy in these bins over time (see Author response image 1). Here it can be seen that in Block 2, there is a more or less stable performance over time with a variation < 10 %. In Block 1, both for visual as well as auditory trials, an improvement over time can be seen. This is especially strong for visual trials, which span a difference of > 20%. Note that the mean performance for the 80-90 trial bin was higher than any mean performance observed in Block 2.
Additionally, the same paradigm has been applied in previous investigations, which also found distractor costs for the here-used auditory stimuli in blocked and non-blocked designs. See:
Mazaheri, A., van Schouwenburg, M. R., Dimitrijevic, A., Denys, D., Cools, R., & Jensen, O. (2014). Region-specific modulations in oscillatory alpha activity serve to facilitate processing in the visual and auditory modalities. NeuroImage, 87, 356–362. https://doi.org/10.1016/j.neuroimage.2013.10.052
Van Diepen, R & Mazaheri, A 2017, 'Cross-sensory modulation of alpha oscillatory activity: suppression, idling and default resource allocation', European Journal of Neuroscience, vol. 45, no. 11, pp. 1431-1438. https://doi.org/10.1111/ejn.13570
Author response image 1.
Accuracy development over time in the MEG experiment. During block 1, a performance increase over time can be observed for visual as well as for auditory stimuli. During Block 2, performance is stable over time. Data are presented as mean ± SEM. N = 27 (one participant was excluded from this analysis, as their trial count in at least one condition was below 90 trials).
The following is the authors’ response to the previous reviews
Reviewer #1 (Public review):
In this study, Brickwedde et al. leveraged a cross-modal task where visual cues indicated whether upcoming targets required visual or auditory discrimination. Visual and auditory targets were paired with auditory and visual distractors, respectively. The authors found that during the cue-to-target interval, posterior alpha activity increased along with auditory and visual frequency-tagged activity when subjects were anticipating auditory targets. The authors conclude that their results disprove the alpha inhibition hypothesis, and instead implies that alpha "regulates downstream information transfer." However, as I detail below, I do not think the presented data irrefutably disproves the alpha inhibition hypothesis. Moreover, the evidence for the alternative hypothesis of alpha as an orchestrator for downstream signal transmission is weak. Their data serves to refute only the most extreme and physiologically implausible version of the alpha inhibition hypothesis, which assumes that alpha completely disengages the entire brain area, inhibiting all neuronal activity.
We thank the reviewer for taking the time to provide additional feedback and suggestions and we improved our manuscript accordingly.
(1) Authors assign specific meanings to specific frequencies (8-12 Hz alpha, 4 Hz intermodulation frequency, 36 Hz visual tagging activity, 40 Hz auditory tagging activity), but the results show that spectral power increases in all of these frequencies towards the end of the cue-to-target interval. This result is consistent with a broadband increase, which could simply be due to additional attention required when anticipating auditory target (since behavioral performance was lower with auditory targets, we can say auditory discrimination was more difficult). To rule this out, authors will need to show a power spectral density curve with specific increases around each frequency band of interest. In addition, it would be more convincing if there was a bump in the alpha band, and distinct bumps for 4 vs 36 vs 40 Hz band.
This is an interesting point with several aspects, which we will address separately
Broadband Increase vs. Frequency-Specific Effects:
The suggestion that the observed spectral power increases may reflect a broadband effect rather than frequency-specific tagging is important. However, Supplementary Figure 11 shows no difference between expecting an auditory or visual target at 44 Hz. This demonstrates that (1) there is no uniform increase across all frequencies, and (2) the separation between our stimulation frequencies was sufficient to allow differentiation using our method.
Task Difficulty and Performance Differences:
The reviewer suggests that the observed effects may be due to differences in task difficulty, citing lower performance when anticipating auditory targets in the EEG study. This issue was explicitly addressed in our follow-up MEG study, where stimulus difficulty was calibrated. In the second block—used for analysis—accuracy between auditory and visual targets was matched (see Fig. 7B). The replication of our findings under these controlled conditions directly rules out task difficulty as the sole explanation. This point is clearly presented in the manuscript.
Power Spectrum Analysis:
The reviewer’s suggestion that our analysis lacks evidence of frequency-specific effects is addressed directly in the manuscript. While we initially used the Hilbert method to track the time course of power fluctuations, we also included spectral analyses to confirm distinct peaks at the stimulation frequencies. Specifically, when averaging over the alpha cluster, we observed a significant difference at 10 Hz between auditory and visual target expectation, with no significant differences at 36 or 40 Hz in that cluster. Conversely, in the sensor cluster showing significant 36 Hz activity, alpha power did not differ, but both 36 Hz and 40 Hz tagging frequencies showed significant effects These findings clearly demonstrate frequency-specific modulation and are already presented in the manuscript.
(2) For visual target discrimination, behavioral performance with and without the distractor is not statistically different. Moreover, the reaction time is faster with distractor. Is there any evidence that the added auditory signal was actually distracting?
We appreciate the reviewer’s observation regarding the lack of a statistically significant difference in behavioral performance for visual target discrimination with and without the auditory distractor. While this was indeed the case in our EEG experiment, we believe the absence of an accuracy effect may be attributable to a ceiling effect, as overall visual performance approached 100%. This high baseline likely masked any subtle influence of the distractor.
To directly address the question of whether the auditory signal was distracting, we conducted a follow-up MEG experiment. In this study, we observed a significant reduction in visual accuracy during the second block when the distractor was present (see Fig. 7B and Suppl. Fig. 1B), providing clear evidence of a distractor cost under conditions where performance was not saturated.
Regarding the faster reaction times observed in the presence of the auditory distractor, this phenomenon is consistent with prior findings on intersensory facilitation. Auditory stimuli, which are processed more rapidly than visual stimuli, can enhance response speed to visual targets—even when the auditory input is non-informative or nominally distracting (Nickerson, 1973; Diederich & Colonius, 2008; Salagovic & Leonard, 2021). Thus, while the auditory signal may facilitate motor responses, it can simultaneously impair perceptual accuracy, depending on task demands and baseline performance levels.
Taken together, our data suggest that the auditory signal does exert a distracting influence, particularly under conditions where visual performance is not at ceiling. The dual effect—facilitated reaction time but reduced accuracy—highlights the complexity of multisensory interactions and underscores the importance of considering both behavioral and neurophysiological measures.
(3) It is possible that alpha does suppress task-irrelevant stimuli, but only when it is distracting. In other words, perhaps alpha only suppresses distractors that are presented simultaneously with the target. Since the authors did not test this, they cannot irrefutably reject the alpha inhibition hypothesis.
The reviewer’s claim that we did not test whether alpha suppresses distractors presented simultaneously with the target is incorrect. As stated in the manuscript and supported by our data (see point 2), auditory distractors were indeed presented concurrently with visual targets, and they were demonstrably distracting. Therefore, the scenario the reviewer suggests was not only tested—it forms a core part of our design.
Furthermore, it was never our intention to irrefutably reject the alpha inhibition hypothesis. Rather, our aim was to revise and expand it. If our phrasing implied otherwise, we have now clarified this in the manuscript. Specifically, we propose that alpha oscillations:
(a) Exhibit cyclic inhibitory and excitatory dynamics;
(b) Regulate processing by modulating transfer pathways, which can result in either inhibition or facilitation depending on the network context.
In our study, we did not observe suppression of distractor transfer, likely due to the engagement of a supramodal system that enhances both auditory and visual excitability. This interpretation is supported by prior findings (e.g., Jacoby et al., 2012), which show increased visual SSEPs under auditory task load, and by Zhigalov et al. (2020), who found no trial-by-trial correlation between alpha power and visual tagging in early visual areas, despite a general association with attention.
Recent evidence (Clausner et al., 2024; Yang et al., 2024) further supports the notion that alpha oscillations serve multiple functional roles depending on the network involved. These roles include intra- and inter-cortical signal transmission, distractor inhibition, and enhancement of downstream processing (Scheeringa et al., 2012; Bastos et al., 2015; Zumer et al., 2014). We believe the most plausible account is that alpha oscillations support both functions, depending on context.
To reflect this more clearly, we have updated Figure 1 to present a broader signal-transfer framework for alpha oscillations, beyond the specific scenario tested in this study.
We have now revised Figure 1 and several sentences in the introduction and discussion, to clarify this argument.
L35-37: Previous research gave rise to the prominent alpha inhibition hypothesis, which suggests that oscillatory activity in the alpha range (~10 Hz) plays a mechanistic role in selective attention through functional inhibition of irrelevant cortical areas (see Fig. 1; Foxe et al., 1998; Jensen & Mazaheri, 2010; Klimesch et al., 2007).
L60-65: In contrast, we propose that functional and inhibitory effects of alpha modulation, such as distractor inhibition, are exhibited through blocking or facilitating signal transmission to higher order areas (Peylo et al., 2021; Yang et al., 2023; Zhigalov & Jensen, 2020; Zumer et al., 2014), gating feedforward or feedback communication between sensory areas (see Fig. 1; Bauer et al., 2020; Haegens et al., 2015; Uemura et al., 2021).
L482-485: This suggests that responsiveness of the visual stream was not inhibited when attention was directed to auditory processing and was not inhibited by occipital alpha activity, which directly contradicts the proposed mechanism behind the alpha inhibition hypothesis.
L517-519: Top-down cued changes in alpha power have now been widely viewed to play a functional role in directing attention: the processing of irrelevant information is attenuated by increasing alpha power in areas involved with processing this information (Foxe, Simpson, & Ahlfors, 1998; Hanslmayr et al., 2007; Jensen & Mazaheri, 2010).
L566-569: As such, it is conceivable that alpha oscillations can in some cases inhibit local transmission, while in other cases, depending on network location, connectivity and demand, alpha oscillation can facilitate signal transmission. This mechanism allows to increase transmission of relevant information and to block transmission of distractors.
(4) In the abstract and Figure 1, the authors claim an alternative function for alpha oscillations; that alpha "orchestrates signal transmission to later stages of the processing stream." In support, the authors cite their result showing that increased alpha activity originating from early visual cortex is related to enhanced visual processing in higher visual areas and association areas. This does not constitute a strong support for the alternative hypothesis. The correlation between posterior alpha power and frequency-tagged activity was not specific in any way; Fig. 10 shows that the correlation appeared on both 1) anticipating-auditory and anticipating-visual trials, 2) the visual tagged frequency and the auditory tagged activity, and 3) was not specific to the visual processing stream. Thus, the data is more parsimonious with a correlation than a causal relationship between posterior alpha and visual processing.
Again, the reviewer raises important points, which we want to address
The correlation between posterior alpha power and frequency-tagged activity was not specific, as it is present both when auditory and visual targets are expected:
If there is a connection between posterior alpha activity and higher-order visual information transfer, then it can be expected that this relationship remains across conditions and that a higher alpha activity is accompanied by higher frequency-tagged activity, both over trials and over conditions. However, it is possible that when alpha activity is lower, such as when expecting a visual target, the signal-to-noise ratio is affected, which may lead to higher difficulty to find a correlation effect in the data when using non-invasive measurements.
The connection between alpha activity and frequency-tagged activity appears both for auditory as well as visual stimuli and The correlation is not specific to the visual processing stream:
While we do see differences between conditions (e.g. in the EEG-analysis, mostly 36 Hz correlated with alpha activity and only in one condition 40 Hz showed a correlation as well), it is true that in our MEG analysis, we found correlations both between alpha activity and 36 Hz as well as alpha activity and 40 Hz.
We acknowledge that when analysing frequency-tagged activity on a trial-by-trial basis, where removal of non-timelocked activity through averaging (which we did when we tested for condition differences in Fig. 4 and 9) is not possible, there is uncertainty in the data. Baseline-correction can alleviate this issue, but it cannot offset the possibility of non-specific effects. We therefore decided to repeat the analysis with a fast-fourier calculated power instead of the Hilbert power, in favour of a higher and stricter frequency-resolution, as we averaged over a time-period and thus, the time-domain was not relevant for this analysis. In this more conservative analysis, we can see that only 36 Hz tagged activity when expecting an auditory target correlated with early visual alpha activity.
Additionally, we added correlation analyses between alpha activity and frequency-tagged activity within early visual areas, using the sensor cluster which showed significant condition differences in alpha activity. Here, no correlations between frequency-tagged activity and alpha activity could be found (apart from a small correlation with 40 Hz which could not be confirmed by a median split; see SUPPL Fig. 14 C). The absence of a significant correlation between early visual alpha and frequency-tagged activity has previously been described by others (Zhigalov & Jensen, 2020) and a Bayes factor of below 1 also indicated that the alternative hypotheses is unlikely.
Nonetheless, a correlation with auditory signal is possible and could be explained in different ways. For example, it could be that very early auditory feedback in early visual cortex (see for example Brang et al., 2022) is transmitted alongside visual information to higher-order areas. Several studies have shown that alpha activity and visual as well as auditory processing are closely linked together (Bauer et al., 2020; Popov et al., 2023). Inference on whether or how this link could play out in the case of this manuscript expands beyond the scope of this study.
To summarize, we believe the fact that 36 Hz activity within early visual areas does not correlate with alpha activity on a trial-by-trial basis, but that 36 Hz activity in other areas does, provides strong evidence that alpha activity affects down-stream signal processing.
We mention this analysis now in our discussion:
L533-536: Our data provides evidence in favour of this view, as we can show that early sensory alpha activity does not covary over trials with SSEP magnitude in early visual areas, but covaries instead over trials with SSEP magnitude in higher order sensory areas (see also SUPPL. Fig. 14).
Reviewer #1 (Recommendations for the authors):
The evidence for the alternative hypothesis, that alpha in early sensory areas orchestrates downstream signal transmission, is not strong enough to be described up front in the abstract and Figure 1. I would leave it in the Discussion section, but advise against mentioning it in the abstract and Figure 1.
We appreciate the reviewer’s concern regarding the inclusion of the alternative hypothesis—that alpha activity in early sensory areas orchestrates downstream signal transmission—in the abstract and Figure 1. While we agree that this interpretation is still developing, recent studies (Keitel et al., 2025; Clausner et al., 2024; Yang et al., 2024) provide growing support for this framework.
In response, we have revised the introduction, discussion, and Figure 1 to clarify that our intention is not to outright dismiss the alpha inhibition hypothesis, but to refine and expand it in light of new data. This revision does not invalidate the prior literature on alpha timing and inhibition; rather, it proposes an updated mechanism that may better account for observed effects.
We have though retained Figure 1, as it visually contextualizes the broader theoretical landscape. while at the same time added further analyses to strengthen our empirical support for this emerging view.
References:
Bastos, A. M., Litvak, V., Moran, R., Bosman, C. A., Fries, P., & Friston, K. J. (2015). A DCM study of spectral asymmetries in feedforward and feedback connections between visual areas V1 and V4 in the monkey. NeuroImage, 108, 460–475. https://doi.org/10.1016/j.neuroimage.2014.12.081
Bauer, A. R., Debener, S., & Nobre, A. C. (2020). Synchronisation of Neural Oscillations and Cross-modal Influences. Trends in cognitive sciences, 24(6), 481–495. https://doi.org/10.1016/j.tics.2020.03.003
Brang, D., Plass, J., Sherman, A., Stacey, W. C., Wasade, V. S., Grabowecky, M., Ahn, E., Towle, V. L., Tao, J. X., Wu, S., Issa, N. P., & Suzuki, S. (2022). Visual cortex responds to sound onset and offset during passive listening. Journal of neurophysiology, 127(6), 1547–1563. https://doi.org/10.1152/jn.00164.2021
Clausner T., Marques J., Scheeringa R. & Bonnefond M (2024). Feature specific neuronal oscillations in cortical layers BioRxiv :2024.07.31.605816. https://doi.org/10.1101/2024.07.31.605816
Diederich, A., & Colonius, H. (2008). When a high-intensity "distractor" is better then a low-intensity one: modeling the effect of an auditory or tactile nontarget stimulus on visual saccadic reaction time. Brain research, 1242, 219–230. https://doi.org/10.1016/j.brainres.2008.05.081
Haegens, S., Nácher, V., Luna, R., Romo, R., & Jensen, O. (2011). α-Oscillations in the monkey sensorimotor network influence discrimination performance by rhythmical inhibition of neuronal spiking. Proceedings of the National Academy of Sciences of the United States of America, 108(48), 19377–19382. https://doi.org/10.1073/pnas.1117190108
Jacoby, O., Hall, S. E., & Mattingley, J. B. (2012). A crossmodal crossover: opposite effects of visual and auditory perceptual load on steady-state evoked potentials to irrelevant visual stimuli. NeuroImage, 61(4), 1050–1058. https://doi.org/10.1016/j.neuroimage.2012.03.040
Keitel, A., Keitel, C., Alavash, M., Bakardjian, K., Benwell, C. S. Y., Bouton, S., Busch, N. A., Criscuolo, A., Doelling, K. B., Dugue, L., Grabot, L., Gross, J., Hanslmayr, S., Klatt, L.-I., Kluger, D. S., Learmonth, G., London, R. E., Lubinus, C., Martin, A. E., … Kotz, S. A. (2025). Brain rhythms in cognition – controversies and future directions. ArXiv. https://doi.org/10.48550/arXiv.2507.15639
Nickerson R. S. (1973). Intersensory facilitation of reaction time: energy summation or preparation enhancement?. Psychological review, 80(6), 489–509. https://doi.org/10.1037/h0035437
Popov, T., Gips, B., Weisz, N., & Jensen, O. (2023). Brain areas associated with visual spatial attention display topographic organization during auditory spatial attention. Cerebral cortex (New York, N.Y. : 1991), 33(7), 3478–3489. https://doi.org/10.1093/cercor/bhac285
Salagovic, C. A., & Leonard, C. J. (2021). A nonspatial sound modulates processing of visual distractors in a flanker task. Attention, perception & psychophysics, 83(2), 800–809. https://doi.org/10.3758/s13414-020-02161-5
Scheeringa, R., Petersson, K. M., Kleinschmidt, A., Jensen, O., & Bastiaansen, M. C. (2012). EEG α power modulation of fMRI resting-state connectivity. Brain connectivity, 2(5), 254–264. https://doi.org/10.1089/brain.2012.0088
Spaak, E., Bonnefond, M., Maier, A., Leopold, D. A., & Jensen, O. (2012). Layer-specific entrainment of γ-band neural activity by the α rhythm in monkey visual cortex. Current biology : CB, 22(24), 2313–2318. https://doi.org/10.1016/j.cub.2012.10.020
Yang, X., Fiebelkorn, I. C., Jensen, O., Knight, R. T., & Kastner, S. (2024). Differential neural mechanisms underlie cortical gating of visual spatial attention mediated by alpha-band oscillations. Proceedings of the National Academy of Sciences of the United States of America, 121(45), e2313304121. https://doi.org/10.1073/pnas.2313304121
Zhigalov, A., & Jensen, O. (2020). Alpha oscillations do not implement gain control in early visual cortex but rather gating in parieto-occipital regions. Human brain mapping, 41(18), 5176–5186. https://doi.org/10.1002/hbm.25183
Zumer, J. M., Scheeringa, R., Schoffelen, J. M., Norris, D. G., & Jensen, O. (2014). Occipital alpha activity during stimulus processing gates the information flow to object-selective cortex. PLoS biology, 12(10), e1001965. https://doi.org/10.1371/journal.pbio.1001965
Note: This preprint has been reviewed by subject experts for Review Commons. Content has not been altered except for formatting.
Learn more at Review Commons
Summary:
The authors present MAVISp, a tool for viewing protein variants heavily based on protein structure information. The authors have done a very impressive amount of curation on various protein targets, and should be commended for their efforts. The tool includes a diverse array of experimental, clinical, and computational data sources that provides value to potential users interested in a given target.
Major comments:
Unfortunately I was not able to get the website to work properly. When selecting a protein target in simple mode, I was greeted with a completely blank page in the app window, and in ensemble mode, there was no transition away from the list of targets at all. I'm using Firefox 140.0.2 (64-bit) on Ubuntu 22.04. I would have liked to be able to explore the data myself and provide feedback on the user experience and utility.
I have some serious concerns about the sustainability of the project and think that additional clarifications in the text could help. Currently is there a way to easily update a dataset to add, remove, or update a component (for example, if a new predictor is published, an error is found in a predictor dataset, or a predictor is updated)? If it requires a new round of manual curation for each protein to do this, I am worried that this will not scale and will leave the project with many out of date entries. The diversity of software tools (e.g., three different pipeline frameworks) also seems quite challenging to maintain.
On the same theme, according to the GitHub repository, the program relies on Python 3.9, which reaches end of life in October 2025. It has been tested against Ubuntu 18.04, which left standard support in May 2023. The authors should update the software to more modern versions of Python to promote the long-term health and maintainability of the project.
I appreciate that the authors have made their code and data available. These artifacts should also be versioned and archived in a service like Zenodo, so that researchers who rely on or want to refer to specific versions can do so in their own future publications.
In the introduction of the paper, the authors conflate the clinical challenges of variant classification with evidence generation and it's quite muddled together. The y should strongly consider splitting the first paragraph into two paragraphs - one about challenges in variant classification/clinical genetics/precision oncology and another about variant effect prediction and experimental methods. The authors should also note that they are many predictors other than AlphaMissense, and may want to cite the ClinGen recommendations (PMID: 36413997) in the intro instead.
Also in the introduction on lines 21-22 the authors assert that "a mechanistic understanding of variant effects is essential knowledge" for a variety of clinical outcomes. While this is nice, it is clearly not the case as we are able to classify variants according to the ACMG/AMP guidelines without any notion of specific mechanism (for example, by combining population frequency data, in silico predictor data, and functional assay data). The authors should revise the statement so that it's clear that mechanistic understanding is a worthy aspiration rather than a prerequisite.
In the structural analysis section (page 5, lines 154-155 and elsewhere), the authors define cutoffs with convenient round numbers. Is there a citation for these values or were these arbitrarily chosen by the authors? I would have liked to see some justification that these assignments are reasonable. Also there seems to be an error in the text where values between -2 and -3 kcal/mol are not assigned to a bin (I assume they should also be uncertain). There are other similar seemingly-arbitrary cutoffs later in the section that should also be explained.
On page 9, lines 294-298 the authors talk about using the PTEN data from ProteinGym, rather than the actual cutoffs from the paper. They get to the latter later on, but I'm not sure why this isn't first? The ProteinGym cutoffs are somewhat arbitrarily based on the median rather than expert evaluation of the dataset and I'm not sure why it's even worth mentioning them when proper classifications are available. Regarding PTEN, it would be quite interesting to see a comparison of the VAMP-seq PTEN data and the Mighell phosphatase assay, which is cited on page 9 line 288 but is not actually a VAMP-seq dataset. I think this section could be interesting but it requires some additional attention.
The authors mention "pathogenicity predictors" and otherwise use pathogenicity incorrectly throughout the manuscript. Pathogenicity is a classification for a variant after it has been curated according to a framework like the ACMG/AMP guidelines (Richards 2015 and amendments). A single tool cannot predict or assign pathogenicity - the AlphaMissense paper was wrong to use this nomenclature and these authors should not compound this mistake. These predictors should be referred to as "variant effect predictors" or similar, and they are able to produce evidence towards pathogenicity or benignity but not make pathogenicity calls themselves. For example, in Figure 4e, the terms "pathogenic" and "benign" should only be used here if these are the classifications the authors have derived from ClinVar or a similar source of clinically classified variants.
Minor comments:
The target selection table on the website needs some kind of text filtering option. It's very tedious to have to find a protein by scrolling through the table rather than typing in the symbol. This will only get worse as more datasets are added.
The data sources listed on the data usage section of the website are not concordant with what is in the paper. For example, MaveDB is not listed.
I found Figure 2 to be a bit confusing in that it partially interleaves results from two different proteins. I think this would be nicer as two separate figures, one on each protein, or just of a single protein.
Figure 3 panel b is distractingly large and I wonder if the authors could do a little bit more with this visualization.
Capitalization is inconsistent throughout the manuscript. For example, page 9 line 288 refers to VampSEQ instead of VAMP-seq (although this is correct elsewhere). MaveDB is referred to as MAVEdb or MAVEDB in various places. AlphaMissense is referred to as Alphamissense in the Figure 5 legend. The authors should make a careful pass through the manuscript to address this kind of issues.
MaveDB has a more recent paper (PMID: 39838450) that should be cited instead of/in addition to Esposito et al.
On page 11, lines 338-339 the authors mention some interesting proteins including BLC2, which has base editor data available (PMID: 35288574). Are there plans to incorporate this type of functional assay data into MAVISp?
General assessment:
This is a nice resource and the authors have clearly put a lot of effort in. They should be celebrated for their achievments in curating the diverse datasets, and the GitBooks are a nice approach. However, I wasn't able to get the website to work and I have raised several issues with the paper itself that I think should be addressed.
Advance:
New ways to explore and integrate complex data like protein structures and variant effects are always interesting and welcome. I appreciate the effort towards manual curation of datasets. This work is very similar in theme to existing tools like Genomics 2 Proteins portal (PMID: 38260256) and ProtVar (PMID: 38769064). Unfortunately as I wasn't able to use the site I can't comment further on MAVISp's position in the landscape.
Audience:
MAVISp could appeal to a diverse group of researchers who are interested in the biology or biochemistry of proteins that are included, or are interested in protein variants in general either from a computational/machine learning perspective or from a genetics/genomics perspective.
My expertise:
I am an expert in high-throughput functional genomics experiments and am an experienced computational biologist with software engineering experience.
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public review):
Summary:
The paper by Boch and colleagues, entitled Comparative Neuroimaging of the Carnivore Brain: Neocortical Sulcal Anatomy, compares and describes the cortical sulci of eighteen carnivore species, and sets a benchmark for future work on comparative brains.
Based on previous observations, electrophysiological, histological and neuroimaging studies and their own observations, the authors establish a correspondence between the cortical sulci and gyri of these species. The different folding patterns of all brain regions are detailed, put into perspective in relation to their phylogeny as well as their potential involvement in cortical area expansion and behavioral differences.
Strengths:
This is a pioneering article, very useful for comparative brain studies and conducted with great seriousness and based on many past studies. The article is well-written and very didactic. The different protocols for brain collection, perfusion, and scanning are very detailed. The images are self-explanatory and of high quality. The authors explain their choice of nomenclature and labels for sulci and gyri on all species, with many arguments. The opening on ecology and social behavior in the discussion is of great interest and helps to put into perspective the differences in folding found at the level of the different cortexes. In addition, the authors do not forget to put their results into the context of the laws of allometry. They explain, for example, that although the largest brains were the most folded and had the deepest folds in their dataset, they did not necessarily have unique sulci, unlike some of the smaller, smoother brains.
Weaknesses:
The article is aware of its limitations, not being able to take into account interindividual variability within each species, inter-hemispheric asymmetries, or differences between males and females. However, this does not detract from their aim, which is to lay the foundations for a correspondence between the brains of carnivores so that navigation within the brains of these species can be simplified for future studies. This article does not include comparisons of morphometric data such as sulci depth, sulci wall surface, or thickness of the cortical ribbon around the sulci.
We thank the reviewer for their overwhelmingly positive evaluation of our work. As noted by the reviewer, our primary aim was to establish a framework for navigating carnivoran brains to lay the foundation for future research. We are pleased that this objective has been successfully achieved.
Individual differences
As the reviewer points out, we do not quantify within-species intraindividual differences, which was a conscious choice. We aimed to emphasise the breadth of species over individuals, as is standard in large-scale comparative anatomy (cf. Heuer et al., 2023, eLife; Suarez et al., 2022, eLife). Following the logic of phylogenetic relationships, the presence of a particular sulcus across related species is also a measure of reliability. We felt safe in this choice, as previous work in both primates and carnivorans has shown that differences across major sulci across individuals are a matter of degree rather than a case of presence or absence (Connolly, 1950, External morphology of the primate brain, C.C. Thomas; Hecht et al., 2019 J Neurosci; Kawamuro 1971 Acta Anat., Kawamuro & Naito, 1977, Acta Anat.).
In our revised manuscript, we now include additional individuals for six different species, representing both carnivoran suborders (Feliformia and Caniformia), and within Caniformia, both Arctoidea and Canidae (see revised Table 1 and main changes in text below). These additions confirm that intra-species variation primarily affects sulcal shape rather than the presence or absence of major sulci. Furthermore, the inclusion of additional individuals helped validate some initial observations, for example, confirming that the brown bear's proreal sulcus is more accurately characterised as a branch of the presylvian sulcus.
Main changes in the revised manuscript:
Results and discussion, p. 13-14: Presylvian sulcus. Rostral to the pseudo-sylvian fissure, the perisylvian sulcus originates from or close to the rostral lateral rhinal fissure (see Supplementary Note 1 and Figure S2 for ventral view). The sulcus extends dorsally, and we observed a gentle caudal curve in the majority of the species (Figures 2-3, white).
There were no major variations across species, but we noted a shortened sulcus in the meerkat and Egyptian mongoose and the presence of a secondary branch at the dorsal end that extended rostrally in the Eurasian badger and South American coati brain. The brown bear exhibited an additional sulcus in the frontal lobe, previously labelled as the proreal sulcus (see, e.g., Sienkiewicz et al., 2019); however, its shape closely resembled the secondary branches of the perisylvian sulcus seen in the South American coati and Eurasian badger. Sienkiewicz et al. (2019) also noted that this sulcus merges with the presylvian sulcus in their specimen, consistent with our findings in the left hemisphere of the brown bear and bilaterally in the Ussuri brown bear (see Supplementary Figure S3A, S5A). Given the known gyrencephaly of Ursidae brains with frequent secondary and tertiary sulci (Lyras et al., 2023), we propose that this sulcus represents a branch of the perisylvian sulcus.
General Discussion, p. 23-24:Regarding individual variability in external brain morphology, previous work in primates and carnivorans has shown that differences across individuals typically affect sulcal shape, depth, or extent, but not the presence of major sulci. This has been reported in diverse contexts, including comparisons between captive and (semi-)wild macaque (Sallet et al., 2011; Testard et al., 2022), different dog breeds (Hecht et al., 2019), domestic cats (Kawamura, 1971b), or selectively bred foxes (Hecht et al., 2021). By including additional individuals for selected species, we extend these findings to a broader range of carnivorans. Notably, we observed no major sulcal differences between closely related species, even when specimens were acquired using different extraction and scanning protocols, for example, across felid clades or among wolf-like canids, further suggesting that substantial within-species variation is unlikely. While a full analysis of interindividual variability lies beyond the scope of this study, our findings support the reliability of the major sulcal patterns described.
Interhemispheric differences
Regarding potential inter-hemispheric differences, we have now also created digital atlases of all identified sulci in both hemispheres, which are publicly available at https://git.fmrib.ox.ac.uk/neuroecologylab/carnivore-surfaces. While the manuscript continues to focus primarily on descriptions of the right hemisphere, we now also report observed inter-hemispheric differences where applicable. These differences remain minor and, again, a matter of degree. For example, the complementary quantitative analyses investigating covariation between sulcal length and behavioural traits conducted in the right hemisphere were replicated in the left (Supplementary Figure S6 and related Supplementary tables S1-S3).
Main changes in the revised manuscript:
Materials and Methods, p. 33: We focused on the major lateral and dorsal sulci of the carnivoran brain, but the medial wall and ventral view of the sulci are also described. For consistency, we started by labelling the right hemispheres on the mid-thickness surfaces; these are the hemispheres presented in the manuscript. An exception was made for the jungle cat, for which only the left hemisphere was available and is therefore shown. We aimed to facilitate interspecies comparisons and the exploration of previously undescribed carnivoran brains. To this end, we first created standardized criteria (henceforth referred to as recipes) for identifying each sulcus, drawing from existing literature on carnivoran neuroanatomy, particularly in paleoneurology (Lyras et al., 2023), and our own observations. In addition, we created digital sulcal masks for both hemispheres, which allowed us to test whether the same patterns were observable bilaterally and to further facilitate future research building on our framework. For the Egyptian mongoose, only the right hemisphere was available, and thus, a bilateral comparison was not possible for this species. Anatomical nomenclature primarily follows the recommendations of Czeibert et al (2018); if applicable, alternative names of sulci are provided once.
Materials and Methods, p. 34-35: We first briefly illustrated the gyri of the carnivoran brain with a focus on gyri that are not present in some species as a consequence of absent sulci to complement our observations. We then summarised the key differences and similarities in sulcal anatomy between species and related them to their ecology and behaviour. To complement this qualitative description, we conducted an initial quantitative analysis of sulcal length data from both hemispheres.
To test whether sulcal length covaries with behavioural traits, we fit linear models predicting the relative length of the three target sulci (cruciate, postcruciate, proreal) as a function of forepaw dexterity (low vs.
high) and sociality (solitary vs cooperative hunting). We measured the absolute length of each sulcus using the wb_command -border-length function from the Connectome Workbench toolkit (Marcus et al., 2011) applied to the manually defined sulcal masks (i.e., border files). Relative sulcal length was calculated by dividing the length of each target sulcus by that of a reference sulcus in the same hemisphere, reducing interspecies variation in brain or sulcal size. Reference sulci were required to be present in all species within a hemisphere and excluded if they were a target sulcus, part of the same functional system (e.g., somatosensory/motor), or anatomically atypical (e.g., the pseudosylvian fissure). This resulted in seven reference sulci for the proreal sulcus (ansate, coronal, marginal, presylvian, retrosplenial, splenial, suprasylvian) and four for the cruciate and postcruciate sulci (marginal, retrosplenial, splenial, suprasylvian). For each target-reference pair, we fit the following linear model: relative length ~ forepaw dexterity + sociality. Models were run separately for left and right hemispheres, with the left serving as a replication test. Associations were considered meaningful if the predictor reached statistical significance (p ≤ .05) in ≥ 75% of reference sulcus models per hemisphere. Additional individuals were not included in the analysis.
Data and code availability statement, p. 35-36: Generated surfaces of all species and T1-like contrast images of post-mortem samples obtained by the C Generated surfaces of all species and T1-like contrast images of post-mortem samples obtained by the Copenhagen Zoo and the Zoological Society of London (see Table 1) are available at the Digital Brain Zoo of the University of Oxford (Tendler et al., 2022) (https://open.win.ox.ac.uk/DigitalBrainBank/#/datasets/zoo). For all other species, except the domestic cat, the cortical surface reconstructions are available through the same resource. In-vivo data for the domestic cat is available upon request.
We created, extracted and analysed sulcal length data using the Connectome Workbench toolkit (Marcus et al., 2011), R 4.4.0 (R Core Team, 2023) and Python 3.9.7. Sulcal masks, along with the associated midthickness cortical surface reconstructions for all 32 animals, species-specific behavioural data, and the code used to extract sulcal lengths and perform the statistical analyses are available at: https://git.fmrib.ox.ac.uk/neuroecologylab/carnivore-surfaces.
Further brain measures
We feel that sulci depth, sulci wall surface, or thickness of the cortical ribbon are measures that vary more across individuals, and we have therefore not included them in the study. In addition, these are measures that are not generally used as betweenspecies comparative measures, whereas sulcal patterning is (cf. Amiez et al., 2019, Nat Comms; Connolly, 1950; Miller et al., 2021, Brain Behav Evol; Radinsky 1975, J Mammal; Radinsky 1969, Ann N Y Acad Sci; Welker & Campos 1963 J. Comp Neurol).
We, therefore, added them as suggestions for future directions, building on our work.
Major changes in the revised manuscript:
Limitations and future directions, p. 25-26: Our findings represent a critical first step for linking brains within and across species for interspecies insights. The present analyses are based on multiple individuals pooled into families and genera, primarily focusing on single representatives per species. Additional individuals for selected species confirmed that intra-species variation is a matter of degree rather than a case of presence or absence of major sulci, but we do not provide an extensive account of the possible range of sulcal shape or other anatomical features. Future studies will aim to systematically investigate interindividual variability in sulcal shape, depth, surface area, or thickness of the cortical ribbon surrounding the sulci, and will extend to more detailed investigations of the medial part of the cortex, as well as the subcortical structures and the cerebellum.The present framework and resulting database also provides the foundation to guide and facilitate future investigations of inter- and intra-species variation in regional brain size.
Reviewer #2 (Public review):
Summary:
The authors have completed MRI-based descriptions of the sulcal anatomy of 18 carnivoran species that vary greatly in behaviour and ecology. In this descriptive study, different sulcal patterns are identified in relation to phylogeny and, to some extent, behaviour. The authors argue that the reported differences across families reflect behaviour and electrophysiology, but these correlations are not supported by any analyses.
Strengths:
A major strength of this paper is using very similar imaging methods across all specimens. Often papers like this rely on highly variable methods so that consistency reduces some of the variability that can arise due to methodology.
The descriptive anatomy was accurate and precise. I could readily follow exactly where on the cortical surface the authors referring. This is not always the case for descriptive anatomy papers, so I appreciated the efforts the authors took to make the results understandable for a broader audience.
I also greatly appreciate the authors making the images open access through their website.
Weaknesses:
Although I enjoyed many aspects of this manuscript, it is lacking in any quantitative analyses that would provide more insights into what these variations in sulcal anatomy might mean. The authors do discuss inter-clade differences in relation to behaviour and older electrophysiology papers by Welker, Campos, Johnson, and others, but it would be more biologically relevant to try to calculate surface areas or volumes of cortical fields defined by some of these sulci. For example, something like the endocast surface area measurements used by Sakai and colleagues would allow the authors to test for differences among clades, in relation to brain/body size, or behaviour. Quantitative measurements would also aid significantly in supporting some of the potential correlations hinted at in the Discussion.
Although quantitative measurements would be helpful, there are also some significant concerns in relation to the specimens themselves. First, almost all of these are captive individuals. We know that environmental differences can alter neocortical development and humans and nonhuman animals and domestication affects neocortical volume and morphology. Whether captive breeding affects neocortical anatomy might not be known, but it can affect other brain regions and overall brain size and could affect sulcal patterns. Second, despite using similar imaging methods across specimens, fixation varied markedly across specimens. Fixation is unlikely to affect the ability to recognize deep sulci, but variations in shrinkage could nevertheless affect overall brain size and morphology, including the ability to recognize shallow sulci. Third, the sample size = 1 for every species examined. In humans and nonhuman animals, sulcal patterns can vary significantly among individuals. In domestic dogs, it can even vary greatly across breeds. It, therefore, remains unclear to what extent the pattern observed in one individual can be generalized for a species, let alone an entire genus or family. The lack of accounting for inter-individual variability makes it difficult to make any firm conclusions regarding the functional relevance of sulcal patterns.
We thank the reviewer for their assessment of our work. The primary aim of this study was to establish a framework for navigating carnivoran brains by providing a comprehensive overview of all major neocortical sulci across eighteen different species. Given the inconsistent nomenclature in the literature and the lack of standardized criteria (“recipes”) for identifying the major sulci, we specifically focused on homogenizing the terminology and creating recipes for their identification. In addition to generating digital cortical surfaces for all brains, we have now also added sulcal masks to further support future research building on this framework. We are pleased that our primary objective is seen as successfully achieved and are delighted to report that, following the reviewer’s recommendations, we have further expanded the dataset by including eight additional species and a second individual for six species, yielding a total of 32 carnivorans from eight carnivoran families (see revised Table 1 for a detailed list).
The present dataset constitutes the most comprehensive collection of fissiped carnivoran brains to date, encompassing a wide range of land-dwelling species from eight families. It includes diverse representatives, such as both social and solitary mongooses, weasel-like and non-weasel mustelids, and a broad spectrum of canids including wolf-like, fox-like, and more basal forms. Further expanding this already extensive dataset has even led to novel discoveries, such as the felid-specific diagonal sulcus and the unique occipito-temporal sulcal configuration shared by herpestids and hyaenids.
Major changes in the revised manuscript:
Results and discussion, p. 4-5: We labelled the neocortical sulci of twenty-six carnivoran species (see Figure 1) based on reconstructed surfaces and developed standardised criteria (“recipes”) for identifying each major sulcus. For each sulcus, we also created corresponding digital masks. Our study included eleven Feliformia and fifteen Caniformia species from eight different carnivoran families. Within the suborder Caniformia, we examined eight Canidae and seven Arctoidea species. In addition, we describe relative intra-species variation in sulcal shape based on supplementary specimens from six species (see Table 1).
Overall, of the carnivorans studied, Canidae brains exhibited the largest number of unique major sulci, while the brown bear brain was the most gyrencephalic, with the deepest folds and many secondary sulci (see Figures 2-3; brains are arranged by descending number of major sulci). The brown bear was also the largest animal in the sample. The brains of the smaller species, such as the fennec fox, meerkat or ferret, were the most lissencephalic, with the sulci having fewer undulations or indentations compared to the other species. A similar trend has also been observed in the sulci of the prefrontal cortex in primates (Amiez et al., 2023, 2019). The meerkat and Egyptian mongoose exhibited the smallest number of major sulci but possessed, along with the striped hyena, a unique configuration of sulci in the occipito-temporal cortex. In the following, we describe each sulcus' appearance, the recipes on how to identify them, and provide an overview of the most significant differences across species.
Results and discussion, p. 11: Diagonal sulcus. The diagonal sulcus is oriented nearly perpendicularly to the rostral portion of the suprasylvian sulcus (Figure 2, Supplementary Figure S2, red). We identified it in all Felidae and in the striped hyena, but it was absent in Herpestidae and all Caniformia species.
In our sample, the sulcus showed moderate variation in shape and continuity. In the caracal and the second sand cat, it appeared as a detached continuation of the rostral suprasylvian sulcus (Supplementary Figure S3). In the Amur and Persian leopards, the diagonal sulcus merged with the rostral ectosylvian sulcus on the right hemisphere, forming a continuous or bifurcated groove. Similar individual variation has been described in domestic cats (Kawamura, 1971b).
We respectfully disagree with the reviewer on two accounts, where we believe the revieweris not judging the scope of the current work
(1) Intra-individual differences & potential confounding factors
The first is with respect to individual differences relationships. To the best of our knowledge, differences between captive and wild animals, or indeed between individuals, do not affect the presence or absence of any major sulci. No differences in sulcal patterns were detected between captive and (semi-)wild macaques (cf. Sallet et al., 2011, Science; Testard et al., 2022, Sci Adv), different dog breeds (Hecht et al., 2019 J Neurosci) or foxes selectively bred to simulate domestication, compared to controls (Hecht et al., 2021 J. Neurosci).
By including additional individuals for selected species in the revised version of our manuscript, we confirm and extend these findings to a broader range of carnivorans. Indeed, we also did not observe major differences between closely related species, even when specimens were collected using different extraction and scanning protocols - for example, across felid clades or wolf-like canids - making substantial individual variation within a species even less likely. Thus, while a comprehensive analysis of interindividual variability is beyond the scope of this study, our observations support the robustness of the major sulcal patterns described here. Moreover, the inclusion of additional individuals also helped validate some initial observations, for example, confirming that the brown bear's proreal sulcus is more accurately characterised as a branch of the presylvian sulcus.
We do, however, agree with the reviewer that building up a database like ours benefits from providing as much information about the samples as possible to enable these issues to be tested. We, therefore, made sure to include as detailed information as possible, including whether the animals were from captive or wild populations, in our manuscript.
Main changes in the revised manuscript:
Results and discussion, p. 13-14: Presylvian sulcus. There were no major variations across species, but we noted a shortened sulcus in the meerkat and Egyptian mongoose and the presence of a secondary branch at the dorsal end that extended rostrally in the Eurasian badger and South American coati brain. The brown bear exhibited an additional sulcus in the frontal lobe, previously labelled as the proreal sulcus (see, e.g., Sienkiewicz et al., 2019); however, its shape closely resembled the secondary branches of the perisylvian sulcus seen in the South American coati and Eurasian badger. Sienkiewicz et al. (2019) also noted that this sulcus merges with the presylvian sulcus in their specimen, consistent with our findings in the left hemisphere of the brown bear and bilaterally in the Ussuri brown bear (see Supplementary Figure S3A, S5A). Given the known gyrencephaly of Ursidae brains with frequent secondary and tertiary sulci (Lyras et al., 2023), we propose that this sulcus represents a branch of the perisylvian sulcus.
Results and discussion, p. 23-24: Regarding individual variability in external brain morphology, previous work in primates and carnivorans has shown that differences across individuals typically affect sulcal shape, depth, or extent, but not the presence of major sulci. This has been reported in diverse contexts, including comparisons between captive and (semi-)wild macaque (Sallet et al., 2011; Testard et al., 2022), different dog breeds (Hecht et al., 2019), domestic cats (Kawamura, 1971b), or selectively bred foxes (Hecht et al., 2021). By including additional individuals for selected species, we extend these findings to a broader range of carnivorans. Notably, we observed no major sulcal differences between closely related species, even when specimens were acquired using different extraction and scanning protocols, for example, across felid clades or among wolf-like canids, further suggesting that substantial within-species variation is unlikely. While a full analysis of interindividual variability lies beyond the scope of this study, our findings support the reliability of the major sulcal patterns described.
Limitations and future directions, p. 25-26: Our findings represent a critical first step for linking brains within and across species for interspecies insights. The present analyses are based on multiple individuals pooled into families and genera, primarily focusing on single representatives per species. Additional individuals for selected species confirmed that intra-species variation is a matter of degree rather than a case of presence or absence of major sulci, but we do not provide an extensive account of the possible range of sulcal shape or other anatomical features.
Future studies will aim to systematically investigate interindividual variability in sulcal shape, depth, surface area, or thickness of the cortical ribbon surrounding the sulci, and will extend to more detailed investigations of the medial part of the cortex, as well as the subcortical structures and the cerebellum.The present framework and resulting database also provides the foundation to guide and facilitate future investigations of inter- and intra-species variation in regional brain size.
(2) Quantification of structure/function relationships
The second is in the quantification of structure/function relationships. We believe the cortical surfaces, detailed sulci descriptions, and atlases themselves are the main deliverables of this project. We felt it prudent to include some qualitative descriptions of the relationship between sulci as we observed them and behaviours as known from the literature, as a way to illustrate the possibilities that this foundational work opens up. This approach also allowed us to confirm and extend previous findings based on observations from a less diverse range of carnivoran species and families (Radinsky 1968 J Comp Neurol; Radinsky 1969, Ann N Y Acad Sci; Welker & Campos 1963 J Comp Neurol; Welker & Seidenstein, 1959 J Comp Neurol).
However, a full statistical framework for analysis is beyond the scope of this paper. Our group has previously worked on methods to quantitatively compare brain organization across species - indeed, we have developed a full framework for doing so (Mars et al., 2021, Annu Rev Neurosci), based on the idea that brains that differ in size and morphology should be compared based on anatomical features in a common feature space. Previously, we have used white matter anatomy (Mars et al., 2018, eLife) and spatial transcriptomics (Beauchamp et al., 2021, eLife). The present work presents the foundation for this approach to be expanded to sulcal anatomy, but the full development of it will be the topic of future communications.
Nevertheless, we now include a preliminary quantitative analysis of the relationship between the relative length of specific sulci and the two behavioural traits of interest. These analyses, which complement the qualitative observations in Figure 5, show that the relative length of the proreal sulcus was consistently greater in highly social, cooperatively hunting species, while no effect of forepaw dexterity was found (Supplementary Table S1). In contrast, both the cruciate and postcruciate sulci were significantly longer in species with high forepaw dexterity, but not related to sociality (Supplementary Tables S2–S3). These findings were consistent across reference sulci used to compute relative sulcal length and replicated in the left hemisphere (see Supplementary Figure S6).
We also would like to emphasize that we strongly believe that looking at measures of brain organization at a more detailed level than brain size or relative brain size is informative. Although studies correlating brain size with behavioural variables are prominent in the literature, they often struggle to distinguish between competing behavioural hypotheses (Healy, 2021, Adaptation and the Brain, OUP). In contrast, connectivity has a much more direct relationship to behavioural differences across species (Bryant et al., 2024, JoN), as does sulcal anatomy (Amiez et al., 2019, Nat Comms; Miller et al., 2021, Brain Behav Evol). Using our sulcal framework, we observed lineage-specific variations that would be overlooked by analyses focused solely on brain size. Moreover, such measures are less sensitive to the effects of fixation since that will affect brain size but not the presence or absence of a sulcus.
Main changes in the revised manuscript:
Results and discussion, p. 16-17: In the raccoon, red panda, coati, and ferret, considerably larger portions of the postcruciate gyrus S1 area appeared to be allocated to representing the forepaw and forelimbs (McLaughlin et al., 1998; Welker and Campos, 1963; Welker and Seidenstein, 1959) when compared to the domestic cat or dog (Dykes et al., 1980; Pinto Hamuy et al., 1956). This aligns with the observation that all species in the present sample with more complex or elongated postcruciate and cruciate sulci configurations display a preference for using their forepaws when manipulating their environment (see e.g., Iwaniuk et al., 1999; Iwaniuk and Whishaw, 1999; Radinsky, 1968; and Figure 5A). Complementary quantitative analyses further support this link, revealing a positive relationship between the relative length of the cruciate and postcruciate sulci and high forepaw dexterity (see Supplementary Figure S6, Tables S2-S3). This is suggestive of a potential link between sulcal morphology and a behavioural specialization in Arctoidea, consistent with earlier observations in otter species (Radinsky, 1968).
Results and discussion, p. 21: A distinct proreal sulcus was observed in the frontal lobe of the domestic dog, the African wild dog, wolf, dingo, and bush dog. This may indicate an expansion of frontal cortex in these animals compared to the other species in our sample (Figure 5-6). This aligns with findings from a comprehensive study comparing canid endocasts revealing an expanded proreal gyrus in these animals compared to the fennec fox, red fox and other species of the genus Vulpes (Lyras and Van Der Geer, 2003). The canids with a proreal sulcus also exhibit complex social structures compared to the primarily solitary living foxes (Nowak, 2005; Wilson and Mittermeier, 2009; Wilson, 2000, and see Figure 5).Despite living in social groups, the bat-eared fox, an insectivorous canid, does not possess a proreal sulcus. Its foraging behaviour is best described as spatially or communally coordinated rather than truly cooperative (Macdonald and Sillero-Zubiri, 2004), suggesting that the relationship between sulcal morphology and sociality may be specific to species engaging in active cooperative hunting. Supplementary quantitative analyses also confirm an increase in the relative length of the proreal sulcus
in cooperatively hunting species Moreover, a previous investigation of Canidae and Felidae brain evolution, using endocasts of extant and extinct species, also suggested a link between the emergence of pack structures and the proreal sulcus in Canidae (Radinsky, 1969). Despite being highly social and living in large social groups (i.e., mobs), meerkats appear to have a relatively small frontal lobe and no proreal sulcus compared to the social Canids (Figure 5), which would suggest that if the presence of a proreal sulcus correlates with complex social behaviour, this is canid-specific.
General discussion, p. 22-23: Our results revealed several interesting patterns of local variation in sulcal morphology between and within different lineages, and successfully replicate and expand upon prior observations based on more limited sets of species (Radinsky, 1969, 1968; Welker and Campos, 1963; Welker and Seidenstein, 1959). For example, Arctoidea showed relatively complex sulcal anatomy in the somatosensory cortex but low complexity in the occipito-temporal regions. In Canidae and Felidae, we found more complex occipito-temporal sulcal patterns indicative of changes in the amount of cortex devoted to visual and auditory processing in these regions. These observations may be linked to social or ecological factors, such as how the animals interact with objects or each other and their varied foraging strategies. Another example was the differential relative expansion of the neocortex surrounding the cruciate sulcus, which was particularly complex in Arctoidea species that are known to use their paws to manipulate their environment. Consistent with this observation, complementary quantitative analyses of both hemispheres revealed that species with high forepaw dexterity tended to have longer cruciate and postcruciate sulci. Although it has been argued that the cruciate sulcus appeared independently in different lineages and its exact relationship to the location of primary motor areas varies (Radinsky, 1971), our results provide a detailed exploration of the relationship between brain morphology and behavioural preferences across such a range of species.
Materials and Methods, p. 33: We focused on the major lateral and dorsal sulci of the carnivoran brain, but the medial wall and ventral view of the sulci are also described. For consistency, we started by labelling the right hemispheres on the mid-thickness surfaces; these are the hemispheres presented in the manuscript. An exception was made for the jungle cat, for which only the left hemisphere was available and is therefore shown. We aimed to facilitate interspecies comparisons and the exploration of previously undescribed carnivoran brains. To this end, we first created standardized criteria (henceforth referred to as recipes) for identifying each sulcus, drawing from existing literature on carnivoran neuroanatomy, particularly in paleoneurology (Lyras et al., 2023), and our own observations.In addition, we created digital sulcal masks for both hemispheres, which allowed us to test whether the same patterns were observable bilaterally and to further facilitate future research building on our framework. For the Egyptian mongoose, only the right hemisphere was available, and thus, a bilateral comparison was not possible for this species. Anatomical nomenclature primarily follows the recommendations of Czeibert et al (2018); if applicable, alternative names of sulci are provided once.
Materials and Methods, p. 34-35: We first briefly illustrated the gyri of the carnivoran brain with a focus on gyri that are not present in some species as a consequence of absent sulci to complement our observations. We then summarised the key differences and similarities in sulcal anatomy between species and related them to their ecology and behaviour. To complement this qualitative description, we conducted an initial quantitative analysis of sulcal length data from both hemispheres. To test whether sulcal length covaries with behavioural traits, we fit linear models predicting the relative length of the three target sulci (cruciate, postcruciate, proreal) as a function of forepaw dexterity (low vs.high) and sociality (solitary vs cooperative hunting). We measured the absolute length of each sulcus using the wb_command -border-length function from the Connectome Workbench toolkit (Marcus et al., 2011) applied to the manually defined sulcal masks (i.e., border files). Relative sulcal length was calculated by dividing the length of each target sulcus by that of a reference sulcus in the same hemisphere, reducing interspecies variation in brain or sulcal size. Reference sulci were required to be present in all species within a hemisphere and excluded if they were a target sulcus, part of the same functional system (e.g., somatosensory/motor), or anatomically atypical (e.g., the pseudosylvian fissure). This resulted in seven reference sulci for the proreal sulcus (ansate, coronal, marginal, presylvian, retrosplenial, splenial, suprasylvian) and four for the cruciate and postcruciate sulci (marginal, retrosplenial, splenial, suprasylvian). For each target-reference pair, we fit the following linear model: relative length ~ forepaw dexterity + sociality. Models were run separately for left and right hemispheres, with the left serving as a replication test. Associations were considered meaningful if the predictor reached statistical significance (p ≤ .05) in ≥ 75% of reference sulcus models per hemisphere. Additional individuals were not included in the analysis.
Data and code availability statement, p. 35-36: Generated surfaces of all species and T1-like contrast images of post-mortem samples obtained by the C Generated surfaces of all species and T1-like contrast images of post-mortem samples obtained by the Copenhagen Zoo and the Zoological Society of London (see Table 1) are available at the Digital Brain Zoo of the University of Oxford (Tendler et al., 2022) (https://open.win.ox.ac.uk/DigitalBrainBank/#/datasets/zoo). For all other species, except the domestic cat, the cortical surface reconstructions are available through the same resource. In-vivo data for the domestic cat is available upon request.
We created, extracted and analysed sulcal length data using the Connectome Workbench toolkit (Marcus et al., 2011), R 4.4.0 (R Core Team, 2023) and Python 3.9.7. Sulcal masks, along with the associated midthickness cortical surface reconstructions for all 32 animals, species-specific behavioural data, and the code used to extract sulcal lengths and perform the statistical analyses are available at:
https://git.fmrib.ox.ac.uk/neuroecologylab/carnivore-surfaces.
Reviewer #1 (Recommendations for the authors):
I was convinced by your model of labels in the temporal region and the nomenclature used, thanks to your argument concerning the primary auditory area in ferrets located in the gyrus called ectosylvian even though they have no ectosylvian sulcus. While this region raises questions, it seems to me that you make a good case for your labelling.
However, I don't understand your arguments in the occipital region regarding the ectomarginal sulcus. In the bear, for example, I don't understand why the caudal part of the marginal sulcus is not referred to as ectomarginal? You say that this sulci is specific to canids.
Whether in the paragraph describing the ectomarginal sulcus, the marginal sulcus, in the paragraphs on the gyri, or in the paragraph concerning the potential relationship to function, I don't see any argument to support your hypothesis. Especially as there is no information in the literature on the functions in this area of the bear brain as in that of the dog or other related species.
You just mention that in Canidae, the ectomarginal "runs between the suprasylvian and marginal sulcus", and I don't see why this is an argument.
Could you explain in more detail your choice of label and the specificity you claim to have in the canids of this region?
We have now expanded our rationale in the revised manuscript, particularly in the section describing the marginal sulcus, which directly follows the description of the ectomarginal sulcus. In brief, across our sample, including Ursidae and Canidae, we observed variation in whether the caudal marginal sulcus was detached or continuous, or extended further caudally vs ventrally, but no separate additional sulcus resembling the ectomarginal sulcus was seen in any species outside the canid family. We therefore reserve the label ectomarginal sulcus for the distinct structure consistently observed in Canidae and avoid applying it to the detached caudal marginal sulcus observed in Ursidae.
Main changes in the revised manuscript:
Results and discussion, p. 10-11: In several species, including the dingo, domestic cat, brown bear and South American coati and further supplementary individuals (Supplementary figure S3B), the caudal portion of the marginal sulcus was detached in one or both hemispheres, which is a frequently reported occurrence (England, 1973; Kawamura, 1971a; Kawamura and Naito, 1978). Potentially due to the similar caudal bend, some authors have labelled the (detached) caudal portion of the marginal sulcus in Ursidae as the ectomarginal sulcus (Lyras et al., 2023, but see e.g., Sienkiewicz et al., 2019);
The (detached) caudal marginal sulcus in Ursidae continues the course of the marginal sulcus caudally and/or ventrally and is topologically continuous with it. In contrast, the ectomarginal sulcus in Canidae is an entirely separate sulcus that runs between the suprasylvian and marginal sulci, forming a small, additional arch that is rarely connected to the marginal sulcus (Kawamura and Naito, 1978). This distinction is illustrated, for example, in the dingo and grey wolf. In the dingo, we observed both a detached caudal extension of the marginal sulcus and a distinct ectomarginal sulcus. In both grey wolf specimens, the marginal sulcus extended ventrally in a way that resembled the brown bear, but they also exhibited a clearly separate ectomarginal sulcus, confirming that the two features are not equivalent. In contrast, in the brown bear and Ussuri brown bear (Supplementary Figure S3B), we observed variation in whether the marginal sulcus was detached or continuous, but no separate sulcus resembling the ectomarginal sulcus seen in Canidae.
Reviewer #2 (Recommendations for the authors):
Although I indicated this already, I stress that the lack of quantification is problematic. In its current format, this is a classic descriptive study suitable for an anatomy journal, but even then, the conclusions are highly speculative. I would advise including some quantification of sulcal lengths or depths and surface areas or volumes of individual regions and relate all of those to overall brain size and potential clade differences. Figure 5 hints at some of these putative correlations, but is not an analysis. Some of these correlations are discussed in the manuscript, but without quantification, it is simply more descriptions and some speculative associations that largely parallel and corroborate findings from Radinsky's papers. In addition to quantification, the authors should consider a more fulsome explanation of the potential confounds and limitations of their data. As alluded to above, there are many sources of variation that were not sufficiently discussed but are critically important for interpreting any putative differences among and within clades.
We would like to reiterate that the primary aim of our study was to establish a comprehensive sulcal framework for carnivoran brains. The behavioural and ecological associations were secondary and exploratory, arising from a first application of this framework, and will require further investigation in future studies.
We already acknowledged in the initial version of the manuscript that many of our observations were consistent with those previously reported by Radinsky in more limited sets of species. However, we recognise that this point may not have come across clearly. We carefully revised our manuscript to further emphasise that our findings replicate and extend Radinsky’s work in a larger cross-species comparison, showing that our framework also successfully replicates and expands prior work.
As detailed in the public reviews, we did not measure overall or relative brain sizes. However, in the revised version of the manuscript, we have now quantified the relationship between sulcal length and its association with forepaw dexterity and sociality to complement the qualitative observations in Figure 5. Although preliminary, we believe that these analyses further showcase the strength of our sulcal framework and its potential for future investigations.
We also revised our discussion section to highlight the potential for future studies to build on our framework to systematically investigate interindividual variability in sulcal shape, depth, surface area, or thickness of the cortical ribbon surrounding the sulci. We also added that our framework and accompanying dataset can facilitate and guide future investigations into both inter- and intra-species variation in regional brain size.
Main changes in the revised manuscript:
General discussion, p. 22-23: Our results revealed several interesting patterns of local variation in sulcal morphology between and within different lineages, and successfully replicate and expand upon prior observations based on more limited sets of species (Radinsky, 1969, 1968; Welker and Campos, 1963; Welker and Seidenstein, 1959). For example, Arctoidea showed relatively complex sulcal anatomy in the somatosensory cortex but low complexity in the occipito-temporal regions. In Canidae and Felidae, we found more complex occipito-temporal sulcal patterns indicative of changes in the amount of cortex devoted to visual and auditory processing in these regions. These observations may be linked to social or ecological factors, such as how the animals interact with objects or each other and their varied foraging strategies. Another example was the differential relative expansion of the neocortex surrounding the cruciate sulcus, which was particularly complex in Arctoidea species that are known to use their paws to manipulate their environment. Consistent with this observation, complementary quantitative analyses of both hemispheres revealed that species with high forepaw dexterity tended to have longer cruciate and postcruciate sulci. Although it has been argued that the cruciate sulcus appeared independently in different lineages and its exact relationship to the location of primary motor areas varies (Radinsky, 1971), our results provide a detailed exploration of the relationship between brain morphology and behavioural preferences across such a range of species.
Limitations and future directions, p. 25-26: Our findings represent a critical first step for linking brains within and across species for interspecies insights. The present analyses are based on multiple individuals pooled into families and genera, primarily focusing on single representatives per species. Additional individuals for selected species confirmed that intra-species variation is a matter of degree rather than a case of presence or absence of major sulci, but we do not provide an extensive account of the possible range of sulcal shape or other anatomical features. Future studies will aim to systematically investigate interindividual variability in sulcal shape, depth, surface area, or thickness of the cortical ribbon surrounding the sulci, and will extend to more detailed investigations of the medial part of the cortex, as well as the subcortical structures and the cerebellum. The present framework and resulting database also provides the foundation to guide and facilitate future investigations of inter- and intra-species variation in regional brain size.
Another point that I did not see raised in the Discussion, but would be important and useful to include is that the authors are lacking specimens for several clades that could show additional differences in neocortical anatomy. For example, no hyaenids or viverrids were represented and an otter and badger are not necessarily representative of all mustelids, the majority of which are weasel-like. One could even argue that the meerkat is not necessarily representative of all herpestids given its behaviour and ecology. Of course, there are also pinnipeds, but they are divergent in many ways, and restricting the analyses to fissiped carnivorans is completely reasonable. Please note that I am not suggesting that the authors go back and try to procure even more species; rather they should emphasize that this is an incomplete survey of fissiped carnivorans.
The reviewer’s comments prompted us to further expand our carnivoran brain collection to include a broader range of species, representatives, and individual specimens. Notably, the collection now includes a hyaenid representative, the striped hyena. In addition to the otter and badger, we have added a weasel-like mustelid, the ferret, as well as the solitary Egyptian mongoose to complement the highly social meerkat within Herpestidae. Our felid dataset has also been expanded to include additional small and large wild cats, such as the sand cat and the Bengal tiger. As described above, these additions have led to the discovery of novel sulcal patterns, including the felid-specific diagonal sulcus.
We now also specify the fissiped families currently missing from the collection, which can be readily incorporated using our existing sulcal framework. The same applies to pinniped species, which we are currently investigating to support broader macro-level comparisons across the order.
Main changes in the revised manuscript:
General discussion, p. 23: Comparative neuroimaging requires balancing the level of anatomical detail with the breadth of species. The present sample represents the most comprehensive collection of fissiped carnivoran brains to date, encompassing a wide range of land-dwelling species from eight families. It includes diverse representatives, such as both social and solitary mongooses, weasel-like and non-weasel mustelids, and a broad array of canids, including wolf-like, fox-like, and more basal forms of canids. The framework and detailed protocols developed in this study are designed to facilitate navigation of additional fissiped species, such as Viverridae, Eupleridae, Mephitidae, Nandiniidae, and
Prionodontidae. Moreover, the approach can be readily extended to aquatic carnivorans, enabling broader macro-level comparisons across the order.
Apart from these broader issues, I also found some of the figures difficult to interpret in many instances. For example, the colour scheme used to highlight sulci is not colourblind friendly for Figures 2 and 3. It was also difficult for me to glean much information from Figure 6. I understand that functional regions of the cortex are shown for those species that were subject to electrophysiological studies in the past, but I could not work out how to transfer that data to the other brains. One suggestion for improving this would be to highlight putative cortical regions on the other brains in a lighter shade of the same colours.
We have carefully revised our figures to improve clarity and accessibility, particularly for individuals with colour vision deficiencies. Specifically, we have added numerical labels alongside the coloured sulci labels in Figures 2 and 3, as well as in all related supplementary figures (see examples on the following pages). For sulci that merge, such as the marginal, ansate, and coronal sulci, we have used colour combinations that are distinguishable across all major types of colour-blindness. Figure 4 has also been updated with a colour-blind-friendly palette and additional numerical labels for the gyri to further enhance interpretability.
Regarding Figure 6, we have updated the colour palette to ensure accessibility and have labelled all landmark sulci discussed in the main text using acronyms (e.g., the postcruciate sulcus as the boundary between S1 and M1). This is intended to facilitate the transfer of information between brains and guide orientation for readers less familiar with these structures. While we appreciate the suggestion to highlight putative cortical regions on other brains, we have opted not to do so. Our concern is that such visual cues, even when rendered in lighter shades, may be misinterpreted as established rather than hypothetical regional boundaries. We believe this more conservative approach appropriately reflects the current evidence base and avoids unintentionally overstating the certainty of functional homologies.
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public review):
Summary:
Recruitment of neutrophils to the lungs is known to drive susceptibility to infection with M. tuberculosis. In this study, the authors present data in support of the hypothesis that neutrophil production of the cytokine IL-17 underlies the detrimental effect of neutrophils on disease. They claim that neutrophils harbor a large fraction of Mtb during infection, and are a major source of IL-17. To explore the effects of blocking IL-17 signaling during primary infection, they use IL-17 blocking antibodies, SR221 (an inverse agonist of Th17 differentiation), and celecoxib, which they claim blocks Th17 differentiation, and observe modest improvements in bacterial burdens in both WT and IFN-γ deficient mice using the combination of IL-17 blockade with celecoxib during primary infection. Celecoxib enhances control of infection after BCG vaccination.
Thank you for the summary.
Strengths:
The most novel finding in the paper is that treatment with celecoxib significantly enhances control of infection in BCG-vaccinated mice that have been challenged with Mtb. It was already known that NSAID treatments can improve primary infection with Mtb.
Thank you.
Weaknesses:
The major claim of the manuscript - that neutrophils produce IL-17 that is detrimental to the host - is not strongly supported by the data. Data demonstrating neutrophil production of IL17 lacks rigor.
Our response: Neutrophil production of IL-17 is supported by two independent methods/ techniques in the current version:
(1) Through Flow cytometry- a large fraction of Ly6G<sup>+</sup>CD11b<sup>+</sup> cells from the lungs of Mtb-infected mice were also positive for IL-17 (Fig. 3C).
(2) IFA co-staining of Ly6G <SUP>+</SUP> cells with IL-17 in the lung sections from Mtb-infected mice (Fig. 3 E_G and Fig. 4H, Fig. 5I). For most of these IFA data, we provide quantified plots to show IL17<SUP>+</SUP>Ly6G<SUP>+</SUP> cells.
(3) Most importantly, conditions that inhibited IL-17 levels and controlled infection also showed a decline in IL-17 staining in Ly6G<SUP>+</SUP> cells.
Our efforts on IL-17 ELISPOT assay were not very successful and it needs further standardization.
Several independent publications support the production of IL-17 by neutrophils (Li et al. 2010; Katayama et al. 2013; Lin et al. 2011). For example, neutrophils have been identified as a source of IL-17 in human psoriatic lesions (Lin et al. 2011), in neuroinflammation induced by traumatic brain injury (Xu et al. 2023) and in several mouse models of infectious and autoimmune inflammation (Ferretti et al. 2003; Hoshino et al. 2008) (Li et al. 2010).
The experiments examining the effects of inhibitors of IL-17 on the outcome of infection are very difficult to interpret. First, treatment with IL-17 inhibitors alone has no impact on bacterial burdens in the lung, either in WT or IFN-γ KO mice. This suggests that IL-17 does not play a detrimental role during infection. Modest effects are observed using the combination of IL-17 blocking drugs and celecoxib, however, the interpretation of these results mechanistically is complicated. Celecoxib is not a specific inhibitor of Th17. Indeed, it affects levels of PGE2, which is known to have numerous impacts on Mtb infection separate from any effect on IL-17 production, as well as other eicosanoids.
The reviewer correctly says that Celecoxib is not a specific inhibitor of Th17. However, COX2 inhibition does have an effect on IL-17 levels, and numerous reports support this observation (Paulissen et al. 2013; Napolitani et al. 2009; Lemos et al. 2009).
(1) The detrimental role of IL-17 is obvious in the IFNγ KO experiment, where IL-17 neutralization led to a significant improvement in the lung pathology.
(2) In the highly susceptible IFNγ KO mice, IL-17 neutralization alone extended the survival of mice by ~10 days.
(3) IL-17 production independent of IL-23 is known to require PGE2 (Paulissen et al. 2013; Polese et al. 2021). In either WT or IFNγ KO mice, in contrast to IL-17 levels, we observed a decline in IL-23 levels. The PGE2 dependence of IL-17 production is obvious in the WT mice, where celecoxib abrogated IL-17 production.
(4) While deciding the impact of celecoxib or IL17 inhibition, looking at the cumulative readout of lung CFU, spleen CFU, Ly6G<sup>+</sup> cell recruitment, Ly6G<sup>+</sup> cell-resident Mtb pool and overall pathology, the effects are quite significant.
(5) Finally, in the revised manuscript, we provide additional results on the effect of SR2211 in BCG-vaccinated animals. It shows the direct impact of IL-17 inhibition on the BCG vaccine efficacy in WT mice.
Finally, the human data simply demonstrates that neutrophils and IL-17 both are higher in patients who experience relapse after treatment for TB, which is expected and does not support their specific hypothesis.
We disagree with the above statement. It also contradicts reviewers’ own assessments in one of the comments below, where a protective role of IL-17 is referred to. The literature lacks consensus in terms of a protective or pathological role of IL-17 in TB. Therefore, it was not expected to see higher IL-17 in patients who experienced relapse, death, or failed treatment outcomes. We do not have evidence from human subjects whether neutrophil-derived IL-17 has a similar pathological role as observed in mice. However, higher IL-17 in failed outcome cases confirm the central theme that IL-17 is pathological in both human and mouse models.
The use of genetic ablation of IL-17 production specifically in neutrophils and/or IL-17R in mice would greatly enhance the rigor of this study.
The reviewer’s point is well-taken. Having a genetic ablation of IL-17 production, specifically in the neutrophils, would be excellent. At present, however, we lack this resource. For the revised manuscript, we include the data with SR2211, a direct inhibitor of RORgt and, therefore, IL-17, in BCG-vaccinated mice.
The authors do not address the fact that numerous studies have shown that IL-17 has a protective effect in the mouse model of TB in the context of vaccination.
Yes, there are a few articles that talk about the protective effect of IL-17 in the mouse model of TB in the context of vaccination (Khader et al. 2007; Desel et al. 2011; Choi et al. 2020). This part was discussed in the original manuscript (in the Introduction section). For the revised manuscript, we also provide results from the experiment where we blocked IL-17 production by inhibiting RORgt using SR2211 in BCG-vaccinated mice. The results clearly show IL-17 as a negative regulator of BCG-mediated protective immunity. We believe some of the reasons for the observed differences could be 1) in our study, we analysed IL-17 levels in the lung homogenates at late phases of infection, and 2) most published studies rely on ex vivo stimulation of immune cells to measure cytokine production, whereas we actually measured the cytokine levels in the lung homogenates. We will elaborate on these points in the revised version.
Finally, whether and how many times each animal experiment was repeated is unclear.
We provide the details of the number of experiments in the revised version. Briefly, the BCG vaccination experiment (Figure 1) and BCG vaccination with Celecoxib treatment experiment (Figure 6) were performed twice and thrice, respectively. The IL-17 neutralization experiment (Figure 4) and the SR2211 treatment experiment (Figure 5) were done once. We will add another SR2211 experiment data in the revised version.
Reviewer #2 (Public review):
Summary:
In this study, Sharma et al. demonstrated that Ly6G+ granulocytes (Gra cells) serve as the primary reservoirs for intracellular Mtb in infected wild-type mice and that excessive infiltration of these cells is associated with severe bacteremia in genetically susceptible IFNγ/- mice. Notably, neutralizing IL-17 or inhibiting COX2 reversed the excessive infiltration of Ly6G+Gra cells, mitigated the associated pathology, and improved survival in these susceptible mice. Additionally, Ly6G+Gra cells were identified as a major source of IL-17 in both wild-type and IFNγ-/- mice. Inhibition of RORγt or COX2 further reduced the intracellular bacterial burden in Ly6G+Gra cells and improved lung pathology.
Of particular interest, COX2 inhibition in wild-type mice also enhanced the efficacy of the BCG vaccine by targeting the Ly6G+Gra-resident Mtb population.
Thank you for the summary.
Strengths:
The experimental results showing improved BCG-mediated protective immunity through targeting IL-17-producing Ly6G+ cells and COX2 are compelling and will likely generate significant interest in the field. Overall, this study presents important findings, suggesting that the IL-17-COX2 axis could be a critical target for designing innovative vaccination strategies for TB.
Thank you for highlighting the overall strengths of the study.
Weaknesses:
However, I have the following concerns regarding some of the conclusions drawn from the experiments, which require additional experimental evidence to support and strengthen the overall study.
Major Concerns:
(1) Ly6G+ Granulocytes as a Source of IL-17: The authors assert that Ly6G+ granulocytes are the major source of IL17 in wild-type and IFN-γ KO mice based on colocalization studies of Ly6G and IL-17. In Figure 3D, they report approximately 500 Ly6G+ cells expressing IL-17 in the Mtb-infected WT lung. Are these low numbers sufficient to drive inflammatory pathology? Additionally, have the authors evaluated these numbers in IFN-γ KO mice?
Thank you for pointing out the numbers in Fig. 3D It was our oversight to label the axis as No. of. For the observation that Ly6G<sup>+</sup> Gra are the major source of IL-17 in TB, we have used two separate strategies- a) IFA and b) FACS IL17<SUP>+</SUP> Ly6G<SUP>+</SUP> Gra/lung. For this data, only a part of the lung was used. For the revised manuscript, we provide the number of these cells at the whole lung level from Mtb-infected WT mice. Unfortunately, we did not evaluate these numbers in IFN-γ KO mice through FACS..
Our efforts to perform the IL-17 ELISpot assay on the sorted Ly6G<SUP>+</SUP>Gra from the lungs of Mtbinfected WT mice were unsuccessful. However, we provide a quantified representation of IFA of the tissue sections to stress upon the role of Ly6G<SUP>+</SUP> cells in IL-17 production in TB pathogenesis.
(2) Role of IL-17-Producing Ly6G Granulocytes in Pathology: The authors suggest that IL-17producing Ly6G granulocytes drive pathology in WT and IFN-γ KO mice. However, the data presented only demonstrate an association between IL-17<SUP>+</SUP> Ly6G cells and disease pathology. To strengthen their conclusion, the authors should deplete neutrophils in these mice to show that IL-17 expression, and consequently the pathology, is reduced.
Thank you for this suggestion. Neutrophil depletion studies in TB remain inconclusive. In some studies, neutrophil depletion helps the pathogen (Rankin et al. 2022; Pedrosa et al. 2000; Appelberg et al. 1995), and in others, it helps the host (Lovewell et al. 2021; Mishra et al. 2017). One reason for this variability is the stage of infection when neutrophil depletion was done. However, another crucial factor is the heterogeneity in the neutrophil population. There are reports that suggest neutrophil subtypes with protective versus pathological trajectories (Nwongbouwoh Muefong et al. 2022; Lyadova 2017; Hellebrekers, Vrisekoop, and Koenderman 2018; Leliefeld et al. 2018). Depleting the entire population using anti-Ly6G could impact this heterogeneity and may impact the inferences drawn.
A better approach would be to characterise this heterogeneous population, efforts towards which could be part of a separate study. Another direct approach could be Ly6G<SUP>+</SUP>-specific deletion of IL-17 function as part of a separate study.
For the revised manuscript, we provide results from the SR2211 experiment in BCG-vaccinated mice and other results to show the role of IL-17-producing Ly6G<SUP>+</SUP> Gra in TB pathology.
(3) IL-17 Secretion by Mtb-Infected Neutrophils: Do Mtb-infected neutrophils secrete IL-17 into the supernatants? This would serve as confirmation of neutrophil-derived IL-17. Additionally, are Ly6G<SUP>+</SUP> cells producing IL-17 and serving as pathogenic agents exclusively in vivo? The authors should provide comments on this.
Secretion of IL-17 by Mtb-infected neutrophils in vitro has been reported earlier (Hu et al. 2017). Our efforts to do a neutrophil IL-17 ELISPOT assay were not successful, and we are still standardising it. Whether there are a few neutrophil roles exclusively seen under in vivo conditions is an interesting proposition.
(4) Characterization of IL-17-Producing Ly6G+ Granulocytes: Are the IL-17-producing Ly6G+ granulocytes a mixed population of neutrophils and eosinophils, or are they exclusively neutrophils? Sorting these cells followed by Giemsa or eosin staining could clarify this.
This is a very important point. While usually eosinophils do not express Ly6G markers in laboratory mice, under specific contexts, including infections, eosinophils can express Ly6G. Since we have not characterized these potential Ly6G<SUP>+</SUP> sub-populations, that is one of the reasons we refer to the cell types as Ly6G<SUP>+</SUP> granulocytes, which do not exclude Ly6G<SUP>+</SUP> eosinophils. A detailed characterization of these subsets could be taken up as a separate study.
Reviewer #3 (Public review):
Summary:
The authors examine how distinct cellular environments differentially control Mtb following BCG vaccination. The key findings are that IL17-producing PMNs harbor a significant Mtb load in both wild-type and IFNg<sup>-/-</sup> mice. Targeting IL17 and Cox2 improved disease and enhanced BCG efficacy over 12 weeks and neutrophils/IL17 are associated with treatment failure in humans. The authors suggest that targeting these pathways, especially in MSMD patients may improve disease outcomes.
Thank you.
Strengths:
The experimental approach is generally sound and consists of low-dose aerosol infections with distinct readouts including cell sorting followed by CFU, histopathology, and RNA sequencing analysis. By combining genetic approaches and chemical/antibody treatments, the authors can probe these pathways effectively.
Understanding how distinct inflammatory pathways contribute to control or worsen Mtb disease is important and thus, the results will be of great interest to the Mtb field
Thank you.
Weaknesses:
A major limitation of the current study is overlooking the role of non-hematopoietic cells in the IFNg/IL17/neutrophil response. Chimera studies from Ernst and colleagues (Desvignes and Ernst 2009) previously described this IDO-dependent pathway following the loss of IFNg through an increased IL17 response. This study is not cited nor discussed even though it may alter the interpretation of several experiments.
Thank you for pointing out this earlier study, which we concede, we missed discussing. We disagree on the point that results from that study may alter the interpretation of several experiments in our study. On the contrary, the main observation that loss of IFNγ causes severe IL-17 levels is aligned in both studies.
IDO1 is known to alter T-helper cell differentiation towards Tregs and away from Th17 (Baban et al. 2009). It is absolutely feasible for the non-hematopoietic cells to regulate these events. However, that does not rule out the neutrophil production of IL-17 and the downstream pathological effect shown in this study. We have discussed and cited this study in the revised manuscript.
Several of the key findings in mice have previously been shown (albeit with less sophisticated experimentation) and human disease and neutrophils are well described - thus the real new finding is how intracellular Mtb in neutrophils are more refractory to BCG-mediated control. However, given there are already high levels of Mtb in PMNs compared to other cell types, and there is a decrease in intracellular Mtb in PMNs following BCG immunization the strength of this finding is a bit limited.
The reviewer’s interpretation of the BCG-refractory Mtb population in the neutrophil is interesting. The reviewer is right that neutrophils had a higher intracellular Mtb burden, which decreased in the BCG-vaccinated animals. Thus, on that account, the reviewer rightly mentions that BCG is able to control Mtb even in neutrophils. However, BCG almost clears intracellular burden from other cell types analysed, and therefore, the remnant pool of intracellular Mtb in the lungs of BCG-vaccinated animals could be mostly those present in the neutrophils. This is a substantial novel development in the field and attracts focus towards innate immune cells for vaccine efficacy.
References:
Appelberg, R., A. G. Castro, S. Gomes, J. Pedrosa, and M. T. Silva. 1995. 'SuscepBbility of beige mice to Mycobacterium avium: role of neutrophils', Infect Immun, 63: 3381-7.
Baban, B., P. R. Chandler, M. D. Sharma, J. Pihkala, P. A. Koni, D. H. Munn, and A. L. Mellor. 2009. 'IDO acBvates regulatory T cells and blocks their conversion into Th17-like T cells', J Immunol, 183: 2475-83.
Choi, H. G., K. W. Kwon, S. Choi, Y. W. Back, H. S. Park, S. M. Kang, E. Choi, S. J. Shin, and H. J. Kim. 2020. 'AnBgen-Specific IFN-gamma/IL-17-Co-Producing CD4(+) T-Cells Are the Determinants for ProtecBve Efficacy of Tuberculosis Subunit Vaccine', Vaccines (Basel), 8.
Cruz, A., A. G. Fraga, J. J. Fountain, J. Rangel-Moreno, E. Torrado, M. Saraiva, D. R. Pereira, T. D. Randall, J. Pedrosa, A. M. Cooper, and A. G. Castro. 2010. 'Pathological role of interleukin 17 in mice subjected to repeated BCG vaccinaBon afer infecBon with Mycobacterium tuberculosis', J Exp Med, 207: 1609-16.
Desel, C., A. Dorhoi, S. Bandermann, L. Grode, B. Eisele, and S. H. Kaufmann. 2011. 'Recombinant BCG DeltaureC hly+ induces superior protecBon over parental BCG by sBmulaBng a balanced combinaBon of type 1 and type 17 cytokine responses', J Infect Dis, 204: 1573-84.
Desvignes, L., and J. D. Ernst. 2009. 'Interferon-gamma-responsive nonhematopoieBc cells regulate the immune response to Mycobacterium tuberculosis', Immunity, 31: 974-85.
Ferreg, S., O. Bonneau, G. R. Dubois, C. E. Jones, and A. Trifilieff. 2003. 'IL-17, produced by lymphocytes and neutrophils, is necessary for lipopolysaccharide-induced airway neutrophilia: IL-15 as a possible trigger', J Immunol, 170: 2106-12.
Hellebrekers, P., N. Vrisekoop, and L. Koenderman. 2018. 'Neutrophil phenotypes in health and disease', Eur J Clin Invest, 48 Suppl 2: e12943.
Hoshino, A., T. Nagao, N. Nagi-Miura, N. Ohno, M. Yasuhara, K. Yamamoto, T. Nakayama, and K. Suzuki. 2008. 'MPO-ANCA induces IL-17 producBon by acBvated neutrophils in vitro via classical complement pathway-dependent manner', J Autoimmun, 31: 79-89.
Hu, S., W. He, X. Du, J. Yang, Q. Wen, X. P. Zhong, and L. Ma. 2017. 'IL-17 ProducBon of Neutrophils Enhances AnBbacteria Ability but Promotes ArthriBs Development During Mycobacterium tuberculosis InfecBon', EBioMedicine, 23: 88-99.
Hult, C., J. T. Magla, H. P. Gideon, J. J. Linderman, and D. E. Kirschner. 2021. 'Neutrophil Dynamics Affect Mycobacterium tuberculosis Granuloma Outcomes and DisseminaBon', Front Immunol, 12: 712457.
Katayama, M., K. Ohmura, N. Yukawa, C. Terao, M. Hashimoto, H. Yoshifuji, D. Kawabata, T. Fujii, Y. Iwakura, and T. Mimori. 2013. 'Neutrophils are essenBal as a source of IL-17 in the effector phase of arthriBs', PLoS One, 8: e62231.
Khader, S. A., G. K. Bell, J. E. Pearl, J. J. Fountain, J. Rangel-Moreno, G. E. Cilley, F. Shen, S. M. Eaton, S. L. Gaffen, S. L. Swain, R. M. Locksley, L. Haynes, T. D. Randall, and A. M. Cooper. 2007. 'IL-23 and IL-17 in the establishment of protecBve pulmonary CD4+ T cell responses afer vaccinaBon and during Mycobacterium tuberculosis challenge', Nat Immunol, 8: 369-77.
Leliefeld, P. H. C., J. Pillay, N. Vrisekoop, M. Heeres, T. Tak, M. Kox, S. H. M. Rooijakkers, T. W. Kuijpers, P. Pickkers, L. P. H. Leenen, and L. Koenderman. 2018. 'DifferenBal anBbacterial control by neutrophil subsets', Blood Adv, 2: 1344-55.
Lemos, H. P., R. Grespan, S. M. Vieira, T. M. Cunha, W. A. Verri, Jr., K. S. Fernandes, F. O. Souto, I. B. McInnes, S. H. Ferreira, F. Y. Liew, and F. Q. Cunha. 2009. 'Prostaglandin mediates IL-23/IL-17induced neutrophil migraBon in inflammaBon by inhibiBng IL-12 and IFNgamma producBon', Proc Natl Acad Sci U S A, 106: 5954-9.
Li, L., L. Huang, A. L. Vergis, H. Ye, A. Bajwa, V. Narayan, R. M. Strieter, D. L. Rosin, and M. D. Okusa. 2010. 'IL-17 produced by neutrophils regulates IFN-gamma-mediated neutrophil migraBon in mouse kidney ischemia-reperfusion injury', J Clin Invest, 120: 331-42.
Lin, A. M., C. J. Rubin, R. Khandpur, J. Y. Wang, M. Riblen, S. Yalavarthi, E. C. Villanueva, P. Shah, M. J. Kaplan, and A. T. Bruce. 2011. 'Mast cells and neutrophils release IL-17 through extracellular trap formaBon in psoriasis', J Immunol, 187: 490-500.
Lovewell, R. R., C. E. Baer, B. B. Mishra, C. M. Smith, and C. M. Sasseg. 2021. 'Granulocytes act as a niche for Mycobacterium tuberculosis growth', Mucosal Immunol, 14: 229-41.
Lyadova, I. V. 2017. 'Neutrophils in Tuberculosis: Heterogeneity Shapes the Way?', Mediators Inflamm, 2017: 8619307.
Mishra, B. B., R. R. Lovewell, A. J. Olive, G. Zhang, W. Wang, E. Eugenin, C. M. Smith, J. Y. Phuah, J. E. Long, M. L. Dubuke, S. G. Palace, J. D. Goguen, R. E. Baker, S. Nambi, R. Mishra, M. G. Booty, C. E. Baer, S. A. Shaffer, V. Dartois, B. A. McCormick, X. Chen, and C. M. Sasseg. 2017. 'Nitric oxide prevents a pathogen-permissive granulocyBc inflammaBon during tuberculosis', Nat Microbiol, 2: 17072.
Napolitani, G., E. V. Acosta-Rodriguez, A. Lanzavecchia, and F. Sallusto. 2009. 'Prostaglandin E2 enhances Th17 responses via modulaBon of IL-17 and IFN-gamma producBon by memory CD4+ T cells', Eur J Immunol, 39: 1301-12.
Nwongbouwoh Muefong, C., O. Owolabi, S. Donkor, S. Charalambous, A. Bakuli, A. Rachow, C. Geldmacher, and J. S. Sutherland. 2022. 'Neutrophils Contribute to Severity of Tuberculosis
Pathology and Recovery From Lung Damage Pre- and Posnreatment', Clin Infect Dis, 74: 175766.
Paulissen, S. M., J. P. van Hamburg, N. Davelaar, P. S. Asmawidjaja, J. M. Hazes, and E. Lubberts. 2013. 'Synovial fibroblasts directly induce Th17 pathogenicity via the cyclooxygenase/prostaglandin E2 pathway, independent of IL-23', J Immunol, 191: 1364-72.
Pedrosa, J., B. M. Saunders, R. Appelberg, I. M. Orme, M. T. Silva, and A. M. Cooper. 2000. 'Neutrophils play a protecBve nonphagocyBc role in systemic Mycobacterium tuberculosis infecBon of mice', Infect Immun, 68: 577-83.
Polese, B., B. Thurairajah, H. Zhang, C. L. Soo, C. A. McMahon, G. Fontes, S. N. A. Hussain, V. Abadie, and I. L. King. 2021. 'Prostaglandin E(2) amplifies IL-17 producBon by gammadelta T cells during barrier inflammaBon', Cell Rep, 36: 109456.
Rankin, A. N., S. V. Hendrix, S. K. Naik, and C. L. Stallings. 2022. 'Exploring the Role of Low-Density Neutrophils During Mycobacterium tuberculosis InfecBon', Front Cell Infect Microbiol, 12: 901590.
Xu, X. J., Q. Q. Ge, M. S. Yang, Y. Zhuang, B. Zhang, J. Q. Dong, F. Niu, H. Li, and B. Y. Liu. 2023. 'Neutrophil-derived interleukin-17A parBcipates in neuroinflammaBon induced by traumaBc brain injury', Neural Regen Res, 18: 1046-51.
Reviewer #1 (Recommendations for the authors):
All figures: Clear information about the number of repeat experiments for each figure must be included.
We have provided the details of the number of repeat experiments in the revised version.
Figure 1: The claim that neutrophils are a dominant cell type infected during Mtb infection of the lungs is undermined by the limited number of markers used to identify cell types. The gating strategy used to initially identify what cells are infected with Mtb divided cells into three categories; granulocytes (Ly6G<SUP>+</SUP> Cd11b<SUP>+</SUP>), CD64+MerTK+ macrophages, or Sca1+CD90.1+CD73+ (mesenchymal stem cells). This strategy leaves out monocyte populations that have been shown to be the dominant infected cells in other strategies (most recently, PMID: 36711606).
Thank you for this important point. We agree that we did not assess the infected monocyte population, specifically the Cd11c<SUP>+</SUP> population. Both CD11c<SUP>Hi</SUP> and CD11c<SUP>Lo</SUP> monocyte cells appear to be important for Mtb infection, in different studies (Lee et al., 2020), (Zheng et al., 2024). Therefore, leaving out the CD11c<SUP>+</SUP> population in our assays was a conscious decision to ensure the clarity of the cell types being studied.
In addition, substantial evidence from multiple studies indicates that Ly6G⁺ granulocytes constitute the predominant infected population in the Mtb-infected lungs of both mice and humans (Lovewell et al., 2021) (Eum et al., 2010). While monocytes may contribute to Mtb infection dynamics, our findings align with a growing body of research emphasizing the significant role of neutrophils as a dominant infected cell type in the lungs during TB pathology.
Figure 1: Putting the data from separate panels together, it appears that very few bacteria are isolated from the three cell types in the lung, suggesting there may be some loss in the preparation steps. Why is the total sorted CFU from neutrophils, macrophages, and MSCs so low, <400 bacteria total, when the absolute CFU is so high? Is it because only a fraction of the lung is being sorted/plated?
Yes, only a fraction of the lung was used for cell sorting and subsequent plating. The CFU plating from sorted cells also does not account for any bacteria growing extracellularly.
Figure 3C: It is difficult to ascertain whether the gating on IL-17<SUP>+</SUP> cells is accurately identifying IL-17 producing cells. It is surprising, based on other published work, that the authors claim that almost half of CD45+CD11b-Ly6G- cells produce IL-17 in WT mice. It would be informative to show cell type-specific production of IL-17 in both WT and IFN-γ KO mice for comparison with the literature. Unstained/isotype controls for IL-17 staining should be shown. With this in mind, it is difficult to interpret the authors' claim that 80% of neutrophils produce IL-17.
Thank you for the points above. We do agree that we were surprised to see ~50% of CD45<SUP>+</SUP> CD11b<SUP>-</SUP>Ly6G<SUP>-</SUP> cells producing IL-17. We have now done multiple experiments to confirm that this number is actually less than 1% (~90 cells) in the uninfected mice and less than 4% (~4000) in the Mtb-infected mice.
Neutrophil-derived IL-17 production in Mtb-infected lungs is supported by two independent techniques in our current study: Flow Cytometry and Immunofluorescence assay. While Neutrophil production of IL-17 is rarely studied in the context of TB, in several other settings it has been widely reported (Gonzalez-Orozco et al., 2019; Li et al., 2010; Ramirez-Velazquez et al., 2013). We consistently get >60% IL-17 positive cells in the CD11b<SUP>+</SUP> Ly6G<SUP>+</SUP> population, specifically in the infected samples.
To specifically address the reviewer’s concerns, we have now used an isotype control for IL17 staining and show the specificity of IL-17A antibody binding. The Author response image 1 is from the uninfected mice, 8 weeks age.
Unfortunately, our efforts to establish an IL-17 ELISPOT assay from neutrophils were not very successful and need further standardisation. The new results are included in Fig. 3C-D and Fig. S2F-G in the revised manuscript.
Author response image 1.
Figure 3 D-H. Quantification of immunofluorescence microscopy should be provided.
In the revised manuscript, we provide the quantification of IFA results.
Figure 4: Effects on neutrophil numbers in IFN-γ Kos do not correlate with CFU reductions, suggesting there may be a neutrophilindependent mechanism.
In the IFN-γ KO, we agree that the effect was less than dramatic. The immune dysfunction in the IFN-γ KO mice is too severe to see a strong reversal in the phenotype through interventions.
While we do not rule out any neutrophil-independent mechanism, in the context of following observations, neutrophil-dependent mechanisms certainly appear to play an important role-
(a) Improved pathology and survival upon IL-17 neutralization, which further improves with the inclusion of celecoxib.
(b) Loss of IL17<sup>+</sup>-Ly6G<sup>+</sup> cells upon IL-17 neutralization, which is further exacerbated when combined with celecoxib.
(c) Significant reduction in PMN number (shown by FACS) without any major impact on Th17 cell population upon IL-17 neutralization.
Finally, we believe some of the observations may become stronger once we characterize the specific sub-population among the Ly6G+ cells that correlates with pathology. For example, as shown in Figure 4I, FACS analysis of the Ly6G<sup>⁺</sup> cell population in Mtb-infected IFNγ<sup>⁻/⁻</sup> mice revealed a substantial subset of CD11b<sup>mid</sup> Ly6G<sup>ʰⁱ</sup> cells, indicative of an immature neutrophil population (Scapini et al., 2016). Efforts are currently underway to identify these important subpopulations.
Figure 4: Differences observed in the spleen cannot be connected to dissemination per se but instead could be a result of enhanced immune control in the spleen.
Thank you for this important point. We have revised this section. The role of neutrophils in Mtb dissemination is an emerging area of research, with growing evidence suggesting that these cells contribute to the spread of Mtb beyond the lungs (Hult et al., 2021). We highlight that the observed correlation could be speculative at this juncture.
Figure 4, 5: IL-17 neutralization alone has no effect on CFU in the lungs of Mtb-infected mice. While the combination of IL-17 neutralization and celecoxib has a very modest effect on CFU, the mechanism behind this observation is unclear. Further, the experiment shown has only 3 mice per group and it is unclear whether this (or any other) mouse experiment was repeated.
For Fig. 4, the experiment was done with 3 mice/group. The IFN KO mice were used to help identify the mechanism. IL-17 neutralisation or Celecoxib treatment alone did not have any significant effect on the bacterial burden (in lungs or isolated PMNs). However, it did show a significant effect on the number of PMNs recruited. Combination of IL-17 neutralisation and celecoxib led to about a one-log decrease in CFU, which is significant.
For Fig. 5, we used SR2211 instead of anti-IL-17 Ab for the experiment. This experiment had WT mice and 5 animals/group. Here, celecoxib and SR2211 alone showed a significant decline in PMN-resident Mtb pool as well as spleen burden. Only in the lungs, the impact of SR2211 alone was not significant.
Figure 6: The decreases in CFU correlate with a decrease in neutrophils; nothing connects this to neutrophil production of IL-17.
We now show quantification of observation in Fig. 5I, where in the WT mice, treatment with Celecoxib reduces the frequency of IL-17-producing Ly6G+ cells. In the revised manuscript, we also show direct evidence of SR2211 activity on BCG vaccine efficacy, which causes a significant decline in the Mtb burden in whole lung or in the isolated PMNs.
Figure 7. The Human data shows that elevated neutrophil levels and elevated IL-17 levels are associated with treatment failure in TB patients. This is expected, and does not
The literature lacks consensus in terms of a protective or pathological role of IL-17 in TB. Therefore, it was not expected to see higher IL-17 in patients who experienced relapse, death, or failed treatment outcomes. We do not have evidence from human subjects whether neutrophil derived IL-17 has a similar pathological role as observed in mice. However, higher IL-17 in failed outcome cases confirm the central theme that IL-17 is pathological in both human and mouse models.
Reviewer #2 (Recommendations for the authors):
(1) Survival of IFN-γ-/- Mice: The survival of IFN-γ-/- mice up to 100 days following a challenge with ~100 CFU of H37Rv is quite unusual. Have the authors checked PDIM expression in their Mtb strain, given that several studies report earlier mortality in these mice?
As shown in Fig. 4F, H37Rv-infected IFN-γ⁻/⁻ mice survived up to a little over 80 days. These figures are not unusual in the light of the following:
(1) In one study, IFNγ⁻/⁻ survived for about 40 days when the hypervirulent Mtb strain was used to infect these mice at 100-200 CFU using nose-only aerosol exposure (Nandi and Behar, 2011)
(2) In yet another study, IFNγ⁻/⁻ mice survived for ~50 days, however, they used H37Rv at 1-3x10<sup>5</sup> CFU to infect through intravenous injection (Kawakami et al., 2004)
Thus, compared with the above observations, where IFN-γ<sup>-/-</sup> mice survived for maximum 50 days due to hypervirulent infection or a very high dose infection, infection with H37Rv at ~100 CFU through the aerosol route and surviving for ~80 days is not unusual. The H37Rv cultures used in our study are always animal-passaged to ensure PDIM integrity.
(2) Granuloma Scoring: The granuloma scores appear to represent the percentage of lesion area. Please clarify and, if necessary, amend this in the manuscript.
The granuloma score is based on the calculation of the number of granulomatous infiltration and their severity. These are not % lesion area. We have added this detail in the revised manuscript.
(3) Pathology Comparison in Figures 4F and 4G: Does the pathology shown in Figure 4G correspond to the same groups as in Figure 4F? The celecoxib group in Figure 4F and the WT group in Figure 4G seem to be missing. Please clarify.
Figures 4F and 4G depict two independent experiments. For the time-to-death experiment, we had to leave the animals. The rest of the panels in Fig. 4 represent animals from the same experiment.
(4) Effect of Celecoxib on Ly6G+ Cells: The authors demonstrated that celecoxib treatment reduces Ly6G+ cells and IL-17-producing Ly6G+ cells. Do Ly6G+ cells express EP2/EP4 receptors? Alternatively, could the reduction in IL-17-producing Ly6G+ cells be due to an improved bactericidal response in other innate cells? The authors should discuss this possibility.
Yes, Ly6G<sup>⁺</sup> granulocytes express EP2/EP4 receptors (Lavoie et al., 2024), which mediate PGE₂ signaling. Prostaglandin E<sub>₂</sub> (PGE<sub>₂</sub>) is known to regulate neutrophil function and can enhance IL-17 production in various immune cells (Napolitani et al., 2009). However, the expression and functional role of EP2/EP4 receptors specifically on Ly6G<sup>⁺</sup> granulocytes in the context of Mtb infection require further investigation.
The alternate suggestion by the reviewer that the reduction in IL-17-producing Ly6G<sup>⁺</sup> cells following celecoxib treatment could be attributed to an improved bactericidal response in other innate immune cells is attractive. While we did not experimentally rule out this possibility, since reduced IL-17 invariably associated with reduced neutrophil-resident Mtb population, a cell-autonomous mechanism operational in Ly6G+ granulocytes is a highly likely mechanism.
(5) Culture Conditions: The methods section indicates that bacteria were cultured in 7H9+ADC. Is there a specific reason why the Oleic acid supplement was not added, given that standard Mtb culture conditions typically use 7H9+OADC supplements? Please comment on this choice.
It is a standard microbiological experimental procedure to use 7H9+ADC for broth culture, while 7H11+OADC for solid culture. Compared to broth culture, solid media are usually more stressful for bacteria because of hypoxia inside the growing colonies. Therefore, the media used are enriched in casein hydrolysate (like 7H11) and oleic acid (OADC).
Reviewer #3 (Recommendations for the authors):
Major suggestion: To really determine the role of neutrophil IL17 will require depletion studies and chimera experiments. These are clearly a major undertaking. I believe making significant re-writes to alter the conclusions or reanalyze any data to determine the role of nonhematopoietic and hematopoietic cells in IL17 is needed. If the conclusions are left as is, further experimentation is needed to fully support those conclusions.
Thank you for the suggestion. We have embarked on the specific deletion studies; however, as mentioned, this is a major undertaking and will take time. As suggested, we have discussed the results in accordance with the strength of evidence currently provided.
Eum, S.Y., J.H. Kong, M.S. Hong, Y.J. Lee, J.H. Kim, S.H. Hwang, S.N. Cho, L.E. Via, and C.E. Barry, 3rd. 2010. Neutrophils are the predominant infected phagocyGc cells in the airways of paGents with acGve pulmonary TB. Chest 137:122-128.
Gonzalez-Orozco, M., R.E. Barbosa-Cobos, P. Santana-Sanchez, L. Becerril-Mendoza, L. Limon-
Camacho, A.I. Juarez-Estrada, G.E. Lugo-Zamudio, J. Moreno-Rodriguez, and V. OrGzNavarrete. 2019. Endogenous sGmulaGon is responsible for the high frequency of IL-17Aproducing neutrophils in paGents with rheumatoid arthriGs. Allergy Asthma Clin Immunol 15:44.
References
Hult, C., J.T. Ma[la, H.P. Gideon, J.J. Linderman, and D.E. Kirschner. 2021. Neutrophil Dynamics Affect Mycobacterium tuberculosis Granuloma Outcomes and DisseminaGon. Front Immunol 12:712457.
Kawakami, K., Y. Kinjo, K. Uezu, K. Miyagi, T. Kinjo, S. Yara, Y. Koguchi, A. Miyazato, K. Shibuya, Y. Iwakura, K. Takeda, S. Akira, and A. Saito. 2004. Interferon-gamma producGon and host protecGve response against Mycobacterium tuberculosis in mice lacking both IL-12p40 and IL-18. Microbes Infect 6:339-349.
Lavoie, J.C., M. Simard, H. Kalkan, V. Rakotoarivelo, S. Huot, V. Di Marzo, A. Cote, M. Pouliot, and N. Flamand. 2024. Pharmacological evidence that the inhibitory effects of prostaglandin E2 are mediated by the EP2 and EP4 receptors in human neutrophils. J Leukoc Biol 115:1183-1189.
Lee, J., S. Boyce, J. Powers, C. Baer, C.M. Sasse[, and S.M. Behar. 2020. CD11cHi monocyte-derived macrophages are a major cellular compartment infected by Mycobacterium tuberculosis. PLoS Pathog 16:e1008621.
Li, L., L. Huang, A.L. Vergis, H. Ye, A. Bajwa, V. Narayan, R.M. Strieter, D.L. Rosin, and M.D. Okusa. 2010. IL-17 produced by neutrophils regulates IFN-gamma-mediated neutrophil migraGon in mouse kidney ischemia-reperfusion injury. J Clin Invest 120:331-342.
Lovewell, R.R., C.E. Baer, B.B. Mishra, C.M. Smith, and C.M. Sasse[. 2021. Granulocytes act as a niche for Mycobacterium tuberculosis growth. Mucosal Immunol 14:229-241.
Nandi, B., and S.M. Behar. 2011. RegulaGon of neutrophils by interferon-gamma limits lung inflammaGon during tuberculosis infecGon. The Journal of experimental medicine 208:22512262.
Napolitani, G., E.V. Acosta-Rodriguez, A. Lanzavecchia, and F. Sallusto. 2009. Prostaglandin E2 enhances Th17 responses via modulaGon of IL-17 and IFN-gamma producGon by memory CD4+ T cells. Eur J Immunol 39:1301-1312.
Ramirez-Velazquez, C., E.C. CasGllo, L. Guido-Bayardo, and V. OrGz-Navarrete. 2013. IL-17-producing peripheral blood CD177+ neutrophils increase in allergic asthmaGc subjects. Allergy Asthma Clin Immunol 9:23.
Sadikot, R.T., H. Zeng, A.C. Azim, M. Joo, S.K. Dey, R.M. Breyer, R.S. Peebles, T.S. Blackwell, and J.W. Christman. 2007. Bacterial clearance of Pseudomonas aeruginosa is enhanced by the inhibiGon of COX-2. Eur J Immunol 37:1001-1009.
Zheng, W., I.C. Chang, J. Limberis, J.M. Budzik, B.S. Zha, Z. Howard, L. Chen, and J.D. Ernst. 2023. Mycobacterium tuberculosis resides in lysosome-poor monocyte-derived lung cells during chronic infecGon. bioRxiv
Zheng, W., I.C. Chang, J. Limberis, J.M. Budzik, B.S. Zha, Z. Howard, L. Chen, and J.D. Ernst. 2024. Mycobacterium tuberculosis resides in lysosome-poor monocyte-derived lung cells during chronic infecGon. PLoS Pathog 20:e1012205.
microscopios y láminas
Se refiere a los microscopios o a las láminas
¿Qué elementos de Anatomía encuentras MÁS DIFÍCILES?
Anatomía? Sugiero que aquí vayan los nombres de las 6 unidades de aprendizaje y que vaya una escala de lickert para determinar la complejidad.
Estimado/a estudiante:
Podría ir en un solo renglón. Esta idea y en las siguientes.
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public review):
Summary:
Syed et al. investigate the circuit underpinnings for leg grooming in the fruit fly. They identify two populations of local interneurons in the right front leg neuromere of ventral nerve cord, i.e. 62 13A neurons and 64 13B neurons. Hierarchical clustering analysis identifies 10 morphological classes for both populations. Connectome analysis reveals their circuit interactions: these GABAergic interneurons provide synaptic inhibition either between the two subpopulations, i.e., 13B onto 13A, or among each other, i.e., 13As onto other 13As, and/or onto leg motoneurons, i.e., 13As and 13Bs onto leg motoneurons. Interestingly, 13A interneurons fall into two categories, with one providing inhibition onto a broad group of motoneurons, being called "generalists", while others project to a few motoneurons only, being called "specialists". Optogenetic activation and silencing of both subsets strongly affect leg grooming. As well aas ctivating or silencing subpopulations, i.e., 3 to 6 elements of the 13A and 13B groups, has marked effects on leg grooming, including frequency and joint positions, and even interrupting leg grooming. The authors present a computational model with the four circuit motifs found, i.e., feed-forward inhibition, disinhibition, reciprocal inhibition, and redundant inhibition. This model can reproduce relevant aspects of the grooming behavior.
Strengths:
The authors succeeded in providing evidence for neural circuits interacting by means of synaptic inhibition to play an important role in the generation of a fast rhythmic insect motor behavior, i.e., grooming. Two populations of local interneurons in the fruit fly VNC comprise four inhibitory circuit motifs of neural action and interaction: feed-forward inhibition, disinhibition, reciprocal inhibition, and redundant inhibition. Connectome analysis identifies the similarities and differences between individual members of the two interneuron populations. Modulating the activity of small subsets of these interneuron populations markedly affects the generation of the motor behavior, thereby exemplifying their important role in generating grooming.
We thank the reviewer for their thoughtful and constructive evaluation of our work.
Weaknesses:
Effects of modulating activity in the interneuron populations by means of optogenetics were conducted in the so-called closed-loop condition. This does not allow for differentiation between direct and secondary effects of the experimental modification in neural activity, as feedforward and feedback effects cannot be disentangled. To do so, open loop experiments, e.g., in deafferented conditions, would be important. Given that many members of the two populations of interneurons do not show one, but two or more circuit motifs, it remains to be disentangled which role the individual circuit motif plays in the generation of the motor behavior in intact animals.
Our optogenetic experiments show a role for 13A/B neurons in grooming leg movements – in an intact sensorimotor system - but we cannot yet differentiate between central and reafferent contributions. Activation of 13As or 13Bs disinhibits motor neurons and that is sufficient to induce walking/grooming. Therefore, we can show a role for the disinhibition motif.
Proprioceptive feedback from leg movements could certainly affect the function of these reciprocal inhibition circuits. Given the synapses we observe between leg proprioceptors and 13A neurons, we think this is likely.
Our previous work (Ravbar et al 2021) showed that grooming rhythms in dusted flies persist when sensory feedback is reduced, indicating that central control is possible. In those experiments, we used dust to stimulate grooming and optogenetic manipulation to broadly silence sensory feedback. We cannot do the same here because we do not yet have reagents to separately activate sparse subsets of inhibitory neurons while silencing specific proprioceptive neurons. More importantly, globally silencing proprioceptors would produce pleiotropic effects and severely impair baseline coordination, making it difficult to distinguish whether observed changes reflect disrupted rhythm generation or secondary consequences of impaired sensory input. Therefore, the reviewer is correct – we do not know whether the effects we observe are feedforward (central), feedback sensory, or both. We have included this in the revised results and discussion section to describe these possibilities and the limits of our current findings.
Additionally, we have used a computational model to test the role of each motif separately and we show that in the results.
Reviewer #2 (Public review):
Summary:
This manuscript by Syed et al. presents a detailed investigation of inhibitory interneurons, specifically from the 13A and 13B hemilineages, which contribute to the generation of rhythmic leg movements underlying grooming behavior in Drosophila. After performing a detailed connectomic analysis, which offers novel insights into the organization of premotor inhibitory circuits, the authors build on this anatomical framework by performing optogenetic perturbation experiments to functionally test predictions derived from the connectome. Finally, they integrate these findings into a computational model that links anatomical connectivity with behavior, offering a systems-level view of how inhibitory circuits may contribute to grooming pattern generation.
Strengths:
(1) Performing an extensive and detailed connectomic analysis, which offers novel insights into the organization of premotor inhibitory circuits.
(2) Making sense of the largely uncharacterized 13A/13B nerve cord circuitry by combining connectomics and optogenetics is very impressive and will lay the foundation for future experiments in this field.
(3) Testing the predictions from experiments using a simplified and elegant model.
We thank the reviewer for their thoughtful and encouraging evaluation of our work.
Weaknesses:
(1) In Figure 4, while the authors report statistically significant shifts in both proximal inter-leg distance and movement frequency across conditions, the distributions largely overlap, and only in Panel K (13B silencing) is there a noticeable deviation from the expected 7-8 Hz grooming frequency. Could the authors clarify whether these changes truly reflect disruption of the grooming rhythm?
We reanalyzed the dataset with Linear Mixed Models. We find significant differences in mean frequencies upon silencing these neurons but not upon activation. The experimental groups are also significantly more variable. We revised these panels with updated analysis. We think these data do support our interpretation that the grooming rhythms are disrupted.
More importantly, all this data would make the most sense if it were performed in undusted flies (with controls) as is done in the next figure.
In our assay conditions, undusted flies groom infrequently. We used undusted flies for some optogenetic activation experiments, where the neuron activation triggers behavior initiation, but we chose to analyze the effect of silencing inhibitory neurons in dusted flies because dust reliably activates mechanosensory neurons and elicits robust grooming behavior enabling us to assess how manipulation of 13A/B neurons alters grooming rhythmicity and leg coordination.
(2) In Figure 4-Figure Supplement 1, the inclusion of walking assays in dusted flies is problematic, as these flies are already strongly biased toward grooming behavior and rarely walk. To assess how 13A neuron activation influences walking, such experiments should be conducted in undusted flies under baseline locomotor conditions.
We agree that there are better ways to assay potential contributions of 13A/13B neurons to walking. We intended to focus on how normal activity in these inhibitory neurons affects coordination during grooming, and we included walking because we observed it in our optogenetic experiments and because it also involves rhythmic leg movements. The walking data is reported in a supplementary figure because we think this merits further study with assays designed to quantify walking specifically. We will make these goals clearer in the revised manuscript and we are happy to share our reagents with other research groups more equipped to analyze walking differences.
(3) For broader lines targeting six or more 13A neurons, the authors provide specific predictions about expected behavioral effects-e.g., that activation should bias the limb toward flexion and silencing should bias toward extension based on connectivity to motor neurons. Yet, when using the more restricted line labeling only two 13A neurons (Figure 4 - Figure Supplement 2), no such prediction is made. The authors report disrupted grooming but do not specify whether the disruption is expected to bias the movement toward flexion or extension, nor do they discuss the muscle target. This is a missed opportunity to apply the same level of mechanistic reasoning that was used for broader manipulations.
Because we cannot unambiguously identify one of the neurons from our sparsest 13A splitGAL4 lines in FANC, we cannot say with certainty which motor neurons they target. That limits the accuracy of any functional predictions.
(4) Regarding Figure 5: The 70ms on/off stimulation with a slow opsin seems problematic. CsChrimson off kinetics are slow and unlikely to cause actual activity changes in the desired neurons with the temporal precision the authors are suggesting they get. Regardless, it is amazing that the authors get the behavior! It would still be important for the authors to mention the optogenetics caveat, and potentially supplement the data with stimulation at different frequencies, or using faster opsins like ChrimsonR.
We were also intrigued by the behavioral consequences of activating these inhibitory neurons with CsChrimson. We appreciate the reviewer’s point that CsChrimson’s slow off-kinetics limit precise temporal control. To address this, we repeated our frequency analysis using a range of pulse durations (10/10, 50/50, 70/70, 110/110, and 120/120 ms on/off) and compared the mean frequency of proximal joint extension/flexion cycles across conditions. We found no significant difference in frequency (LLMS, p > 0.05), suggesting that the observed grooming rhythm is not dictated by pulse period but instead reflects an intrinsic property of the premotor circuit once activated. We now include these results in ‘Figure 5—figure supplement 1’ and clarify in the text that we interpret pulsed activation as triggering, rather than precisely pacing, the endogenous grooming rhythm. We continue to note in the manuscript that CsChrimson’s slow off-kinetics may limit temporal precision. We will try ChrimsonR in future experiments.
Overall, I think the strengths outweigh the weaknesses, and I consider this a timely and comprehensive addition to the field.
Reviewer #3 (Public review):
Summary:
The authors set out to determine how GABAergic inhibitory premotor circuits contribute to the rhythmic alternation of leg flexion and extension during Drosophila grooming. To do this, they first mapped the ~120 13A and 13B hemilineage inhibitory neurons in the prothoracic segment of the VNC and clustered them by morphology and synaptic partners. They then tested the contribution of these cells to flexion and extension using optogenetic activation and inhibition and kinematic analyses of limb joints. Finally, they produced a computational model representing an abstract version of the circuit to determine how the connectivity identified in EM might relate to functional output. The study, in its current form, makes an important but overclaimed contribution to the literature due to a mismatch between the claims in the paper and the data presented.
Strengths:
The authors have identified an interesting question and use a strong set of complementary tools to address it:
(1) They analysed serial‐section TEM data to obtain reconstructions of every 13A and 13B neuron in the prothoracic segment. They manually proofread over 60 13A neurons and 64 13B neurons, then used automated synapse detection to build detailed connectivity maps and cluster neurons into functional motifs.
(2) They used optogenetic tools with a range of genetic driver lines in freely behaving flies to test the contribution of subsets of 13A and 13B neurons.
(3) They used a connectome-constrained computational model to determine how the mapped connectivity relates to the rhythmic output of the behavior.
Weaknesses:
The manuscript aims to reveal an instructive, rhythm-generating role for premotor inhibition in coordinating the multi-joint leg synergies underlying grooming. It makes a valuable contribution, but currently, the main claims in the paper are not well-supported by the presented evidence.
Major points
(1) Starting with the title of this manuscript, "Inhibitory circuits generate rhythms for leg movements during Drosophila grooming", the authors raise the expectation that they will show that the 13A and 13B hemilineages produce rhythmic output that underlies grooming. This manuscript does not show that. For instance, to test how they drive the rhythmic leg movements that underlie grooming requires the authors to test whether these neurons produce the rhythmic output underlying behavior in the absence of rhythmic input. Because the optogenetic pulses used for stimulation were rhythmic, the authors cannot make this point, and the modelling uses a "black box" excitatory network, the output of which might be rhythmic (this is not shown). Therefore, the evidence (behavioral entrainment; perturbation effects; computational model) is all indirect, meaning that the paper's claim that "inhibitory circuits generate rhythms" rests on inferred sufficiency. A direct recording (e.g., calcium imaging or patch-clamp) from 13A/13B during grooming - outside the scope of the study - would be needed to show intrinsic rhythmogenesis. The conclusions drawn from the data should therefore be tempered. Moreover, the "black box" needs to be opened. What output does it produce? How exactly is it connected to the 13A-13B circuit?
We modified the title to better reflect our strongest conclusions: “Inhibitory circuits control leg movements during Drosophila grooming”
Our optogenetic activation was delivered in a patterned (70 ms on/off) fashion that entrains rhythmic movements, but this does not rule out the possibility that the rhythm is imposed externally. In the manuscript, we state that we used pulsed light to mimic a flexion-extension cycle and note that this approach tests whether inhibition is sufficient to drive rhythmic leg movements when temporally patterned. While this does not prove that 13A/13B neurons are intrinsic rhythm generators, it does demonstrate that activating subsets of inhibitory neurons is sufficient to elicit alternating leg movements resembling natural grooming and walking.
Our goal with the model was to demonstrate that it is possible to produce rhythmic outputs with this 13A/B circuit, based on the connectome. The “black box” is a small recurrent neural network (RNN) consisting of 40 neurons in its hidden layer. The inputs are the “dust” levels from the environment (the green pixels in Figure 6I), the “proprioceptive” inputs (“efference copy” from motor neurons), and the amount of dust accumulated on both legs. The outputs (all positive) connect to the 13A neurons, the 13B neurons, and to the motor neurons. We refer to it as the “black box” because we make no claims about the actual excitatory inputs to these circuits. Its function is to provide input, needed to run the network, that reflects the distribution of “dust” in the environment as well as the information about the position of the legs.
The output of the “black box” component of the model might be rhythmic. In fact, in most instances of the model implementation this is indeed the case. However, as mentioned in the current version of the manuscript: “But the 13A circuitry can still produce rhythmic behavior even without those external inputs (or when set to a constant value), although the legs become less coordinated.” Indeed, when we refine the model (with the evolutionary training) without the “black box” (using a constant input of 0.1) the behavior is still rhythmic and sustained. Therefore, the rhythmic activity and behavior can emerge from the premotor circuitry itself without a rhythmic input.
The context in which the 13A and 13B hemilineages sit also needs to be explained. What do we know about the other inputs to the motorneurons studied? What excitatory circuits are there?
We agree that there are many more excitatory and inhibitory, direct and indirect, connections to motor neurons that will also affect leg movements for grooming and walking. 13A neurons provide a substantial fraction of premotor input. For example, 13As account for ~17.1% of upstream synapses for one tibia extensor (femur seti) motor neuron and ~14.6% for another tibia extensor (femur feti) motor neuron. Our goal was to demonstrate what is possible from a constrained circuit of inhibitory neurons that we mapped in detail, and we hope to add additional components to better replicate the biological circuit as behavioral and biomechanical data is obtained by us and others.
Furthermore, the introduction ignores many decades of work in other species on the role of inhibitory cell types in motor systems. There is some mention of this in the discussion, but even previous work in Drosophila larvae is not mentioned, nor crustacean STG, nor any other cell types previously studied. This manuscript makes a valuable contribution, but it is not the first to study inhibition in motor systems, and this should be made clear to the reader.
We thank the reviewer for this important reminder. Previous work on the contribution of inhibitory neurons to invertebrate motor control certainly influenced our research. We have expanded coverage of the relevant history and context in our revised discussion.
(2) The experimental evidence is not always presented convincingly, at times lacking data, quantification, explanation, appropriate rationales, or sufficient interpretation.
We are committed to improving the clarity, rationale, and completeness of our experimental descriptions. We have revisited the statistical tests applied throughout the manuscript and expanded the Methods.
(3) The statistics used are unlike any I remember having seen, essentially one big t-test followed by correction for multiple comparisons. I wonder whether this approach is optimal for these nested, high‐dimensional behavioral data. For instance, the authors do not report any formal test of normality. This might be an issue given the often skewed distributions of kinematic variables that are reported. Moreover, each fly contributes many video segments, and each segment results in multiple measurements. By treating every segment as an independent observation, the non‐independence of measurements within the same animal is ignored. I think a linear mixed‐effects model (LMM) or generalized linear mixed model (GLMM) might be more appropriate.
We thank the reviewer for raising this important point regarding the statistical treatment of our segmented behavioral data. Our initial analysis used independent t-tests with Bonferroni correction across behavioral classes and features, which allowed us to identify broad effects. However, we acknowledge that this approach does not account for the nested structure of the data. To address this, we re-analyzed key comparisons using linear mixed-effects models (LMMs) as suggested by the reviewer. This approach allowed us to more appropriately model within-fly variability and test the robustness of our conclusions. We have updated the manuscript based on the outcomes of these analyses.
(4) The manuscript mentions that legs are used for walking as well as grooming. While this is welcome, the authors then do not discuss the implications of this in sufficient detail. For instance, how should we interpret that pulsed stimulation of a subset of 13A neurons produces grooming and walking behaviours? How does neural control of grooming interact with that of walking?
We do not know how the inhibitory neurons we investigated will affect walking or how circuits for control of grooming and walking might compete. We speculate that overlapping pre-motor circuits may participate because both have similar extension flexion cycles at similar frequencies, but we do not have hard experimental data to support. This would be an interesting area for future research. Here, we focused on the consequences of activating specific 13A/B neurons during grooming because they were identified through a behavioral screen for grooming disruptions, and we had developed high-resolution assays and familiarity with the normal movements in this behavior.
(5) The manuscript needs to be proofread and edited as there are inconsistencies in labelling in figures, phrasing errors, missing citations of figures in the text, or citations that are not in the correct order, and referencing errors (examples: 81 and 83 are identical; 94 is missing in text).
We have proofread the manuscript to fix figure labeling, citation order, and referencing errors.
Reviewing Editor Comments:
In addition to the recommendations listed below, a common suggestion, given the lack of evidence to support that 13A and 13B are rhythm-generating, is to tone down the title to something like, for example, "Inhibitory circuits control leg movements during grooming in Drosophila" (or similar).
We changed the title to Inhibitory circuits control leg movements during Drosophila grooming
Reviewer #1 (Recommendations for the authors):
(1) Naming of movements of leg segments:
The authors refer to movements of leg segments across the leg, i.e., of all joints, as "flexion" and "extension". For example, in Figure 4A and at many other places. This naming is functionally misleading for two reasons: (i) the anatomical organization of an insect leg differs in principle from the organization of the mammalian leg, which the manuscript often refers to. While the organization of a mammalian limb is planar the organization of the insect limb shows a different plane as compared to the body length axis (for detailed accounts see Ritzmann et al. 2004; Büschges & Ache, 2024); (ii) the reader cannot differentiate between places in the text, where "flexion" and "extension" refer to movements of the tibia of the femur-tibia joint, e.g. in the graphical abstract, in Figure 3 and its supplements, and other places, e.g. Figure 4 and its supplements, where these two words refer to movements of leg segments of other joints, e.g. thorax-coxa, coxa-trochanter and tarsal joints. The reviewer strongly suggests naming the movements of the leg segments according to the individual joint and its muscles.
We accept this helpful suggestion. We now include a description of the leg segments and joints in the revised Introduction and refer to which leg segments we mean
“The adult Drosophila leg consists of serially arranged joints—bodywall/thoraco-coxal (Th-C), coxa–trochanter (C-Tr), trochanter–femur (Tr-F), femur–tibia (F-Ti), tibia–tarsus (Ti-Ta)—each powered by opposing flexor and extensor muscles that transmit force through tendons (Soler et al., 2004). The proximal joints, Th-C and C-Tr, mediate leg protraction–retraction and elevation–depression, respectively (Ritzmann et al., 2004; Büschges & Ache, 2025). The medial joint, F-Ti, acts as the principal flexion–extension hinge and is controlled by large tibia extensor motor neurons and flexor motor neurons (Soler et al., 2004; Baek and Mann 2009; Brierley et al., 2012; Azevedo et al., 2024; Lesser et al., 2024). By contrast, distal joints such as Ti-Ta and the tarsomeres contribute to fine adjustments, grasping, and substrate attachment (Azevedo et al., 2024).”
We also clarified femur-tibia joints in the graphical abstract, modified Figure 3 legend and added joints at relevant places.
(2) Figures 3, 4, and 5 with supplements:
The authors optogenetically silence and activate (sub)populations of 13A and 13B interneurons. Changes in frequency of movements and distance between legs or leg movements are interpreted as the effect of these experimental paradigms. No physiological recordings from leg motoneurons or leg muscles are shown. While I understand the notion of the authors to interpret a movement as the outcome of activity in a muscle, it needs to be remembered that it is well known that fast cyclic leg movements, including those for grooming, cannot be used to conclude on the underlying neural activity. Zakotnik et al. (2006) and others provided evidence that such fast cyclic movements can result from the interaction of the rhythmic activity of one leg muscle only, together with the resting tension of its silent antagonist. Given that no physiological recordings are presented, this needs to be mentioned in the discussion, e.g., in the section "Inhibitory Innervation Imbalance.......".
Added studies from Heitler, 1974; Bennet-Clark, 1975; Zakotnik et al., 2006; Page et al., 2008 in discussion.
(3) Introduction and Discussion:
The authors refer extensively to work on the mammalian spinal cord and compare their own work with circuit elements found in the spinal cord. From the perspective of the reviewer this notion is in conflict with acknowledging prior research work on the role of inhibitory network interactions for other invertebrates and lower vertebrates: such are locust flight system (for feedforward inhibition, disinhibition), crustacean stomatogastric nervous system (reciprocal inhibition), clione swimming system (reciprocal inhibition, feedforward inhibition, disinhibition), leech swimming system (reciprocal inhibition, disinhibition, feedforward inhibition), xenopus swimming system (reciprocal inhibition). The next paragraph illustrates this criticism/suggestion for stick insect neural circuits for leg stepping.
(4) Discussion:
"Feedforward inhibition" and "Disinhibition": it is already been described that rhythmic activity of antagonistic insect leg motoneuron pools arises from alternating synaptic inhibition and disinhibition of the motoneurons from premotor central pattern generating networks, e.g., Büschges (1998); Büschges et al. (2004); Ruthe et al. (2024).
We have added these references to the revised Discussion.
(5) Circuit motifs of the simulation, i.e., mutual inhibition between interneurons and onto motoneurons and sensory feedback influences and pathways share similarities to those formerly used by studies simulating rhythmic insect leg movements, for example, Schilling & Cruse 2020, 2023 or Toth et al. 2012. For the reader, it appears relevant that the progress of the new simulation is explained in the light of similarities and differences to these former approaches with respect to the common circuit motifs used.
We now put our work in the context of other models in the Discussion section: “Similar circuit motifs, namely reciprocal inhibitions between pre-motor neurons and the sensory feedback have been modeled before, in particular neuroWalknet, and such simple motifs do not require a separate CPG component to generate rhythmic behavior in these models (Schilling & Cruse 2020, 2023). However, our model is much simpler than the neuroWalknet - it controls a 2D agent operating on an abstract environment (the dust distribution), without physics. In real animals or complex mechanical models such as NeuroMechFly (Lobato-Rios et al), a more explicit central rhythm generation may be advantageous for the coordination across many more degrees of freedom.”
Reviewer #2 (Recommendations for the authors):
I might have missed this, but I couldn't find any mention of how the grooming command pathways, described by previous work from the authors' lab, recruit these predicted grooming pattern-generating neurons. This should be mentioned in the connectome analysis and also discussed later in the discussion.
13A neurons are direct downstream targets of previously described grooming command neurons. Specifically, the antennal grooming command neuron aDN (Hampel et al., 2015) synapses onto two primary 13As (γ and α; 13As-i) that connect to proximal extensor and medial flexor motor neurons, as well as four other 13As (9a, 9c, 9i, 6e) projecting to body wall extensor motor neurons. The 13As-i also form reciprocal connections with 13As-ii, providing a potential substrate for oscillatory leg movements. aDN connects to homologous 13As on both sides, consistent with the bilateral coordination needed for antennal sweeping.
The head grooming/leg rubbing command neuron DNg12 (Guo et al., 2022) synapses directly onto ~50 13As, predominantly those connected to proximal motor neurons.
While sometimes the structural connectivity suggests pathways for generating rhythmic movements, the extensive interconnections among command neurons and premotor circuits indicate that multiple motifs could contribute to the observed behaviors. Further work will be needed to determine how these inputs are dynamically engaged during normal grooming sequences. We have now added it to the discussion.
I encourage the authors to be explicit about caveats wherever possible: e.g., ectopic expression in genetic tools, potential for other unexplored neurons as rhythm generators (rather than 13A/B), given that the authors never get complete silencing phenotypes, CsChrimson kinetics, neurotransmitter predictions, etc.
We now explain these caveats as follows: Ectopic expression is noted in Figure 1—figure supplement 1, and we added the following to the Discussion: “While our experiments with multiple genetic lines labeling 13A/B neurons consistently implicate these cells in leg coordination, ectopic expression in some lines raises the possibility that other neurons may also contribute to this phenotype. In addition, other excitatory and inhibitory neural circuits, not yet identified, may also contribute to the generation of rhythmic leg movements. Future studies should identify such neurons that regulate rhythmic timing and their interactions with inhibitory circuits.”
We also added a caveat regarding CsChrimson kinetics in the Results. Finally, our identification of these neurons as inhibitory is based on genetic access to the GABAergic population (we use GAD-spGAL4 as part of the intersection which targets them), rather than on predictions of neurotransmitter identity.
Reviewer #3 (Recommendations for the authors):
Detailed list of figure alterations:
(1) Figure 1:
(a) Figure 1B and Figure 1 - Figure Supplement 1 lack information on individual cells - how can we tell that the cells targeted are indeed 13A and 13B, and which ones they are? Since off-target expression in neighboring hemilineages isn't ruled out, the interpretation of results is not straightforward.
The neurons labeled by R35G04-DBD and GAD1-AD are identified as 13A and 13B based on their stereotyped cell body positions and characteristic neurite projections into the neuropil, which match those of 13A and 13B neurons reconstructed in the FANC and MANC connectome. While we have not generated flip-out clones in this genotype, we do isolate 13A neurons more specifically later in the manuscript using R35G04-DBD intersected with Dbx-AD, and show single-cell morphology consistent with identified 13A neurons. The purpose of including this early figure was to motivate the study by showing that silencing this population, which includes 13A/13B neurons, strongly reduces grooming in dusted flies.
Regarding Figure 1—Figure Supplement 1:
This figure showed the expression patterns of all lines used throughout the manuscript. Panels C and D illustrated lines with minimal to no ectopic expression. Panels A and B show neurons with posterior cell bodies that may correspond to 13A neurons not reconstructed in our dataset but described in Soffers et al., 2025 and Marin et al., 2025 and we have provided detailed information about all VNC expressions in the figure legend.
(b) Figure 1D lacks explanation of boxplots, asterisks, genotypes/experimental design.
Added.
(c) Figures 1E-F and video 1 lack quantification, scale bars.
Added quantification.
(2) Figure 2:
(a) Figure 2A, Figure 2 - Supplement 3: What are the details of the hierarchical clustering? What metric was used to decide on the number of clusters?
We have used FANC packages to perform NBLAST clustering (Azevedo et al., 2024, Nature). We now include the full protocol in Methods. The details are as follows:
We performed hierarchical clustering on pairwise NBLAST similarity scores computed using navis.nblast_allbyall(). The resulting similarity matrix was symmetrized by averaging it with its transpose, and converted into a distance matrix using the transformation:
distance=(1−similarity)\text{distance} = (1 - \text{similarity})distance=(1−similarity)
This ensures that a perfect NBLAST match (similarity = 1) corresponds to a distance of 0.
Clustering was performed using Ward’s linkage method (method='ward' in scipy.cluster.hierarchy.linkage), which minimizes the total within-cluster variance and is well-suited for identifying compact, morphologically coherent clusters.
We did not predefine the number of clusters. Instead, clusters were visualized using a dendrogram, where branch coloring is based on the default behavior of scipy.cluster.hierarchy.dendrogram(). By default, this function applies a visual color threshold at 70% of the maximum linkage distance to highlight groups of similar elements. In our dataset, this corresponded to a linkage distance of approximately 1–1.5, which visually separated morphologically distinct neuron types (Figures 2A and Figure 2—figure supplement 3A). This threshold was used only as a visual aid and not as a hard cutoff for quantitative grouping.
The Methods section says that the classification "included left-right comparisons". What does that mean? What are the implications of the authors only having proofread a subset of neurons in T1L (see below)?
All adult leg motor neurons and 13A neurons (except one, 13A-ε) have neurite arbors restricted to the local, ipsilateral neuropil associated with the nearest leg. Although 13B neurons have contralateral cell bodies, their projections are also entirely ipsilateral. The Tuthill Lab, with contributions from our group, focused proofreading efforts on the left front neuropil (T1L) in FANC. This is also where the motor neuron to muscle mapping has been most extensively done. We reconstructed/proofread the 13A and 13B neurons from the right side as well (T1R). We see similar clustering based on morphology and connectivity here as well.
Reconstructions lack scale bars and information on orientation (also in other figures), and the figures for the 13B analysis are not consistent with the main figure (e.g., labelling of clusters in panel B along x,y axes).
Added.
(b) Figure 2B: Since the cosine similarity matrix's values should go from -1 to 1, why was a color map used ranging from 0 to 1?
While cosine similarity values can theoretically range from -1 to 1, in our case, all vector entries (i.e., synaptic weights) are non-negative, as they reflect the number of synapses from each 13A neuron to its downstream targets. This means all pairwise cosine similarities fall within the 0 to 1 range.
Why are some neurons not included in this figure, like 1g, 2b, 3c-f (also in Supplement 3)?
The few 13A neurons that don’t connect to motor neurons are not shown in the figure.
(c) Figures 2C and D: the overlaid neurites are difficult to distinguish from one another. If the point here is to show that each 13A neuron class innervates specific motor neurons, then this is not the clearest way of doing that. For instance, the legend indicates that extensors are labelled in red, and that MNs with the highest number of synapses are highlighted in red - does that work? I could not figure out what was going on. On a more general point: if two cells are connected, does that not automatically mean that they should overlap in their projection patterns?
We intended these panels to illustrate that 13A neurons synapse onto overlapping regions of motor neurons, thereby creating a spatial representation of muscle targets. However, we agree that overlapping multiple neurons in a single flat projection makes the figure difficult to interpret. We have therefore removed Figures 2C and 2D.
While neurons must overlap at least somewhere if they form a synaptic connection, the amount of their neurites that overlap can vary, and more extensive overlap suggests more possible connections. Because the synapses are computationally predicted, examining the overlap helps to confirm that these predictions are consistent.
While connected neurons must overlap locally at their synaptic sites, they do not necessarily show extensive or spatially structured overlap of their projections. For example, descending neurons or 13B interneurons may form synapses onto motor neurons without exhibiting a topographically organized projection pattern. In contrast, 13A→MN connectivity is organized in a structured manner: specialist 13A neurons align with the myotopic map of MN dendrites, whereas generalist 13As project more broadly and target MN groups across multiple leg segments, reflecting premotor synergies. This spatial organization—combining both joint-specific and multi-joint representations—was a key finding we wished to highlight, and we have revised the Results text to make this clearer.
(d) Figure 2 - Figure Supplement 1: Why are these results presented in a way that goes against the morphological clustering results, but without explanation? Clusters 1-3 seem to overlap in their connectivity, and are presented in a mixed order. Why is this ignored? Are there similar data for 13B?
The morphological clusters 1–3 do exhibit overlapping connectivity, but this is consistent with both their anatomical similarity and premotor connectivity. Specifically, Cluster 1 neurons connect to SE and TrE motor neurons, Cluster 2 connects only to TrE motor neurons, and Cluster 3 targets multiple motor pools, including SE and TrE (Figure 2—Figure Supplement 1B). This overlap is also reflected in the high pairwise cosine similarity among Clusters 1–3 shown in Figure 2B. Thus, their similar connectivity profiles align with their proximity in the NBLAST dendrogram.
Regarding 13B neurons: there is no clear correlation between morphological clusters and downstream motor targets, as shown in the cosine similarity matrix (Figure 2—figure supplement 3). Moreover, even premotor 13B neurons that fall within the same morphological cluster do not connect to the same set of motor neurons (Figure 3—figure supplement 1F). For example, 13B-2a connects to LTrM and tergo-trochanteral MNs, 13B-2b connects to TiF MNs, and 13B-2g connects to Tr-F, TiE, and tergo-T MNs. Together, these results demonstrate that 13A neurons are spatially organized in a manner that correlates with their motor neuron targets, whereas 13B neurons lack such spatially structured organization, suggesting distinct principles of connectivity for these two inhibitory premotor populations.
(e) Figure 2 - Figure Supplement 2: A comparison is made here between T1R (proofread) and T1L (largely not proofread). A general point is made here that there are "similar numbers of neurons and cluster divisions". First, no quantitative comparison is provided, making it difficult to judge whether this point is accurate. Second, glancing at the connectivity diagram, I can identify a large number of discrepancies. How should we interpret those? Can T1L be proofread? If this is too much of a burden, results should be presented with that as a clear caveat.
The 13A and 13B neurons in the T1L hemisegment are fully proofread (Lesser et al, 2024, current publication); the T1R has been extensively analyzed as well. To compare the clustering and match identities of 13A and 13B neurons on the left and the right, We mirrored the 13A neurons from the left side and used NBLAST to match them with their counterparts on the right.
While individual synaptic counts differ between sides in the FANC dataset (T1L generally showing higher counts), the number of 13A neurons, their clustering, and the overall patterns of connectivity are largely conserved between T1L and T1R.
Importantly, each 13A cluster targets the same subset of motor neurons on both sides, preserving the overall pattern of connectivity. The largest divergence is seen in cluster 9, which shows more variable connectivity.
(f) Figure 2 - Figure Supplements 4 & 5: Why did the authors choose to present the particular cell type in Supplement 4? Why are the cell types in Supplement 5 presented differently? Labels in Supplement 5 are illegible, but I imagine this is due to the format of the file presented to reviewers. Why are there no data for 13B?
We chose to present the particular cell type in Supplement 4 because it corresponds to cell types targeted in the genetic lines used in our behavioral experiments. The 13A neuron shown is also one of the primary neurons in this lineage. This example illustrates its broader connectivity beyond the inhibitory and motor connections emphasized in the main figures.
In Supplement 5, we initially aimed to highlight that the major downstream targets of 13A neurons are motor neurons. We have now removed this figure and instead state in the text that the major downstream targets are MNs.
We did not present 13B neurons in the same format because their major downstream targets are not motor neurons. Instead, we emphasize their role in disinhibition and their connections to 13A neurons, as shown in a specific example in Figure 3—figure supplement 2. This 13B neuron also corresponds to a cell type targeted in the genetic line used in our behavioral experiments.
(3) Figure 3:
(a) Figure 3A: the collection of diagrams is not clear. I'd suggest one diagram with all connections included repeated for each subpanel, with each subpanel highlighting relevant connections and greying out irrelevant ones to the type of connection discussed. The nomenclature should be consistent between the figure and the legend (e.g., feedforward inhibition vs direct MN inhibition in A1.
The intent of Figure 3A is to highlight individual circuit motifs by isolating them in separate panels. Including all connections in every sub panel would likely reduce clarity and make it harder to follow each motif. For completeness, we show the full set of connections together in Panel D. We updated the nomenclature as suggested.
(b) Figure 3B: Why was the medial joint discussed in detail? Do the thicknesses of the lines represent the number of synapses? There should be a legend, in that case. Why are the green edges all the same thickness? Are they indeed all connected with a similarly low number of synapses?
We focused on the medial joint (femur-tibia joint) because it produces alternating flexion and extension of the tibia during both head sweeps and leg rubbing, which are the main grooming actions we analyzed. During head grooming, the tarsus is typically suspended in the air, so the cleaning action is primarily driven by tibial movements generated at the medial joint.
The thickness of the edges represents the number of synapses, and we have now clarified this in the legend. The green edges represent connections from 13B neurons, which were manually added to the graph, as described in the Methods section. 13B neurons are smaller than 13A neurons and form significantly fewer total downstream synapses. For example, the 13B neuron shown in Figure 3—figure supplement 2 makes a total of 155 synapses to all downstream neurons, with only 22 synapses to its most strongly connected partner, a 13A neuron. The relatively sparse connectivity of 13B neurons is shown in thinner or uniform edge weights in this graph.
(C) Figure 3C: This is a potentially important panel, but the connections are difficult to interpret. Moreover, the text says, "This organizational motif applies to multiple joints within a leg as reciprocal connections between generalist 13A neurons suggest a role in coordinating multi-joint movements in synergy". To what extent is this a representative result? The figure also has an error in the legend (it is not labelled as 3C).
This statement is true and based on the connectivity of these neurons. We now added
“Data for 13A-MN connections shown in Figure 2—figure supplement 1 I9, I6, I7, H9, H4, and H5; 13A-13A connections shown in Figure 3—figure supplement 1C.” to the figure legend.
Thanks, we fixed the labelling error.
(d) Figure 3 - Figure Supplement 1: Panel A is very difficult to interpret. Could a hierarchical diagram be used, or some other representation that is easier to digest?
Panel A provides a consolidated view of all upstream and downstream interconnections among individual 13A and 13B neurons, allowing readers to quickly assess which neurons connect to which others without having to examine all subpanels. For a hierarchical representation, we have provided individual neuron-level diagrams in Panels C–F.
(e) Figure 3 - Figure Supplement 2: Why was this cell type selected?
We selected this 13B because it is involved in the disinhibition of 13A neurons and is also present in the genetic line used for our behavioral experiments.
(f) Figure 3 - Figure Supplement 3: The diagram is confusing, with text aligned randomly, and colors lacking some explanations. Legend has odd formatting.
The diagram layout and text alignment are designed to reflect the logical grouping of proprioceptors, 13A neurons, and motor neurons. To improve clarity, we have added node colors, included a written explanation for edge colors, and corrected the formatting of the figure legend.
(4) Figure 4:
(a) Figure 4A: This has no quantification, poor labelling, and odd units (centiseconds?). The colours between the left and right panels also don't align.
We have fixed these issues.
(b) Figure 4D-K: The ranges on the different axes are not the same (e.g., y axis on box plots, x axis on histograms). This obscures the fact that the differences between experimental and control, which in many cases are not big, are not consistent between the various controls. Moreover, the data that are plotted are, as far as I can tell (which is also to say: this should be explained), one value per frame. With imaging at 100Hz, this means that an enormous number of values are used in each analysis. Very small differences can therefore be significant in a statistical sense. However, how different something is between conditions is important (effect size), and this is not taken int account in this manuscript. For instance, in 4D-J, the differences in the mean seem to be minimal. Should that not be taken into consideration? A point in case is panel D in Figure 4 - Figure Supplement 1: even with near identical distributions, a statistically significant difference is detected. The same applies to Figure 4 - Figure Supplements 1-3. Also, what do the boxes and whiskers in the box plots show, exactly?
We have re-plotted all summary panels using linear mixed-effects models (LMMs) as suggested. In the updated plots, each dot represents the mean value for a single animal, and bar height represents the group mean. Whiskers indicate the 95% confidence interval around the group mean. This approach avoids inflating sample size by using per-frame values and provides a more accurate view of both variability and effect size.
(e) Figure 4 - Figure Supplement 1: There are 6 cells labelled in the split line; only 4 are shown in A3. Is cluster 6 a convincing match between EM and MCFO?
We indeed report four neurons targeted by the split-GAL4 line in flip out clones. Generating these clones was technically challenging. In our sample (n=23), we may not have labeled all of the neurons. Alternatively, two neurons may share very similar morphology and connectivity, making it difficult to tell them apart. We have added this clarification to the revised figure legend.
It is interesting to see data on walking in panel K, but why were these analyses not done on any of the other manipulations? What defect produced the reduction in velocity, exactly? How should this be interpreted?
Our primary focus was on grooming, but we did observe changes in walking, so we report illustrative examples. We initially included a panel showing increased walking velocity upon 13A activation, but this effect did not survive FDR correction and was removed in the revised version. We instead included data for 13A silencing which did not affect the frequency of joint movements during walking. However, spatial aspects of walking were affected: the distance between front leg tips during stance was reduced, indicating that although flies continued to walk rhythmically, the positioning of the legs was altered. This suggests that these specific 13A neurons may influence coordination and limb placement during walking without disrupting basic rhythmicity. As reviewer #2 also noted, dust may itself affect walking, so we have chosen not to further pursue this aspect in the current study.
(f) Figure 4 - Figure Supplement 2: panel A is identical to Figure 1 - Figure Supplement 1C. This figure needs particular attention, both in content and style. Why present data on silencing these neurons in C-D, but not in E-F?
We removed the panel Figure 1 - Figure Supplement 1C and kept it in Figure 4 - Figure Supplement 2 A. E-F also shows data on silencing, as C’.
(g) Figure 4 - Figure Supplement 3: In panel B, the authors should more clearly demonstrate the identity of 4b and 4a. Why present such a limited number of parameters in F and G?
The cells shown in panel B represent the best matches we could identify between the light-level expression pattern and EM reconstructions. In panels F and G, we focused on bout duration, as leg position/inter-leg distance and frequency were already presented (in Figure 4). Together, these parameters demonstrate the role of 13B neurons in coordinating leg movements. Maximum angular velocity of proximal joints was not significantly affected and is therefore not included.
(5) Figure 5:
(a) Figure 5B: Lacks a quantification of the periodic nature of the behavior, which is required to compare to experimental conditions, e.g., in panel C.
Added
(b) Figure 5C: Requires a quantification; stimulus dynamics need to be incorporated.
Added
(c) Figure 5D: More information is needed. Does "Front leg" mean "leg rub", and "Head" "head sweep"? How do the dynamics in these behaviors compare to normal grooming behavior?
Yes, head grooming is head sweeps and Front leg grooming is leg rub. Comparison added, shown in 5E-F
(d) Figure 5E: How should we interpret these plots? Do these look like normal grooming/walking?
We have now included the comparison.
(e) Figure 5F: Needs stats to compare it to 5B'.
Done
(6) Figure 6:
(a) Figure 6A: I think the circuit used for the model is lacking the claw/hook extension - 13Bs connection. Any other changes? What is the rationale?
13Bs upstream of these particular 13As do not receive significant connections from claw/hook neurons (there’s only one ~5 synapses connection from one hook extension to one 13B neurons, which we neglected for the modeling purpose).
(b) Figure 6B and C: Needs labels, legend; where is 13B?
In the figure legend we now added: “The 13B neurons in this model do not connect to each other, receive excitatory input from the black box, and only project to the 13As (inhibitory). Their weight matrix, with only two values, is not shown.” We added the colorbar and corrected the color scheme.
(c) Figure 6D-H: plots are very difficult to interpret. Units are also missing (is "Time" correct?).
The units are indeed Time in frames (of simulation). We added this to the figure and the legend. We clarified the units of all variables in these panels. Corrected the color scheme and added their meaning to the legend text.
(d) Figure 6I: I think the authors should consider presenting this in a different format.
(e) Figure 6 J and K (also Figure Supplement): lacks labels.
We added labels for the three joints, increased the size of fonts for clarity, and added panel titles on the top.
More specific suggestions:
(1) It would be helpful if the titles of all figures reflected the take-away message, like in Figure 2.
(2) "Their dendrites occupy a limited region of VNC, suggesting common pre-synaptic inputs" - all dendrites do, so I'd suggest rephrasing to be more precise.
(3) "We propose that the broadly projecting primary neurons are generalists, likely born earlier, while specialists are mostly later-born secondary neurons" - this needs to be explained.
We added the explanation.
We propose that the broadly projecting primary neurons are generalists, likely born earlier, while specialists are mostly later-born secondary neurons. This is consistent with the known developmental sequence of hemilineages, where early-born primary neurons typically acquire larger arbors and integrate across broader premotor and motor targets, whereas later-born secondary neurons often have more spatially restricted projections and specialized roles[18,19,81,82,85]. Our morphological clustering supports this idea: generalist 13As have extensive axonal arbors spanning multiple leg segments, whereas specialist neurons are more narrowly tuned, connecting to a few MN targets within a segment. Thus, both their morphology and connectivity patterns align with the expectation from birth-order–dependent diversification within hemilineages.
(4) "We did not find any correlation between the morphology of premotor 13B and motor connections" - this needs to be explained, as morphology constrains connectivity.
We agree that morphology often constrains connectivity. However, in contrast to 13A neurons—where morphological clusters strongly predict MN connectivity—we did not observe such a correlation for 13B neurons. As we noted in our response to comment 2d, 13B neurons can form synapses onto MNs without exhibiting extensive or spatially structured overlap of their axonal projections with MN dendrites. This suggests that 13B→MN connectivity may be governed by more local, synapse-specific rules rather than by large-scale morphological positioning, in contrast to the spatially organized premotor map we observe for 13As.
(5) "Based on their connectivity, we hypothesized that continuously activating them might reduce extension and increase flexion. Conversely, silencing them might increase extension and reduce flexion." - these clear predictions are then not directly addressed in the results that follow.
We have now expanded this section.
(6) "Thus, 13A neurons regulate both spatial and temporal aspects of leg coordination" "Together, 13A and 13B neurons contribute to both spatial and temporal coordination during grooming" - are these not intrinsically linked? This needs to be explained/justified.
The spatial (leg positioning, joint angles) and temporal (frequency, rhythm) aspects are often linked, but they can be at least partially dissociated. This has been shown in other systems: for example, Argentine ants reduce walking speed on uneven terrain primarily by decreasing stride frequency while maintaining stride length (Clifton et al., 2020), and Drosophila larvae adjust crawling speed mainly by modulating cycle period rather than the amplitude of segmental contractions (Heckscher et al., 2012). Consistent with these findings, we observe that 13A neuron manipulation in dusted flies significantly alters leg positioning without changing the frequency of walking cycles. Thus, leg positioning can be perturbed while the number of extension–flexion cycles per second remains constant, supporting the view that spatial and temporal features are at least partially dissociable.
(7) "Connectome data revealed that 13B neurons disinhibit motor pools (...) One of these 13B neurons is premotor, inhibiting both proximal and tibia extensor MN" - these are not possible at the same time.
We show that the 13B population contains neurons with distinct connectivity motifs:
some inhibit premotor 13A neurons (leading to disinhibition of motor pools), while others directly inhibit motor neurons. The split-GAL4 line we use labels three 13B neurons—two that inhibit the primary 13A neuron 13A-9d-γ (which targets proximal extensor and medial flexor MNs) and one that is premotor, directly inhibiting both proximal and tibia extensor MNs. Although these functions may appear mutually exclusive, their combined action could converge to a similar outcome: disinhibition of proximal extensor and medial flexor MNs while simultaneously inhibiting medial extensor MNs. This suggests that the labeled 13B neurons act in concert to bias the network toward a specific motor state rather than producing contradictory effects.
(8) "we often observed that one leg became locked in flexion while the other leg remained extended, (indicating contribution from additional unmapped left right coordination circuits)." - Are these results not informative? I'd suggest the authors explain the implications of this more, rather than mentioning it within brackets like this.
We agree with the reviewer that these results are highly informative. The observation that one leg can remain locked in flexion while the other stays extended suggests that additional left–right coordination circuits are engaged during grooming. This cross-talk is likely mediated by commissural interneurons downstream of inhibitory premotor neurons, which have not yet been systematically studied. Dissecting these circuits will require a dedicated project combining bilateral connectomic reconstruction, studying downstream targets of these commissural neurons, and functional interrogation, which is beyond the scope of the current study.
(9) "Indeed, we observe that optogenetic activation of specific 13A and 13B neurons triggers grooming movements. We also discover that" - this phrasing suggests that this has already been shown.external
We replaced ‘indeed’ with “Consistent with this connectivity,”
(10) "But the 13A circuitry can still produce rhythmic behavior even without those sensory inputs (or when set to a constant value), although the legs become less coordinated." - what does this mean?
We can train (fine-tune) the model without the descending inputs from the “black box” and the behavior will still be rhythmic, meaning that our modeled 13A circuit alone can produce rhythmic behavior, i.e. the rhythm is not generated externally (by the “black box”). We added Figure 7 to the MS and re-wrote this paragraph. In the revised manuscript we now state: “But the 13A circuitry can still produce rhythmic behavior even without those excitatory inputs from the “black box” (or when set to a constant value), although the legs become less coordinated (because they are “unaware” of each other’s position at any time). Indeed, when we refine the model (with the evolutionary training) without the “black box” (using instead a constant input of 0.1) the behavior is still rhythmic although somewhat less sustained (Figure 7). This confirms that the rhythmic activity and behavior can emerge from the modeled pre-motor circuitry itself, without a rhythmic input.”
(11) "However, to explore the possibility of de novo emergent periodic behavior (without the direct periodic descending input) we instead varied the model's parameters around their empirically obtained values." - why do the authors not show how the model performs without tuning it first? What are the changes exactly that are happening as a result of the tuning? Are there specific connections that are lost? Do I interpret Figure 6B and C correctly when I think that some connections are lost (e.g., an SN-MN connection)? How does that compare to the text, which states that "their magnitudes must be at least 80% of the empirical weights"?
Without the fine-tuning we do not get any behavior (the activation levels saturate). So, we tolerate 20% divergence from the empirically established weights and we keep the signs the same. However, in the previous version we allowed the weights to decrease below 20% of the empirical weight (as long as the sign didn’t change) but not above (the signs were maintained and synapses were not added or removed). We thank the reviewer for observing this important discrepancy. In the current version we ensured that the model’s weights are bounded in both directions (the tolerance = 0.2), but we also partially relaxed the constraint on adjacency matrix re-scaling (see Methods, the “The fine-tuning of the synaptic weights” section, where we now clarify more precisely how the evolving model is fitted to the connectome constraints). We then re-ran the fine-tuning process. The Figure 6B and C is now corrected with the properly constrained model, as well as other panels in the figure. We also applied a better color scheme (now, blue is inhibitory and red is excitatory) for Fig. 6B and C.
(12) "Interestingly, removing 13As-ii-MN connections to the three MNs (second row of the 13A → MN matrices in Figures 6B and C) does not have much effect on the leg movement (data not shown). It seems sufficient for this model to contract only one of the two antagonistic muscles per joint, while keeping the other at a steady state." - this is not clear.
We repeated this test with the newly fine-tuned model and re-wrote the result as follows: “...when we remove just the 13A-i-MN connections (which control the flexors of the right leg) we likewise get a complete paralysis of the leg. However, removing the 13A-ii-MN (which control the extensors of the right leg) has only a modest effect on the leg movement. So, we need the 13A-i neurons to inhibit the flexors (via motor neurons), but not extensors, in order to obtain rhythmic movements.”
(13) The Discussion needs to reference the specific Results in all relevant sections.
We have revised the discussion to explicitly reference the specific results.
(14) "Flexors and extensors should alternate" - there are circumstances in which flexors and extensors should co-contract. For instance, co-contraction modulates joint stiffness for postural stability and helps generate forces required for fast movements.
Thanks for pointing this out. We added “However, flexor–extensor co-contraction can also be functionally relevant, such as for modulating joint stiffness during postural stabilization or for generating large forces required for fast movements (Zakotnik et al., 2006; Günzel et al., 2022; Ogawa and Yamawaki 2025). Some generalist 13A neurons could facilitate co-contraction across different leg segments, but none target antagonistic motor neurons controlling the same joint. Therefore, co-contraction within a single joint would require the simultaneous activation of multiple 13A neurons.”
(15) "While legs alternate between extension and flexion, they remain elevated during grooming. To maintain this posture, some MNs must be continuously activated while their antagonists are inactivated." - this is not necessarily correct. Small limbs, like those of Drosophila, can assume gravity-independent rest angles (10.1523/JNEUROSCI.5510-08.2009).
We added it to discussion
(16) The discussion "Spatial Mapping of premotor neurons in the nerve cord" seems to me to be making obvious points, and does not need to be included.
We have now revised this section to highlight the significance of 13A spatial organization, emphasizing premotor topographic mapping, multi-joint movement modules, and parallels to myotopic, proprioceptive, and vertebrate spinal maps.
(17) Key point, albeit a small one: "Normal activity of these inhibitory neurons is critical for grooming" - the use of the word critical is problematic, and perhaps typical of the tone of the manuscript. These animals still groom when many of these neurons are manipulated, so what does "critical" really mean?
In this instance, we now changed “critical” to “important”. We observed that silencing or activating a large number (>8) 13A neurons or few 13A and B neurons together completely abolishes grooming in dusted flies as flies get paralyzed or the limbs get locked in extreme poses. Therefore we think we have a justification for the statement that these neurons are critical for grooming. These neurons may contribute to additional behaviors, and there may be partially redundant circuits that can also support grooming. We have revised the manuscript with the intention of clarifying both what we have observed and the limits.
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public review):
Summary:
In this manuscript, the authors endeavor to capture the dynamics of emotion-related brain networks. They employ slice-based fMRI combined with ICA on fMRI time series recorded while participants viewed a short movie clip. This approach allowed them to track the time course of four non-noise independent components at an effective 2s temporal resolution at the BOLD level. Notably, the authors report a temporal sequence from input to meaning, followed by response, and finally default mode networks, with significant overlap between stages. The use of ICA offers a data-driven method to identify large-scale networks involved in dynamic emotion processing. Overall, this paradigm and analytical strategy mark an important step forward in shifting affective neuroscience toward investigating temporal dynamics rather than relying solely on static network assessments
Strengths:
(1) One of the main advantages highlighted is the improved temporal resolution offered by slice-based fMRI. However, the manuscript does not clearly explain how this method achieves a higher effective resolution, especially since the results still show a 2s temporal resolution, comparable to conventional methods. Clarification on this point would help readers understand the true benefit of the approach.
(2) While combining ICA with task fMRI is an innovative approach to study the spatiotemporaldynamics of emotion processing, task fMRI typically relies on modeling the hemodynamic response (e.g., using FIR or IR models) to mitigate noise and collinearity across adjacent trials. The current analysis uses unmodeled BOLD time series, which might risk suffering from these issues.
(3) The study's claims about emotion dynamics are derived from fMRI data, which are inherently affected by the hemodynamic delay. This delay means that the observed time courses may differ substantially from those obtained through electrophysiology or MEG studies. A discussion on how these fMRI-derived dynamics relate to - or complement - is critical for the field to understand the emotion dynamics.
(4) Although using ICA to differentiate emotion elements is a convenient approach to tell a story, it may also be misleading. For instance, the observed delayed onset and peak latency of the 'response network' might imply that emotional responses occur much later than other stages, which contradicts many established emotion theories. Given the involvement of largescale brain regions in this network, the underlying reasons for this delay could be very complex.
Concerns and suggestions:
However, I have several concerns regarding the specific presentation of temporal dynamics in the current manuscript and offer the following suggestions.
(1) One selling point of this work regarding the advantages of testing temporal dynamics is the application of slice-based fMRI, which, in theory, should improve the temporal resolution of the fMRI time course. Improving fMRI temporal resolution is critical for a research project on this topic. The authors present a detailed schematic figure (Figure 2) to help readers understand it. However, I have difficulty understanding the benefits of this method in terms of temporal resolution.
(a) In Figure 2A, if we examine a specific voxel in slice 2, the slice acquisitions occur at 0.7s, 2.7s, and 4.7s, which implies a temporal resolution of 2s rather than 0.7s. I am unclear on how the temporal resolution could be 0.7s for this specific voxel. I would prefer that the authors clarify this point further, as it would benefit readers who are not familiar with this technology.
We very much appreciate these concerns as they highlight shortcomings in our explanation of the method. Please note that the main explanation of the method (and comparison with expected HRF and FIR based methods) is done in Janssen et al. (2018, NeuroImage; see further explanations in Janssen et al., 2020). However, to make the current paper more selfcontained, we provided further explanation of the Slice-Based method in Figure 2. With respect to the specific concern of the reviewer, in the hypothetical example used in Figure 2, the temporal resolution of the voxel on slice 2 is 0.7s because it combines the acquisitions from stimulus presentations across all trials. Specifically, given the specific study parameters as outlined in Figures 2A and B, slice 2 samples the state of the brain exactly 0s after stimulus presentation on trial 1 (red color), 0.7s after stimulus presentation on trial 3 (green color), and 1.3s after stimulus presentation on trial 2 (yellow color). Thus after combining data acquisitions across these three 3 stimuli presentations, slice 2 has sampled the state of the brain at timepoints that are multiples of 0.7s starting from stimulus onset. This is why we say that the theoretical maximum temporal resolution is equal to the TR divided by the number of slices (in the example 2/3 = 0.7s, in the actual experiment 3/39 = 0.08s). In the current study we used temporal binning across timepoints to reduce the temporal resolution (to 2 seconds) and improve the tSNR.
We have updated the legend of Figure 3 to more clearly explain this issue.
(b) Even with the claim of an increased temporal resolution (0.7s), the actual data (Figure 3) still appears to have a 2s resolution. I wonder what specific benefit slice-based fMRI brings in terms of testing temporal dynamics, aside from correcting the temporal distortions that conventional fMRI exhibits.
This is a good point. In the current experiment, the TR was 3s, but we extracted the fMRI signal at 2s temporal resolution, which means an increment of 33%. In this study we did not directly compare the impact of different temporal resolutions on the efficacy of detection of network dynamics. Indeed, we agree with the reviewer that there remain many unanswered questions about the issue of temporal resolution of the extracted fMRI signal and the impact on the ability to detect fMRI network dynamics. We think that questions such as those posed by the reviewer should be addressed in future studies that are directly focused on this issue. We have updated our discussion section (page 21-22) to more clearly reflect this point of view.
(2) In task-fMRI, the hemodynamic response is usually estimated using a specific model (e.g., FIR, IR model; see Lindquist et al., 2009). These models are effective at reducing noise and collinearity across adjacent trials. The current method appears to be conducted on unmodeled BOLD time series.
(a) I am wondering how the authors avoid the issues that are typically addressed by these HRF modeling approaches. For example, if we examine the baseline period (say, -4 to 0s relative to stimulus onset), the activation of most networks does not remain around zero, which could be due to delayed influences from the previous trial. This suggests that the current time course may not be completely accurate.
We thank the reviewer for highlighting this issue. Let us start by reiterating what we stated above: That there are many issues related to BOLD signal extraction and fMRI network discovery in task-based fMRI that remain poorly understood and should be addressed in future work. Such work should explore, for example, the impact of using a FIR vs Slice-based method on the discovery of networks in task-fMRI. These studies should also investigate the impact of different types of baselines and baseline durations on the extraction of the BOLD signal and network discovery. For the present purposes, our goal was not to introduce a new technique of fMRI signal extraction, but to show that the slice-based technique, in combination with ICA, can be used to study the brain’s networks dynamics in an emotional task. In other words, while we clearly appreciate the reviewer’s concerns and have several other studies underway that directly address these concerns, we believe that such concerns are better addressed in independent research. See our discussion on page 21-22 that addresses this issue.
(b) A related question: if the authors take the spatial map of a certain network and apply a modeling approach to estimate a time series within that network, would the results be similar to the current ICA time series?
Interesting point. Typically in a modeling approach the expected HRF (e.g., the double gamma function) is fitted to the fMRI data. Importantly, this approach produces static maps of the fit between the expected HRF and the data. By contrast, model-free approaches such as FIR or slice-based methods extract the fMRI signal directly from the data without making apriori assumptions about the expected shape of the signal. These approaches do not produce static maps but instead are capable of extracting the whole-brain dynamics during the execution of a task (event-related dynamics). These data-driven approaches (FIR, SliceBased, etc) are therefore a necessary first step in the analyses of the dynamics of brain activity during a task. The subsequent step involves the analyses of these complex eventrelated brain dynamics. In the current paper we suggest that a straightforward way to do this is to use ICA which produces spatial maps of voxels with similar time courses, and hence, yields insights into the temporal dynamics of whole-brain fMRI networks. As we mentioned above, combining ICA with a high temporal resolution data-driven signal is new and there are many new avenues for research in this burgeoning new field.
(3) Human emotion should be inherently fast to ensure survival, as shown in many electrophysiology and MEG studies. For example, the dynamics of a fearful face can occur within 100ms in subcortical regions (Méndez-Bértolo et al., 2016), and general valence and arousal effects can occur as early as 200ms (e.g., Grootswagers et al., 2020; Bo et al., 2022). In contrast, the time-to-peak or onset timing in the BOLD time series spans a much larger time range due to the hemodynamic delay. fMRI findings indeed add spatial precision to our understanding of the temporal dynamics of emotion, but could the authors comment on how the current temporal dynamics supplement those electrophysiology studies that operate on much finer temporal scales?
We really like this point. One way that EEG and fMRI are typically discussed is that these two approaches are said to be complementary. While EEG is able to provide information on temporal dynamics, but not spatial localization of brain activity, fMRI cannot provide information on the temporal dynamics, but can provide insights into spatial localization. Our study most directly challenges the latter part of this statement. We believe that by using tasks that highlight “slow” cognition, fMRI can be used to reveal not only spatial but also temporal information of brain activity. The movie task that we used presumably relies on a kind of “slow” cognition that takes place on longer time scales (e.g., the construction of the meaning of the scene). Our results show that with such tasks, whole-brain networks with different temporal dynamics can be separated by ICA, at odds with the claim that fMRI is only good for spatial information. One avenue of future research would be to attempt such “slow” tasks directly with EEG and try to find the electrical correlates of the networks detected in the current study.
We hope to have answered the concerns of the reviewer.
(4) The response network shows activation as late as 15 to 20s, which is surprising. Could the authors discuss further why it takes so long for participants to generate an emotional response in the brain?
We thank the reviewer for this question. Our study design was such that there was an initial movie clip that lasted 12.5s, which was then followed by a two-alternative forced-choice decision task (including a button press, 2.5s), and finally followed by a 10s rest period. We extracted the fMRI signal across this entire 25s period (actually 28s because we also took into account some uncertainty in BOLD signal duration). Network discovery using ICA then showed various networks with distinct time courses (across the 25s period), including one network (IC2 response) that showed a peak around 21s (see Figure 3). Given the properties of the spatial map (eg., activity in primary motor areas, Figure 4), as well as the temporal properties of its timecourse (e.g., peak close to the response stage of the task), we interpreted this network as related to generating the manual response in the two-alternative forced-choice decision task. Further analyses showed that this aspect of the task (e.g., deciding the emotion of the character in the movie clip) was also sensitive to the emotional content of the earlier movie clip (Figure 6 and 7).
We have further clarified this aspect of our results (see pages 16-17). We thank the reviewer for pointing this out.
(5) Related to 4. In many theories, the emotion processing stages-including perception, valuation, and response-are usually considered iterative processes (e.g., Gross, 2015), especially in real-world scenarios. The advantage of the current paradigm is that it incorporates more dynamic elements of emotional stimuli and is closer to reality. Therefore, one might expect some degree of dynamic fluctuation within the tested brain networks to reflect those potential iterative processes (input, meaning, response). However, we still do not observe much brain dynamics in the data. In Figure 5, after the initial onset, most network activations remain sustained for an extended period of time. Does this suggest that emotion processing is less dynamic in the brain than we thought, or could it be related to limitations in temporal resolution? It could also be that the dynamics of each individual trial differ, and averaging them eliminates these variations. I would like to hear the authors' comments on this topic.
We thank the reviewer for this interesting question. We are assuming the reviewer is referring to Figure 3 and not Figure 5. Indeed what Figure 3 shows is the average time course of each detected network across all subjects and trial types. This figure therefore does not directly show the difference in dynamics between the different emotions. However, as we show in further analyses that examine how emotion modulates specific aspects of the fMRI signal dynamics (time to peak, peak value, duration) of different networks, there are differences in the dynamics of these networks depending on the emotion (Figure 6 and 7). Thus, our results show that different emotions evoked by movie clips differ in their dynamics. Obviously, generalizing this to say that in general, different emotions have different brain dynamics is not straightforward and would require further study (probably using other tasks, and other emotions). We have updated the discussion section as well as the caption of Figure 3 to better explain this issue (see also comments by reviewer 2).
(6) The activation of the default mode network (DMN), although relatively late, is very interesting. Generally, one would expect a deactivation of this network during ongoing external stimulation. Could this suggest that participants are mind-wandering during the later portion of the task?
Very good point. Indeed this is in line with our interpretation. The late activity of the default mode network could reflect some further processing of the previous emotional experience. More work is required to clarify this further in terms of reflective, mind-wandering or regulatory processing. We have updated our discussion section to better highlight this issue (see page 19).
We thank the reviewer for their really insightful comments and suggestions!
Reviewer #2 (Public review):
Summary:
This manuscript examined the neural correlates of the temporal-spatial dynamics of emotional processing while participants were watching short movie clips (each 12.5 s long) from the movie "Forrest Gump". Participants not only watched each film clip, but also gave emotional responses, followed by a brief resting period. Employing fMRI to track the BOLD responses during these stages of emotional processing, the authors found four large-scale brain networks (labeled as IC0,1,2,4) were differentially involved in emotional processing. Overall, this work provides valuable information on the neurodynamics of emotional processing.
Strengths:
This work employs a naturalistic movie watching paradigm to elicit emotional experiences. The authors used a slice-based fMRI method to examine the temporal dynamics of BOLD responses. Compared to previous emotional research that uses static images, this work provides some new data and insights into how the brain supports emotional processing from a temporal dynamics view.
Thank you!
Weaknesses:
Some major conclusions are unwarranted and do not have relevant evidence. For example, the authors seemed to interpret some neuroimaging results to be related to emotion regulation. However, there were no explicit instructions about emotional regulation, and there was no evidence suggesting participants regulated their emotions. How to best interpret the corresponding results thus requires caution.
We thank the reviewer for pointing this out. We have updated the limitations section of our Discussion section (page 20) to better qualify our interpretations.
Relatedly, the authors argued that "In turn, our findings underscore the utility of examining temporal metrics to capture subtle nuances of emotional processing that may remain undetectable using standard static analyses." While this sentence makes sense and is reasonable, it remains unclear how the results here support this argument. In particular, there were only three emotional categories: sad, happy, and fear. These three emotional categories are highly different from each other. Thus, how exactly the temporal metrics captured the "subtle nuances of emotional processing" shall be further elaborated.
This is an important point. We also discuss this limitation in the “limitations” section of our Discussion (page 20). We again thank the reviewer for pointing this out.
The writing also contained many claims about the study's clinical utility. However, the authors did not develop their reasoning nor elaborate on the clinical relevance. While examining emotional processing certainly could have clinical relevance, please unpack the argument and provide more information on how the results obtained here can be used in clinical settings.
We very much appreciate this comment. Note that we did not intend to motivate our study directly from a clinical perspective (because we did not test our approach on a clinical population). Instead, our point is that some researchers (e.g., Kuppens & Verduyn 2017; Waugh et al., 2015) have conceptualized emotional disorders frequently having a temporal component (e.g., dwelling abnormally long on negative thoughts) and that our technique could be used to examine if temporal dynamics of networks are affected in such disorders. However, as we pointed out, this should be verified in future work. We have updated our final paragraph (page 22) to more clearly highlight this issue. We thank the reviewer for pointing this out.
Importantly, how are the temporal dynamics of BOLD responses and subjective feelings related? The authors showed that "the time-to-peak differences in IC2 ("response") align closely with response latency results, with sad trials showing faster response latencies and earlier peak times". Does this mean that people typically experience sad feelings faster than happy or fear? Yet this is inconsistent with ideas such that fear detection is often rapid, while sadness can be more sustained. Understandably, the study uses movie clips, which can be very different from previous work, mostly using static images (e.g., a fearful or a sad face). But the authors shall explicitly discuss what these temporal dynamics mean for subjective feelings.
Excellent point! Our results indeed showed that sad trials had faster reaction times compared to happy and fearful trials, and that this result was reflected in the extracted time-to-peak measures of the fMRI data (see Figure 8D). To us, this primarily demonstrates that, as shown in other studies (e.g., Menon et al., 1997), that gross differences detected in behavioral measures can be directly recovered from temporal measures in fMRI data, which is not trivial. However, we do not think we are allowed to make interpretations of the sort suggested by the reviewer (and to be clear: we do not make such interpretations in the paper). Specifically, the faster reaction times on sad trials likely reflect some audio/visual aspect of the movie clips that result in faster reaction times instead of a generalized temporal difference in the subjective experience of sad vs happy/fearful emotions. Presumably the speed with which emotional stimuli influence the brain depends on the context. Perhaps future studies that examine emotional responses while controlling for the audio/visual experience could shed further light on this issue. We have updated the discussion section to address the reviewer’s concern.
We thank the reviewer for the interesting points which have certainly improved our manuscript!
Reviewer #1 (Recommendations for the authors):
Minor:
(1) Please add the unit to the y-axis in Figure 7, if applicable.
Done. We have added units.
(2) Adding a note in the legend of Figure 3 regarding the meaning of the amplitude of the timeseries would be helpful.
Done. We have added a sentence further explaining the meaning of the timecourse fluctuations.
Related references:
(1) Lindquist, M. A., Loh, J. M., Atlas, L. Y., & Wager, T. D. (2009). Modeling the hemodynamic response function in fMRI: efficiency, bias, and mis-modeling. Neuroimage, 45(1), S187-S198.
(2) Méndez-Bértolo, C., Moratti, S., Toledano, R., Lopez-Sosa, F., Martínez-Alvarez, R., Mah, Y. H., ... & Strange, B. A. (2016). A fast pathway for fear in human amygdala. Nature neuroscience, 19(8), 1041-1049.
(3) Bo, K., Cui, L., Yin, S., Hu, Z., Hong, X., Kim, S., ... & Ding, M. (2022). Decoding the temporal dynamics of affective scene processing. NeuroImage, 261, 119532.
(4) Grootswagers, T., Kennedy, B. L., Most, S. B., & Carlson, T. A. (2020). Neural signatures of dynamic emotion constructs in the human brain. Neuropsychologia, 145, 106535.
(5) Gross, J. J. (2015). The extended process model of emotion regulation: Elaborations, applications, and future directions. Psychological inquiry, 26(1), 130-137.
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public review):
Summary:
The study by Gupta et al. investigates the role of mast cells (MCs) in tuberculosis (TB) by examining their accumulation in the lungs of M. tuberculosis-infected individuals, non-human primates, and mice. The authors suggest that MCs expressing chymase and tryptase contribute to the pathology of TB and influence bacterial burden, with MC-deficient mice showing reduced lung bacterial load and pathology.
Strengths:
(1) The study addresses an important and novel topic, exploring the potential role of mast cells in TB pathology.
(2) It incorporates data from multiple models, including human, non-human primates, and mice, providing a broad perspective on MC involvement in TB.
(3) The finding that MC-deficient mice exhibit reduced lung bacterial burden is an interesting and potentially significant observation.
Weaknesses:
(1) The evidence is inconsistent across models, leading to divergent conclusions that weaken the overall impact of the study.
The strength of the study is the use of multiple models including mouse, nonhuman primate as well as human samples. The conclusions have now been refined to reflect the complexity of the disease and the use of multiple models.
(2) Key claims, such as MC-mediated cytokine responses and conversion of MC subtypes in granulomas, are not well-supported by the data presented.
To address the reviewer’ s comments we will carry out further experimentation to strengthen the link between MC subtypes and cytokine responses.
(3) Several figures are either contradictory or lack clarity, and important discrepancies, such as the differences between mouse and human data, are not adequately discussed.
We will further clarify the figures and streamline the discussions between the different models used in the study.
(4) Certain data and conclusions require further clarification or supporting evidence to be fully convincing.
We will either provide clarification or supporting evidence for some of the key conclusions in the paper.
Reviewer #2 (Public review):
Summary:
The submitted manuscript aims to characterize the role of mast cells in TB granuloma. The manuscript reports heterogeneity in mast cell populations present within the granulomas of tuberculosis patients. With the help of previously published scRNAseq data, the authors identify transcriptional signatures associated with distinct subpopulations.
Strengths:
(1) The authors have carried out a sufficient literature review to establish the background and significance of their study.
(2) The manuscript utilizes a mast cell-deficient mouse model, which demonstrates improved lung pathology during Mtb infection, suggesting mast cells as a potential novel target for developing host-directed therapies (HDT) against tuberculosis.
Weaknesses:
(1) The manuscript requires significant improvement, particularly in the clarity of the experimental design, as well as in the interpretation and discussion of the results. Enhanced focus on these areas will provide better coherence and understanding for the readers.
The strength of the study is the use of multiple models including mouse, nonhuman primate as well as human samples. The conclusions have now been refined to reflect the complexity of the disease and the use of multiple models.
(2) Throughout the manuscript, the authors have mislabelled the legends for WT B6 mice and mast cell-deficient mice. As a result, the discussion and claims made in relation to the data do not align with the corresponding graphs (Figure 1B, 3, 4, and S2). This discrepancy undermines the accuracy of the conclusions drawn from the results.
We apologize for the discrepancy which will be corrected in the revised manuscript
(3) The results discussed in the paper do not add a significant novel aspect to the field of tuberculosis, as the majority of the results discussed in Figure 1-2 are already known and are a re-validation of previous literature.
This is the first study which has used mouse, NHP and human TB samples from Mtb infection to characterize and validate the role of MC in TB. We believe the current study provides significant novel insights into the role of MC in TB.
(4) The claims made in the manuscript are only partially supported by the presented data. Additional extensive experiments are necessary to strengthen the findings and enhance the overall scientific contribution of the work.
We will either provide clarification or supporting evidence for some of the key conclusions in the paper.
Reviewer #1 (Recommendations for the authors):
In the study by Gupta et al., the authors report an accumulation of mast cells (MCs) expressing the proteases chymase and tryptase in the lungs of M. tuberculosis-infected individuals and non-human primates, as compared to healthy controls and latently infected individuals. They also MCs appear to play a pathological role in mice. Notably, MC-deficient mice show reduced lung bacterial burden and pathology during infection.
While the topic is of interest, the study is overall quite preliminary, and many conclusions are not wellsupported by the presented data. The reliance on three different models, each suggesting divergent outcomes, weakens the ability to draw definitive conclusions. Specifically, the claim that "MCs (...) mediate cytokine responses to drive pathology and promote Mtb susceptibility and dissemination during TB" is not substantiated by the data.
Major comments
(1) In human samples, the authors conclude that "While MCTCs accumulated in early immature granulomas within TB lesions, MCCs accumulated in late granulomas in TB patients" and that MCTs "likely convert first to MCTCs in early granulomas before becoming MCCs in late mature granulomas with necrotic cores." However, Figure 1B shows the opposite. Furthermore, the assertion that MCTs "convert" into MCTCs is not justified by the data.
Corrections have been made to the figures to ensure clarity for the reader. We demonstrate accumulation of tryptase-expressing MCs in healthy individuals, while the dual tryptase and chymaseexpressing MCs were seen in early granulomas, and only chymase-associated MCs were observed in late granulomas depicting more pathology of the disease. We have removed the line as advised by the reviewer.
(2) In Figure 2 I and J, the panels do not demonstrate co-expression of chymase and tryptase in clusters 0, 1, and 3 in PTB samples, which contradicts the histology data. This discrepancy is left unaddressed and raises concerns about the conclusions drawn from Figures 1 and 2.
We thank the reviewer for pointing this out. We revisited the data and now show the coexpression of the dual expressing cells in the data (Figure 2H). This discrepancy stemmed from the crossspecies nature of the dataset. It turns out the there is a considerable diversity in sequence similarity and tryptase function between human and NHPs (Trivedi et al., 2007). We explain this in the section now (line 313-364). Briefly, while humans express TPSG1 (encoding tryptase) and TPSD1 (encoding tryptase) and have the same gene name in NHP, the gene name for more widely expressed TPSAB1(encoding / tryptase) is different for NHP and the gene names are not shared as they are still predicated putative protein. The putative genes from NHP that map to human TPSAB1 is LOC699599 for M. mulatta and LOC102139613 for M. fasicularis, respectively. Thus, looking for TPSAB1 gene yielded no result in our previous analysis but examining these orthologous gene names, now phenocopy the results we see in the histology data. To strengthen our findings, we have now analyzed an additional single-cell dataset from the lungs of NHP M. fasicularis (Figure 2J-L) and found the co-expression of chymase and tryptase, adding an important validation to our histological findings.
(3) Figure 2 serves more as a resource and contributes little to the core findings of the study. It might be better suited as supplementary material.
We thank the reviewer for the suggestion; however, we believe that Figure 2 serves as an independent validation in a different species (NHP), showing heterogeneity in MCs across species in a TB model. The figure adds value as there are only a handful of studies (Tauber et al., 2023, Derakhshan et al., 2022, Cildir et al., 2021) but none in TB, describing MCs at single cell level, of which one is published from our group showing MC cluster in Mtb infected macaques (Esaulova et al., 2021). We feel strongly that dissecting MCs as specifically done here provides an important insight into the transcriptional heterogeneity of these cells linked to disease states. We have also added an additional NHP lung single cell dataset (Gideon et al., 2022) to complement our analysis, thus adding another validation, strengthening these findings. So, we believe in retaining the figure as an integral part of the main paper.
(4) In lines 275-277, the data referenced should be shown to support the claims.
We thank the reviewer for the suggestion. The text originally noted by the reviewer now appears in the revised manuscript at line 370-372 and the corresponding data has now been included as supplementary Figure S3.
(5) In Figure 3B, the difference between the two mouse strains becomes non-significant by day 150 pi, weakening the overall conclusion that MCs contribute to the bacterial burden.
At 100 dpi, MC-deficient mice exhibit lower Mtb CFU in both the lung and spleen, indicating improved protection. By 150 dpi, lung CFU differences are no longer significant; however, dissemination to the spleen remains reduced in MC-deficient mice. Thus, the overall conclusion that MCs contribute to increased bacterial burden remains valid, particularly with respect to dissemination. This conclusion is further supported by new data showing that adoptive transfer of MCs into B6 Mtb-infected mice increased Mtb dissemination to the spleen (Figure 5E).
(6) Figures 3D and E are not particularly convincing.
Figures 3D and 3E illustrate lung inflammation in MC-deficient mice compared to wild-type which more distinctly show that MC-deficient mice exhibit significantly less inflammation at 150 dpi, supporting the role of MCs in driving lung.
(7) In Figures 4 and S3, the color coding in panels A-F appears incorrect but is accurate in G. This inconsistency is confusing.
We thank the reviewer for noting this. The color coding has been corrected to ensure consistency across all figures.
(8) In the mouse model, MCs seem to disappear during infection, in contrast to observations in human and macaque samples. This discrepancy is not discussed in the paper.
We thank the reviewer for this important observation. In response, we performed a new analysis of lung MCs at baseline in wild-type and MC-deficient mice. Our data show that naïve wild-type lungs contain a small population of MCs, which is further reduced in MC-deficient mice. Following Mtb infection, MCs progressively accumulate in wild-type mice, whereas this accumulation is significantly impaired in MC-deficient mice. These new data are now included in Figure (Figure 4A) and also updated in the text (line 395-403).
(9) In lines 306-307, data should be shown to support the claims.
We thank the reviewer for the suggestion. The text originally noted by the reviewer now appears in the revised manuscript at line 399-400 and the corresponding data has now been included as supplementary Figure S4.
Minor comments
(1) What does "granuloma-associated" cells mean in samples from healthy controls?
We thank the reviewer for this point. The language has been revised to accurately refer to cells in the lung parenchyma in the Figure 1, rather than “granuloma associated” cells.
(2) In line 229, it is unclear what "these cells" refers to.
The phrase “these cells” refers to tryptase-expressing mast cells. This has now been clarified in the revised manuscript (line 276-277).
(3) The citation of Figure 3A in lines 284-285 is misplaced in the text and should be corrected.
The figure citation has been corrected in the text in the revised manuscript (lines 376-379).
Reviewer #2 (Recommendations for the authors):
(1) The data presented in Figure 1 seems to be a re-validation of the already known aspects of mast cells in TB granulomas. While distinct roles for mast cells in regulating Mtb infection have been reported, the manuscript appears to be a failed opportunity to characterize the transcriptional signatures of the distinct subsets and identify their role in previously reported processes towards controlling TB disease progression.
We thank the reviewer for the insight. While it was not our intent to investigate the bulk transcriptome, owing to the high number of cells required to get enough RNA for transcriptomic sequencing, it is technically challenging due to the low abundance of mast cells during TB infection (Figure 2). The motivation for Figure 2, that we utilized a more sensitive transcriptomic analysis to find the different transcriptional states in the distinct TB disease states. We believe that this analysis captures the essence of what the reviewer and provides meaningful insights into mast cell heterogeneity during TB.
(2) The experiments lack uniformity with respect to the strains of Mtb used for experimentation. For eg: Mtb strain HN878 was used for aerosol infection of mice while Mtb CDC1551 was used for macaques. If there were experimental constraints with respect to the choice, the same should be mentioned.
We thank the reviewer for this comment. The Mtb strain usage has been consistent within each species: HN878 for mice and CDC1551 for non-human primates (NHPs), in line with prior studies from our lab. The species-specific choice reflects the differences in pathogenicity of these strains in mice versus NHPs. CDC1551, which exhibits lower virulence, allows the development of a macaque model that recapitulates human latent to chronic TB when administered via aerosol at low to moderate doses (Kaushal et al., 2015; Sharan et al., 2021; Singh et al., 2025). In contrast, the more virulent HN878 strain leads to severe disease and high mortality in NHPs and is therefore not suitable for these models. Using CDC1551 in macaques provides a controlled and clinically relevant platform to study immunological and pathophysiological mechanisms of TB, justifying its use in the current study. This explanation has now been added to the manuscript method section (lines 109-114).
(3) Line 84- 85, the authors state that "Chymase positive MCs contribute to immune pathology and reduced Mtb control". Previous reports including Garcia-Rodriguez et al., 2021 associate high MCTCs with improved lung function. Additionally, in the macaques model of latent TB infection reported in the manuscript, the number of chymase-expressing MCs seems to significantly decrease. The authors should justify the same.
We thank the reviewer for this comment. In Garcia-Rodriguez et al., 2021, chymase-expressing MCs accumulate in fibrotic lung lesions. Fibrosis is a result of excessive inflammation in TB infection and is associated with lung damage. Similarly, in idiopathic pulmonary fibrosis, higher density and percentage of chymase-expressing MCs correlate positively with fibrosis severity (Andersson et al., 2011). In our study, although fibrosis was not directly assessed, chymase-positive MCs increased in late lung granulomas, consistent with advanced inflammatory disease. Therefore, our conclusion that chymaseproducing MCs contribute to lung pathology is justified and aligns with prior observations.
(4) The manuscript would benefit from a brief description of the experimental conditions for the previously published scRNAseq data used in the current study.
We thank the reviewer for the suggestion, and the information has been included in the final manuscript (lines 294-297) and represented as Figure 2A.
(5) The authors have not mentioned the criteria used to categorize early and late granulomas in TB patients. A lucid description of the same is necessary.
Based on reviewer’s comment the detailed categorization of early and late granulomas in TB patients is now included in the revised manuscript (line 256-260). Early granulomas: Discrete conglomerates of immune cells and resident stromal cells with defined borders and absence of central necrosis, and Late granulomas: Large and dense clusters of immune cells and resident cells with an evident necrotic center containing bacteria and dead neutrophils and lymphocytic infiltrating cells on the periphery of the necrotic center. MCs were measured in the periphery and inside early granulomas, while in the late granulomas, they were mainly quantified in the periphery.
(6) The authors mention that "While MCTCs accumulated in early immature granulomas within TB lesions, MCCs accumulated in late granulomas in TB patients". While this is evident from the representative, the quantification in Figure 1B seems to indicate otherwise.
We thank the reviewer for pointing this out. The labeling in the quantitative analysis shown in Figure 1B has been corrected in the revised manuscript to accurately reflect the accumulation of MC<sub>TC</sub>s in early granulomas and MC<sub>C</sub>s in late granulomas.
(7) The labelling followed in Figures 3, 4 and S2 do not match with the discussion. Such errors should be rectified to minimize any ambiguity within the text of the manuscript.
We thank the reviewer for noting this. The color coding has been corrected to ensure consistency across all figures.
(8) The mast cell deficient mice model has a differential number of immune cells at the site of granuloma as reported in the manuscript. This could contribute to the altered mycobacterial survival and inflammation cytokine production in the lung and hence might not be a direct effect of mast cell depletion. The authors can consider reconstituting mast cell populations to analyze the mast cell function.
We thank the reviewers for this suggestion. In the revised manuscript, we have adoptively transferred MCs into WT mice before Mtb challenge to assess if this would increase inflammation and Mtb CFU in the lung and spleen. Our results show that while lung inflammation was not impacted, we found that the dissemination to the spleen and the frequency of neutrophils in the lung were increased in WT mice that received MCs (Figure 5, lines 429-443).
(9) Line 295- 297, the authors state "MCs continued to accumulate in the lung up to 100 dpi in CgKitWsh mice, following which the MC numbers decreased at later stages". However, the quantification in Figure 4A does not reflect the same. This should be addressed.
In response to the reviewers' comments, we conducted a new analysis of lung MCs at baseline, comparing wild-type and MC-deficient mice. The revised data show that MC-deficient mice have fewer mast cells at baseline compared to B6 mice. Furthermore, mast cell numbers increase during infection, peaking at 100 days post-infection (dpi) and subsequently stabilize by 150 dpi. The revised data has been included in Figure 4A and text line 395-403.
(10) Additionally, while the scRNAseq data reflects a lower production of TNF in pulmonary TB granulomas, the mice deficient in mast cells are discussed to have a lower production of proinflammatory cytokines.
Mast cells increasing and contributing to the TB pathogenesis is the theme of the paper and as such we see and increase in the IFNG pathway genes and similar reduction in the production of pro- inflammatory cytokines. The relative decrease in the TNF pathway gene expression can be reconciled by the fact that less TNF gene expression in PTB could also represent loss of Mtb control and increased pathogenesis (Yuk et al., 2024), which is maintained in the LTBI/HC clusters. Higher bacterial burden of Mtb can also decrease the host TNF production, which is in line with what we observe here (Olsen et al., 2016, Reed et al., 2004, Kurtz et al., 2006).
(11) The authors have not annotated Figure 2 I and J in the text while describing their results and interpretation.
We thank the reviewer for noting this and the figure 2 has been revised and the results as pointed out have been added to the revised manuscript.
(12) In line 284, the authors have discussed the results pertaining to Figure 3B, however, mentioned it as Figure 3A in the text.
We thank the reviewer for noting this and the corrections have been made in the revised manuscript (lines 379-384).
References
ANDERSSON, C. K., ANDERSSON-SJOLAND, A., MORI, M., HALLGREN, O., PARDO, A., ERIKSSON, L., BJERMER, L., LOFDAHL, C. G., SELMAN, M., WESTERGREN-THORSSON, G. & ERJEFALT, J. S. 2011. Activated MCTC mast cells infiltrate diseased lung areas in cystic fibrosis and idiopathic pulmonary fibrosis. Respir Res, 12, 139.
CILDIR, G., YIP, K. H., PANT, H., TERGAONKAR, V., LOPEZ, A. F. & TUMES, D. J. 2021. Understanding mast cell heterogeneity at single cell resolution. Trends Immunol, 42, 523-535.
DERAKHSHAN, T., BOYCE, J. A. & DWYER, D. F. 2022. Defining mast cell differentiation and heterogeneity through single-cell transcriptomics analysis. J Allergy Clin Immunol, 150, 739-747.
ESAULOVA, E., DAS, S., SINGH, D. K., CHORENO-PARRA, J. A., SWAIN, A., ARTHUR, L., RANGEL-MORENO, J., AHMED, M., SINGH, B., GUPTA, A., FERNANDEZ-LOPEZ, L. A., DE LA LUZ GARCIA-HERNANDEZ, M., BUCSAN, A., MOODLEY, C., MEHRA, S., GARCIA-LATORRE, E., ZUNIGA, J., ATKINSON, J., KAUSHAL, D., ARTYOMOV, M. N. & KHADER, S. A. 2021. The immune landscape in tuberculosis reveals populations linked to disease and latency. Cell Host Microbe, 29, 165-178 e8.
GARCIA-RODRIGUEZ, K. M., BINI, E. I., GAMBOA-DOMINGUEZ, A., ESPITIA-PINZON, C. I., HUERTA-YEPEZ, S., BULFONE-PAUS, S. & HERNANDEZ-PANDO, R. 2021. Differential mast cell numbers and characteristics in human tuberculosis pulmonary lesions. Sci Rep, 11, 10687.
GIDEON, H. P., HUGHES, T. K., TZOUANAS, C. N., WADSWORTH, M. H., 2ND, TU, A. A., GIERAHN, T. M., PETERS, J. M., HOPKINS, F. F., WEI, J. R., KUMMERLOWE, C., GRANT, N. L., NARGAN, K., PHUAH, J. Y., BORISH, H. J., MAIELLO, P., WHITE, A. G., WINCHELL, C. G., NYQUIST, S. K., GANCHUA, S. K. C., MYERS, A., PATEL, K. V., AMEEL, C. L., COCHRAN, C. T., IBRAHIM, S., TOMKO, J. A., FRYE, L. J., ROSENBERG, J. M., SHIH, A., CHAO, M., KLEIN, E., SCANGA, C. A., ORDOVAS-MONTANES, J., BERGER, B., MATTILA, J. T., MADANSEIN, R., LOVE, J. C., LIN, P. L., LESLIE, A., BEHAR, S. M., BRYSON, B., FLYNN, J. L., FORTUNE, S. M. & SHALEK, A. K. 2022. Multimodal profiling of lung granulomas in macaques reveals cellular correlates of tuberculosis control. Immunity, 55, 827846 e10.
KAUSHAL, D., FOREMAN, T. W., GAUTAM, U. S., ALVAREZ, X., ADEKAMBI, T., RANGEL-MORENO, J., GOLDEN, N. A., JOHNSON, A. M., PHILLIPS, B. L., AHSAN, M. H., RUSSELL-LODRIGUE, K. E., DOYLE, L. A., ROY, C. J., DIDIER, P. J., BLANCHARD, J. L., RENGARAJAN, J., LACKNER, A. A., KHADER, S. A. & MEHRA, S. 2015. Mucosal vaccination with attenuated Mycobacterium tuberculosis induces strong central memory responses and protects against tuberculosis. Nat Commun, 6, 8533.
KURTZ, S., MCKINNON, K. P., RUNGE, M. S., TING, J. P. & BRAUNSTEIN, M. 2006. The SecA2 secretion factor of Mycobacterium tuberculosis promotes growth in macrophages and inhibits the host immune response. Infect Immun, 74, 6855-64.
OLSEN, A., CHEN, Y., JI, Q., ZHU, G., DE SILVA, A. D., VILCHEZE, C., WEISBROD, T., LI, W., XU, J., LARSEN, M., ZHANG, J., PORCELLI, S. A., JACOBS, W. R., JR. & CHAN, J. 2016. Targeting Mycobacterium tuberculosis Tumor Necrosis Factor Alpha-Downregulating Genes for the Development of Antituberculous Vaccines. mBio, 7.
REED, M. B., DOMENECH, P., MANCA, C., SU, H., BARCZAK, A. K., KREISWIRTH, B. N., KAPLAN, G. & BARRY, C. E., 3RD 2004. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature, 431, 84-7.
SHARAN, R., SINGH, D. K., RENGARAJAN, J. & KAUSHAL, D. 2021. Characterizing Early T Cell Responses in Nonhuman Primate Model of Tuberculosis. Front Immunol, 12, 706723.
SINGH, D. K., AHMED, M., AKTER, S., SHIVANNA, V., BUCSAN, A. N., MISHRA, A., GOLDEN, N. A., DIDIER, P. J., DOYLE, L. A., HALL-URSONE, S., ROY, C. J., ARORA, G., DICK, E. J., JR., JAGANNATH, C., MEHRA, S., KHADER, S. A. & KAUSHAL, D. 2025. Prevention of tuberculosis in cynomolgus macaques by an attenuated Mycobacterium tuberculosis vaccine candidate. Nat Commun, 16, 1957.
TAUBER, M., BASSO, L., MARTIN, J., BOSTAN, L., PINTO, M. M., THIERRY, G. R., HOUMADI, R., SERHAN, N., LOSTE, A., BLERIOT, C., KAMPHUIS, J. B. J., GRUJIC, M., KJELLEN, L., PEJLER, G., PAUL, C., DONG, X., GALLI, S. J., REBER, L. L., GINHOUX, F., BAJENOFF, M., GENTEK, R. & GAUDENZIO, N. 2023. Landscape of mast cell populations across organs in mice and humans. J Exp Med, 220.
TRIVEDI, N. N., TONG, Q., RAMAN, K., BHAGWANDIN, V. J. & CAUGHEY, G. H. 2007. Mast cell alpha and beta tryptases changed rapidly during primate speciation and evolved from gamma-like transmembrane peptidases in ancestral vertebrates. J Immunol, 179, 6072-9.
YUK, J. M., KIM, J. K., KIM, I. S. & JO, E. K. 2024. TNF in Human Tuberculosis: A Double-Edged Sword. Immune Netw, 24, e4.
Reviewer #1 (Public review):
Summary:
In this descriptive study, Tateishi et al. report a Tn-seq based analysis of genetic requirements for growth and fitness in 8 clinical strains of Mycobacterium intracellulare Mi), and compare the findings with a type strain ATCC13950. The study finds a core set of 131 genes that are essential in all nine strains, and therefore are reasonably argued as potential drug targets. Multiple other genes required for fitness in clinical isolates have been found to be important for hypoxic growth in the type strain.
Strengths:
The study has generated a large volume of Tn-seq datasets of multiple clinical strains of Mi from multiple growth conditions, including from mouse lungs. The dataset can serve as an important resource for future studies on Mi, which despite being clinically significant remains a relatively understudied species of mycobacteria.
Weaknesses:
The primary claim of the study that the clinical strains are better adapted for hypoxic growth is yet to be comprehensively investigated. However, this reviewer thinks such an investigation would require a complex experimental design and perhaps forms an independent study.
Comments on revisions:
The revised manuscript has responded to the previous concerns of the reviewers, albeit modestly. The overemphasis on hypoxic adaptation of the clinical isolates persist as a key concern in the paper. The authors have compared the growth-curve of each of the clinical and ATCC strains under normal and hypoxic conditions (Fig. 8), but don't show how mutations in some of the genes identified in Tn-seq would impact the growth phenotype under hypoxia. They largely base their arguments on previously published results.
As I mentioned previously, the paper will be better without over-interpreting the TnSeq data in the context of hypoxia.
Other points:
The y-axis legends of plots in Fig.8c are illegible.
The statements in lines 376-389 are convoluted and need some explanation. If the clinical strains enter the log phase sooner than ATCC strain under hypoxia, then how come their growth rates (fig. 8c) are lower? Aren't they are expected to grow faster?
Author response:
The following is the authors’ response to the previous reviews.
Reviewer #1 (Public review):
Summary:
In this descriptive study, Tateishi et al. report a Tn-seq based analysis of genetic requirements for growth and fitness in 8 clinical strains of Mycobacterium intracellulare Mi), and compare the findings with a type strain ATCC13950. The study finds a core set of 131 genes that are essential in all nine strains, and therefore are reasonably argued as potential drug targets. Multiple other genes required for fitness in clinical isolates have been found to be important for hypoxic growth in the type strain.
Strengths:
The study has generated a large volume of Tn-seq datasets of multiple clinical strains of Mi from multiple growth conditions, including from mouse lungs. The dataset can serve as an important resource for future studies on Mi, which despite being clinically significant remains a relatively understudied species of mycobacteria.
Thank you for the comment on the significance of our manuscript on the basic research of non-tuberculous mycobacteria.
Weaknesses:
The primary claim of the study that the clinical strains are better adapted for hypoxic growth is yet to be comprehensively investigated. However, this reviewer thinks such an investigation would require a complex experimental design and perhaps forms an independent study
Thank you for the comment on the issue of the claim of better adaptation for hypoxic growth in the clinical strains being not completely revealed. We agree the reviewer’s comment that comprehensive investigation of adaptation for hypoxic growth in the clinical strains should be a future project in terms of the complexity of an experimental design.
Reviewer #4 (Public review):
Summary:
In this study Tateishi et al. used TnSeq to identify 131 shared essential or growth defect-associated genes in eight clinical MAC-PD isolates and the type strain ATCC13950 of Mycobacterium intracellulare which are proposed as potential drug targets. Genes involved in gluconeogenesis and the type VII secretion system which are required for hypoxic pellicle-type biofilm formation in ATCC13950 also showed increased requirement in clinical strains under standard growth conditions. These findings were further confirmed in a mouse lung infection model.
Strengths:
This study has conducted TnSeq experiments in reference and 8 different clinical isolates of M. intracellulare thus producing large number of datasets which itself is a rare accomplishment and will greatly benefit the research community
Thank you for the comment on the significance of our manuscript on the basic research of non-tuberculous mycobacteria.
Weaknesses:
(1) A comparative growth study of pure and mixed cultures of clinical and reference strains under hypoxia will be helpful in supporting the claim that clinical strains adapt better to such conditions. This should be mentioned as future directions in the discussion section along with testing the phenotype of individual knockout strains.
Thank you for the comment on the idea of a comparative growth assay of pure and mixed cultures of clinical and reference strains under hypoxia. We appreciate the idea that showing the phenomenon of advantage of bacterial growth of the clinical strains under hypoxia in mixed culture with the ATCC strain would be important to strengthen the claim of better adaptation for hypoxic growth in the clinical strains. However, co-culture conditions introduce additional variables, including inter-strain competition or synergy, which can obscure the specific contributions of hypoxic adaptation in each strain. Therefore, we consider that our current approach using monoculture growth curves under defined oxygen conditions offers a clearer interpretation of strain-specific hypoxic responses.
Following the comment, we have added the mention of the mixed culture experiment and the growth assay using individual knockout strains as future directions (page 35 lines 614-632 in the revised manuscript).
“We have provided the data suggesting the preferential hypoxic adaptation in clinical strains compared to the ATCC type strain by the growth assay of individual strains. To strengthen our claim, several experiments are suggested including mixed culture experiments of clinical and reference strains under hypoxia. However, co-culture conditions introduce additional variables, including inter-strain competition or synergy, which can obscure the specific contributions of hypoxic adaptation in each strain. Therefore, we took the current approach using monoculture growth curves under defined oxygen conditions, which offers a clearer interpretation of strainspecific hypoxic responses. Furthermore, one of the limitations of this study is the lack of validation of TnSeq results with individual gene knockouts. Contrary to the case of Mtb, the technique of constructing knockout mutants of slow-growing NTM including M. intracellulare has not been established long time. We have just recently succeeded in constructing the vector plasmids for making knockout mutants of M intracellulare (Tateishi. Microbiol Immunol. 2024). Growth assay of individual knockout strains of genes showing increased genetic requirements such as pckA, glpX, csd, eccC5 and mycP5 in the clinical strains is suggested to provide the direct involvement of these genes on the preferential hypoxic adaptation in clinical strains. We have a future plan to construct knockout mutants of these genes to confirm the involvement of these genes on preferential hypoxic adaptation.”
Reference
Tateishi, Y., Nishiyama, A., Ozeki, Y. & Matsumoto, S. Construction of knockoutmutants in Mycobacterium intracellulare ATCC13950 strain using a thermosensitive plasmid containing negative selection marker rpsL<sup>+</sup>. Microbiol Immunol 68, 339-347 (2024).
(2) Authors should provide the quantitative value of read counts for classifying a gene as "essential" or "non-essential" or "growth-defect" or "growthadvantage". Merely mentioning "no insertions in all or most of their TA sites" or "unusually low read counts" or "unusually high low read counts" is not clear
Thank you for the comment on the issue of not providing the quantitative value of read counts for classifying the gene essentiality. In this study, we used an Hidden Markov Model (HMM) to predict gene essentiality. The HMM does not classify the 4 gene essentiality uniquely by the quantitative number of read counts but uses a probabilistic model to estimate the state at each TA based on the read counts and consistency with adjacent sites (Ioerger. Methods Mol Biol 2022).
The HMM uses consecutive data of read counts and calculates transition probability for predicting gene essentiality across the genome. The HMM allows for the clustering of insertion sites into distinct regions of essentiality across the entire genome in a statistically rigorous manner, while also allowing for the detection of growth-defect and growth-advantage regions. The HMM can smooth over individual outlier values (such as an isolated insertion in any otherwise empty region, or empty sites scattered among insertion in a non-essential region) and make a call for a region/gene that integrates information over multiple sites. The gene-level calls are made based on the majority call among the TA sites within each gene. The HMM automatically tunes its internal parameters (e.g. transition probabilities) to the characteristics of the input datasets (saturation and mean insertion counts) and can work over a broad range of saturation levels (as low as 20%) (DeJesus. BMC Bioinformatics 2013). Thus, HMM can represent the more nuanced ways the growth of an organism might be affected by the disruption of its genes (https://orca1.tamu.edu/essentiality/Tn-HMM/index.html)
Thus, the prediction of gene essentiality by the HMM does not rely on the quantitative threshold of Tn insertion reads independently at each TA site, but rather it is the most probable states for the whole sequence taken together (computed using Vitebri algorithm). Of the statistical methods, the HMM is a standard method for predicting gene essentiality in TnSeq (Ioerger TR. Methods Mol Biol. 2022) since a substantial number of TnSeq studies adopt this method for predicting gene essentiality (Akusobi. mBio 2025, DeJesus. mBio 2017, Dragset mSystems 2019, Mendum. BCG Genomics 2019). The HMM can be applied in many bioinformatics fields such as profiling functional protein families, identifying functional domains, sequence motif discoveries and gene prediction.
Taken together, we do not have the quantitative value of read counts for classifying gene essentiality by an HMM because the statistical methods for predicting gene essentiality do not uniquely use the quantitative value of read counts but use the transition of the read counts across the genome.
Reference
Ioerger TR. Analysis of Gene Essentiality from TnSeq Data Using Transit. Methods Mol Biol. 2022 ; 2377: 391–421. doi:10.1007/978-1-0716-1720-5_22.
DeJesus MA, Ioerger TR (2013) A Hidden Markov Model for identifying essential and 5 growth-defect regions in bacterial genomes from transposon insertion sequencing data. BMC Bioinformatics 14:303 [PubMed: 24103077]
Website by Ioerger: A Hidden Markov Model for identifying essential and growthdefect regions in bacterial genomes from transposon insertion sequencing data. https://orca1.tamu.edu/essentiality/Tn-HMM/index.html
Akusobi. C. et al. Transposon-sequencing across multiple Mycobacterium abscessus isolates reveals significant functional genomic diversity among strains. mBio 6, e0337624 (2025).
DeJesus, M.A. et al. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio 8, e02133-16 (2017).
Dragset, M.S., et al. Global assessment of Mycobacterium avium subsp. hominissuis genetic requirement for growth and virulence. mSystems 4, e00402-19 (2019). Mendum T.A., et al. Transposon libraries identify novel Mycobacterium bovis BCG genes involved in the dynamic interactions required for BCG to persist during in vivo passage in cattle. BMC Genomics 20, 431 (2019)
(3) One of the major limitations of this study is the lack of validation of TnSeq results with individual gene knockouts. Authors should mention this in the discussion section.
Thank you for the comment on the issue of the lack of validation of TnSeq results by using individual knockout mutants. We agree that the lack of validation of TnSeq results is one of the limitations of this study. We have just recently succeeded in constructing the vector plasmids for making knockout mutants of M intracellulare (Tateishi. Microbiol Immunol. 2024). We will proceed to the validation experiment of TnSeq-hit genes by constructing knockout mutants.
Following the comment, we have added the description in the Discussion (page 35 lines 622-632 in the revised manuscript) as follows: “Furthermore, one of the limitations of this study is the lack of validation of TnSeq results with individual gene knockouts. Contrary to the case of Mtb, the technique of constructing knockout mutants of slow-growing NTM including M. intracellulare has not been established long time. We have just recently succeeded in constructing the vector plasmids for making knockout mutants of M intracellulare (Tateishi. Microbiol Immunol 2024). Growth assay of individual knockout strains of genes showing increased genetic requirements such as pckA, glpX, csd, eccC5 and mycP5 in the clinical strains is suggested to provide the direct involvement of these genes on the 6 preferential hypoxic adaptation in clinical strains. We have a future plan to construct knockout mutants of these genes to confirm the involvement of these genes on preferential hypoxic adaptation.”
Reference
Tateishi, Y., Nishiyama, A., Ozeki, Y. & Matsumoto, S. Construction of knockout mutants in Mycobacterium intracellulare ATCC13950 strain using a thermosensitive plasmid containing negative selection marker rpsL + . Microbiol Immunol 68, 339-347 (2024).
Reviewer #5 (Public review):
Summary:
In the research article, "Functional genomics reveals strain-specific genetic requirements conferring hypoxic growth in Mycobacterium intracellulare" Tateshi et al focussed their research on pulmonary disease caused by Mycobacterium avium-intracellulare complex which has recently become a major health concern. The authors were interested in identifying the genetic requirements necessary for growth/survival within host and used hypoxia and biofilm conditions that partly replicate some of the stress conditions experienced by bacteria in vivo. An important finding of this analysis was the observation that genes involved in gluconeogenesis, type VII secretion system and cysteine desulphurase were crucial for the clinical isolates during standard culture while the same were necessary during hypoxia in the ATCC type strain.
Strength of the study:
Transposon mutagenesis has been a powerful genetic tool to identify essential genes/pathways necessary for bacteria under various in vitro stress conditions and for in vivo survival. The authors extended the TnSeq methodology not only to the ATCC strain but also to the recently clinical isolates to identify the differences between the two categories of bacterial strains. Using this approach they dissected the similarities and differences in the genetic requirement for bacterial survival between ATCC type strains and clinical isolates. They observed that the clinical strains performed much better in terms of growth during hypoxia than the type strain. These in vitro findings were further extended to mouse 7 infection models and similar outcomes were observed in vivo further emphasising the relevance of hypoxic adaptation crucial for the clinical strains which could be explored as potential drug targets.
Thank you for the comment on the significance of our manuscript on the basic research of non-tuberculous mycobacteria.
Weakness:
The authors have performed extensive TnSeq analysis but fail to present the data coherently. The data could have been well presented both in Figures and text. In my view this is one of the major weakness of the study.
Thank you for the comment on the issue of data presentation. Our point-by-point response to the Reviewer’s comments is shown below.
Reviewer #5 (Recommendations for the authors):
Major comments:
(1) The result section could have been better organized by splitting into multiple sections with each section focusing on a particular aspect.
Thank you for the comment on the organization of the section. We have split into multiple sections with each section focusing on a particular aspect as follows:
(1) Common essential and growth-defect-associated genes representing the genomic diversity of M. intracellulare strains (page 6 lines 102-103 in the revised manuscript)
(2) The sharing of strain-dependent and accessory essential and growth-defectassociated genes with genes required for hypoxic pellicle formation in the type strain ATCC13950 (page 8 lines 129-131 in the revised manuscript)
(3) Partial overlap of the genes showing increased genetic requirements in clinical MAC-PD strains with those required for hypoxic pellicle formation in the type strain ATCC13950 (page 9 lines 151-153 in the revised manuscript)
(4) Minor role of gene duplication on reduced genetic requirements in clinical MACPD strains (page 11 lines 184-185 in the revised manuscript)
(5) Identification of genes in the clinical MAC-PD strains required for mouse lung infection (page 12 lines 210-211 in the revised manuscript) 8
(6) Effects of knockdown of universal essential or growth-defect-associated genes in clinical MAC-PD strains (page 17 lines 305-306 in the revised manuscript)
(7) Differential effects of knockdown of accessory/strain-dependent essential or growth-defect-associated genes among clinical MAC-PD strains (page 19 lines 325- 326 in the revised manuscript)
(8) Preferential hypoxic adaptation of clinical MAC-PD strains evaluated with bacterial growth kinetics (page 21 lines 365-366 in the revised manuscript)
(9) The pattern of hypoxic adaptation not simply determined by genotypes (page 22 line 386 in the revised manuscript)
(2) The different strains that were used in the study, how they were isolated and some information on their genotypes could have been mentioned in brief in the main text and a table of different strains included as a supplementary table
Thank you for the comment on the information on the clinically isolated strains used in this study. All clinical strains were isolated from sputum of MAC-PD patients (Tateishi. BMC Microbiol. 2021, BMC Microbiol. 2023). Sputum samples were treated by the standard method for clinical isolation of mycobacteria with 0.5% (w/v) Nacetyl-L-cysteine and 2% (w/v) sodium hydroxide and plated on 7H10/OADC agar plates. Single colonies were picked up for use in experiments as isolated strains.
Following the comment, we have added the description on the information of the strains (page 37 lines 652-660 in the revised manuscript). “All eleven clinical strains from MAC-PD patients in Japan were isolated from sputum (Tateishi. BMC Microbiol 2021, BMC Microbiol 2023). Sputum samples were treated by the standard method for clinical isolation of mycobacteria with 0.5% (w/v) N-acetyl-L-cysteine and 2% (w/v) sodium hydroxide and plated on 7H10/OADC agar. Single colonies were picked up for use in experiments as isolated strains. Of these strains, ATCC13950, M.i.198, M.i.27, M018, M005 and M016 belong to the typical M. intracellulare (TMI) genotype and M001, M003, M019, M021 and MOTT64 belong to the M. paraintracellulare-M. indicus pranii (MP-MIP) genotype (Fig. 1, new Supplementary Table 1)”
Moreover, we have added the Supplementary Table showing the information on genotypes of each strain and the purpose of the use of study strains as new Supplementary Table 1
References
Tateishi, Y. et al. Comparative genomic analysis of Mycobacterium intracellulare: implications for clinical taxonomic classification in pulmonary Mycobacterium aviumintracellulare complex disease. BMC Microbiol 21, 103 (2021). Tateishi, Y. et al. Virulence of Mycobacterium intracellulare clinical strains in a mouse model of lung infection - role of neutrophilic inflammation in disease severity. BMC Microbiol 23, 94 (2023).
(3) As stated by the previous reviews, an explanation for the variation in the Tn insertion across different strains has not been provided and how they derive conclusions when the Tn frequency was not saturating.
Thank you for the comment on how to predict gene essentiality from our TnSeq data under the variation in the Tn insertion reads with suboptimal levels of saturation without reaching full saturation of Tn insertion.
As for the overcome of the Tn insertion variation, we normalized data by using Beta-Geometric correction (BGC), a non-linear normalization method. BGC normalizes the datasets to fit an “ideal” geometric distribution with a variable probability parameter ρ, and BGC improves resampling by reducing the skew. On TRANSIT software, we set the replicate option as Sum to combine read counts. And we normalized the datasets by Beta-Geometric correction (BGC) to reduce variabilities and performed resampling analysis by using normalized datasets to compare the genetic requirements between strains.
Following the comment, we have explained the variation in the Tn insertion across different strains in the manuscript (pages 39-40, lines 700-708 in the revised manuscript). “The number of Tn insertion in our datasets varied between 1.3 to 5.8 million among strains. To reduce the variation in the Tn insertion across strains, we adopt a non-linear normalization method, Beta-Geometric correction (BGC). BGC normalizes the datasets to fit an “ideal” geometric distribution with a variable probability parameter ρ, and BGC improves resampling by reducing the skew. On TRANSIT software, we set the replicate option as Sum to combine read counts. And we normalized the datasets by BGC and performed resampling analysis by using normalized datasets to compare the genetic requirements between strains.”
As for the issue of saturation levels of Tn insertion in our Tn mutant libraries, we made a description in the Discussion in the 1st version of the revised manuscript (pages 33-35 lines 592-613 in the 2nd version of the revised manuscript). The saturation of our Tn mutant libraries became 62-79% as follows: ATCC13950: 67.6%, M001: 72.9%, M003: 63.0%, M018: 62.4%, M019: 74.5%, M.i.27: 76.6%, M.i.198: 68.0%, MOTT64: 77.6%, M021: 79.9% by combining replicates. That is, we calculated gene essentiality from the Tn mutant libraries with 62-79% saturation in each strain. The levels of saturation of transposon libraries in our study are similar to the very recent TnSeq anlaysis by Akusobi where 52-80% saturation libraries (so-called “high-density” transposon libraries) are used for HMM and resampling analyses (Supplemental Methods Table 1[merged saturation] in Akusobi. mBio. 2025). The saturation of Tn insertion in individual replicates of our libraries is also comparable to that reported by DeJesus (Table S1 in mBio 2017). Thus, we consider that our TnSeq method of identifying essential genes and detecting the difference of genetic requirements between clinical MAC-PD strains and ATCC13950 is acceptable.
As for the identification of essential or growth-defect-associated genes by an HMM analysis, we do not consider that we made a serious mistake for the classification of essentiality by an HMM method in most of the structural genes that encode proteins. Because, as DeJesus shows, the number essential genes identified by TnSeq are comparable in large genes possessing more than 10 TA sites between 2 and 14 TnSeq datasets, most of which seem to be structural genes (Supplementary Fig 2 in mBio 2017). If the reviewer intends to regard our libraries far less saturated due to the smaller replicates (n = 2 or 3) than the previous DeJesus’ and Rifat’s reports using 10-14 replicates obtained to acquire so-called “high-density” transposon libraries (DeJesus. mBio 2017, Rifat. mBio 2021), there is a possibility that not all genes could be detected as essential due to the incomplete 11 covering of Tn insertion at nonpermissive TA sites, especially the small genes including small regulatory RNAs. Even if this were the case, it would not detract from the findings of our current study
As for the identification of genetic requirements by a resampling analysis, we consider that our data is acceptable because we compared the normalized data between strains whose saturation levels are similar to the previous report by Akusobi with “high-density” transposon libraries as mentioned above.
References
DeJesus, M.A., Ambadipudi, C., Baker, R., Sassetti, C. & Ioerger, T.R. TRANSIT--A software tool for Himar1 TnSeq analysis. PLoS Comput Biol 11, e1004401 (2015). Akusobi. C. et al. Transposon-sequencing across multiple Mycobacterium abscessus isolates reveals significant functional genomic diversity among strains. mBio 6, e0337624 (2025).
DeJesus, M.A. et al. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio 8, e02133-16 (2017).
Rifat, D., Chen L., Kreiswirth, B.N. & Nuermberger, E.L.. Genome-wide essentiality analysis of Mycobacterium abscessus by saturated transposon mutagenesis and deep sequencing. mBio 12, e0104921 (2021).
(4) ATCC strain is missing in the mouse experiment.
Thank you for the comment on the necessity of setting ATCC13950 as a control strain of mouse TnSeq experiment. To set ATCC13950 as a control strain in mouse infection experiments would be ideal. However, we have proved that ATCC13950 is eliminated within 4 weeks of infection in mice (Tateishi. BMC Microbiol 2023). To perform TnSeq, it is necessary to collect colonies at least the number of TA sites mathematically (Realistically, colonies with more than the number of TA sites are needed to produce biologically robust data.). That means, it is impossible to perform in vivo TnSeq study using ATCC13950 due to the inability to harvest sufficient number of colonies.
To make these things understood clearly, we have added the description of being unable to perform in vivo TnSeq in ATCC13950 in the result section (page 13 lines 221-222 in the revised manuscript).
“(It is impossible to perform TnSeq in lungs infected with ATCC13950 because ATCC13950 is eliminated within 4 weeks of infection) (Tateishi. BMC Microbiol 2023)”
Reference
Tateishi, Y. et al. Virulence of Mycobacterium intracellulare clinical strains in a mouse model of lung infection - role of neutrophilic inflammation in disease severity. BMC Microbiol 23, 94 (2023).
(5) The viability assays done in 96 well plate may not be appropriate given that mycobacterial cultures often clump without vigorous shaking. How did they control evaporation for 10 days and above?
Thank you for the comment on the issue of viability assay in terms of bacterial clumping. As described in the Methods (page 44 lines 778-781 in the revised manuscript), we have mixed the culture containing 250 μL by pipetting 40 times to loosen clumping every time before sampling 4 μL for inoculation on agar plates to count CFUs. By this method, we did not observe macroscopic clumping or pellicles like of Mtb or M. bovis BCG as seen in statistic culture.
We used inner wells for culture of bacteria in hypoxic growth assay. To control evaporation of the culture, we filled the distilled water in the outer wells and covered the plates with plastic lids. We cultured the plates with humidification at 37°C in the incubator.
(6) Fig. 7a many time points have only two data points and in few cases. The Y axis could have been kept same for better comparison for all strains and conditions.
Thank you for the comments on the data presentation of hypoxic growth assay in original Fig. 7a (new Fig 8a). The reason of many time points with only two data points is the close values of data in individual replicates. For example, the log10- transformed values of CFUs in ATCC13950 under aerobic culture are 4.716, 4.653, 4.698 at day 5, 4.949, 5.056, 4.954 at day 6, and 5.161, 5.190, 5.204 at day 8. We have added the numerical data of CFUs used for drawing growth curves as new Supplementary Table 19. Therefore, the data itself derives from three independent replicates.
Following the comment, we have revised the data presentation in new Fig 8a (original Fig. 7a) by keeping the same maximal value of Y axis across all graphs. In addition, we have revised the legend to designate clearly how we obtained the data of growth curves as follows (page 63 lines 1107-1108 in the revised manuscript): “Data on the growth curves are the means of three biological replicates from one experiment. Data from one experiment representative of three independent 13 experiments (N = 3) are shown.”
(7) The relevance of 7b is not well discussed and a suitable explanation for the difference in the profiles of M001 and MOTT64 between aerobic and hypoxia is not provided. Data representation should be improved for 7c with appropriate spacing.
Thank you for the comments on the relevance of original Fig. 7b (new Fig. 8b). In order to compare the pattern of logarithmic growth curves between strains quantitatively, we focused on time and slope at midpoint. The time at midpoint is the timing of entry to logarithmic growth phase. The earlier the strain enters logarithmic phase, the smaller the value of the time at midpoint becomes.
The two strains belonging to the MP-MIP subgroup, MOTT64 and M001 showed similar time at midpoint under aerobic conditions. However, the time at midpoint was significantly different between MOTT64 and M001 under hypoxia, the latter showing great delay of timing of entry to logarithmic phase. In contrast to the majority of the clinical strains that showed reduced growth rate at midpoint under hypoxia, neither strain showed such phenomenon under hypoxia. Although the implication in clinical situations has not been proven, strains without slow growth under hypoxia may have different (possibly strain-specific) mechanisms of hypoxic adaptation corresponding to the growth phenotypes under hypoxia.
Following the comment, we have added the explanation on the difference in the profiles of M001 and MOTT64 between aerobic and hypoxia in the Discussion (page 31 lines 552-557, page 32 lines 562-567 in the revised manuscript). “The two strains belonging to the MP-MIP subgroup, MOTT64 and M001 showed similar time at midpoint under aerobic conditions. However, the time at midpoint was significantly different between MOTT64 and M001 under hypoxia, the latter showing great delay of timing of entry to logarithmic phase. In contrast to the majority of the clinical strains that showed slow growth at midpoint under hypoxia, neither strain showed such phenomenon.”.
” Our inability to construct knockdown strains in M001 and MOTT64 prevented us from clarifying the factors that discriminate against the pattern of hypoxic adaptation. Although the implication in clinical situations has not been proven, strains without slow growth under hypoxia may have different (possibly strainspecific) mechanisms of hypoxic adaptation corresponding to the growth phenotypes under hypoxia.”
Following the comment, we have made the space between new Fig. 8b and 14 new Fig. 8c (original Fig. 7b and Fig. 7c).
(8) Fig. 8a, the antibiotic sensitivity at early and later time points do not seem to correlate. Any explanation?
Thank you for the comment on the uncorrelation of data of growth inhibition in knockdown strains of universal essential genes between early and later time points. The diminished effects of growth inhibition observed at Day 7 in knockdown strains may be due to the “escape” clones of knockdown strains under long-term culture by adding anhydrotetracycline (aTc) that induces sgRNA. As described in the Methods (pages 42-43 lines 754-758), we added aTc repeatedly every 48 h to maintain the induction of dCas9 and sgRNAs in experiments that extended beyond 48 h (Singh. Nucl Acid Res 2016). Such phenomenon has been reported by McNeil (Antimicrob Agent Chem. 2019) showing the increase in CFUs by day 9 with 100 ng/mL aTc with bacterial growth being detected between 2 and 3 weeks. These phenotypes of “escape” mutants is considered to be attributed to the promotor responsiveness to aTc.
Nevertheless, except for gyrB in M.i.27, the effect of growth inhibition at Day 7 in knockdown strains of universal essential genes was 10-1 or less of comparative growth rates of knockdown strains to vector control strains (y-axis of original Fig. 8). In this study, we judged the positive level of growth inhibition as 10-1 or less of comparative growth rates of knockdown strains to vector control strains (y-axis of new Fig. 7). Thus, we consider that the CRISPR-i data overall validated the essentiality of these genes.
References
Singh A.K., et al. Investigating essential gene function in Mycobacterium tuberculosis using an efficient CRISPR interference system, Nucl Acid Res 44, e143 (2016) McNeil M.B. &, Cook, G.M. Utilization of CRISPR interference to validate MmpL3 as a drug target in Mycobacterium tuberculosis. Antimicrob Agent Chem 63, e00629-19 (2019)
(9) Fig. 8b and c very data representation could have been improved. Some strains used in 7 are missing. The authors refer to technical challenge with respect to M001. Is it the same for others as well (MOTT64). The interpretation of data in result and discussion section is difficult to follow. Is the data subjected to statistical analysis?
Thank you for the comment on data presentation in original Fig. 8b (new Fig 7b). As 15 mentioned in the Discussion (page 18 lines 316-31 in the revised manuscript), the reason of missing M001 and MOTT64 in CRISPR-i experiment in original Fig. 7 (new Fig. 8) was we were unable to construct the knockdown strains in M001 and MOTT64. We consider these are the same technical challenges between M001 and MOTT64.
Following the comment, we have added the explanation of the technical challenge with respect to M001 and MOTT64 in the Discussion (page 32 lines 561- 566 in the revised manuscript). ”Our inability to construct knockdown strains in M001 and MOTT64 prevented us from clarifying the factors that discriminate against the pattern of hypoxic adaptation. Although the implication in clinical situations has not been proven, strains without slow growth under hypoxia may have different (possibly strain-specific) mechanisms of hypoxic adaptation corresponding to the growth phenotypes under hypoxia.”
As for the interpretation of growth suppression in knockdown experiments described in original Fig. 8 (new Fig. 7), We judged the positive level of growth inhibition as 10-1 or less of comparative growth rates of knockdown strains to vector control strains (y-axis of new Fig. 7). We interpreted the results based on whether the level of growth inhibition was positive or not (i.e. the comparative growth rates of knockdown strains to vector control strains became below 10-1 or not). Since our aim was to investigate whether knockdown of the target genes in each strain leads to growth inhibition, we did not perform statistical analysis between strains or target genes.
The major weakness of the study is the organization and data representation. It became very difficult to connect the role of gluconeogenesis, secretion system and others identified by authors to hypoxia, pellicle formation. The authors may consider rephrasing the results and discussion sections.
Thank you for the comments on the issue of organization and data presentation. Following the comment, we have revised the manuscript to indicate the relevance of the role of gluconeogenesis, secretion system and others defined by us more clearly (page 23 lines 404-408 in the revised manuscript).
“Because the profiles of genetic requirements reflect the adaptation to the environment in which bacteria habits, it is reasonable to assume that the increase of genetic requirements in hypoxia-related genes such as gluconeogenesis (pckA, glpX), type VII secretion system (mycP5, eccC5) and cysteine desulfurase (csd) play an important role on the growth under hypoxia-relevant conditions in vivo.”
Following the comments, we have exchanged the order of data presentation as follows: in vitro TnSeq (pages 6-12 lines 102-208 in the revised manuscript) , Mouse TnSeq (pages 12-17 lines 210-303 in the revised manuscript), Knockdown experiment (pages 17-21 lines 305-363 in the revised manuscript), Hypoxic growth assay (pages 21-23 lines 365-408 in the revised manuscript).
In association with the exchange of the order of data presentation, we have changed the order of the contents of the Discussion as follows: Preferential carbohydrate metabolism under hypoxia such as pckA and glpX (pages 24-26 lines 424-466 in the revised manuscript), Cysteine desulfurase gene (csd) (pages 26-27 lines 467-482 in the revised manuscript), Conditional essential genes in vivo such as type VII secretion system (pages 27-28 lines 483-497 in the revised manuscript), Knockdown experiment (pages 28-30 lines 498-536 in the revised manuscript), Hypoxic growth pattern (pages 30-32 lines 537-571 in the revised manuscript), Failure of assay using PckA inhibitors (pages 32-33 lines 572-578 in the revised manuscript), Transformation efficiencies (page 33 lines 579-591 in the revised manuscript), Saturation of Tn insertion (pages 33-35 lines 592-613 in the revised manuscript), Suggested future experiment plan (pages 35-36 lines 614-632 in the revised manuscript).
Po roce 2020 došlo k násobnému nárůstu, který odráží především rozšíření programů SFŽP v oblasti energetických úspor a modernizace zdrojů tepla v domácnostech – zejména v souvislosti s implementací programu Nová zelená úsporám. 20152016201720182019202020212022202320240102030OdvětvíDávky pomoci v hmotné nouziDávky státní sociální podpory a dávky pěstounské péčeKomunální služby a územní rozvojOchrana ovzduší a klimatuOstatní činnost v oblasti bydlení, komunálních služeb a úz. rozv.Rozvoj bydlení a bytové hospodářstvíSlužby sociální prevenceZáležitosti těžebního průmyslu a energetikyVýdaje [mld. Kč].cls-1 {fill: #3f4f75;} .cls-2 {fill: #80cfbe;} .cls-3 {fill: #fff;}plotly-logomark {"x":{"data":[{"x":[2015,2016,2017,2018,2019,2020,2021,2022,2023,2024],"y":[3.1362012145199998,2.9167721326199998,2.42229314202,1.8933877991400001,1.5792528450799999,1.6272916878099999,1.76658297259,1.84017972437,1.694480889,1.6739637439999999],"text":["Rok: 2015 <br>Odvětví: Dávky pomoci v hmotné nouzi <br>Výdaje: 3.14 mld. Kč","Rok: 2016 <br>Odvětví: Dávky pomoci v hmotné nouzi <br>Výdaje: 2.92 mld. Kč","Rok: 2017 <br>Odvětví: Dávky pomoci v hmotné nouzi <br>Výdaje: 2.42 mld. Kč","Rok: 2018 <br>Odvětví: Dávky pomoci v hmotné nouzi <br>Výdaje: 1.89 mld. Kč","Rok: 2019 <br>Odvětví: Dávky pomoci v hmotné nouzi <br>Výdaje: 1.58 mld. Kč","Rok: 2020 <br>Odvětví: Dávky pomoci v hmotné nouzi <br>Výdaje: 1.63 mld. Kč","Rok: 2021 <br>Odvětví: Dávky pomoci v hmotné nouzi <br>Výdaje: 1.77 mld. Kč","Rok: 2022 <br>Odvětví: Dávky pomoci v hmotné nouzi <br>Výdaje: 1.84 mld. Kč","Rok: 2023 <br>Odvětví: Dávky pomoci v hmotné nouzi <br>Výdaje: 1.69 mld. Kč","Rok: 2024 <br>Odvětví: Dávky pomoci v hmotné nouzi <br>Výdaje: 1.67 mld. Kč"],"type":"scatter","mode":"lines","line":{"width":5.6692913385826778,"color":"rgba(17,49,68,1)","dash":"solid"},"hoveron":"points","name":"Dávky pomoci v hmotné nouzi","legendgroup":"Dávky pomoci v hmotné nouzi","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[2015,2016,2017,2018,2019,2020,2021,2022,2023,2024],"y":[9.1874478112700011,9.2896525793799984,8.6527129472500004,7.7153884478100005,7.1066980742899997,6.9721704018900006,6.64058688196,8.5408560970200007,17.890107087770001,20.330845674189998],"text":["Rok: 2015 <br>Odvětví: Dávky státní sociální podpory a dávky pěstounské péče <br>Výdaje: 9.19 mld. Kč","Rok: 2016 <br>Odvětví: Dávky státní sociální podpory a dávky pěstounské péče <br>Výdaje: 9.29 mld. Kč","Rok: 2017 <br>Odvětví: Dávky státní sociální podpory a dávky pěstounské péče <br>Výdaje: 8.65 mld. Kč","Rok: 2018 <br>Odvětví: Dávky státní sociální podpory a dávky pěstounské péče <br>Výdaje: 7.72 mld. Kč","Rok: 2019 <br>Odvětví: Dávky státní sociální podpory a dávky pěstounské péče <br>Výdaje: 7.11 mld. Kč","Rok: 2020 <br>Odvětví: Dávky státní sociální podpory a dávky pěstounské péče <br>Výdaje: 6.97 mld. Kč","Rok: 2021 <br>Odvětví: Dávky státní sociální podpory a dávky pěstounské péče <br>Výdaje: 6.64 mld. Kč","Rok: 2022 <br>Odvětví: Dávky státní sociální podpory a dávky pěstounské péče <br>Výdaje: 8.54 mld. Kč","Rok: 2023 <br>Odvětví: Dávky státní sociální podpory a dávky pěstounské péče <br>Výdaje: 17.89 mld. Kč","Rok: 2024 <br>Odvětví: Dávky státní sociální podpory a dávky pěstounské péče <br>Výdaje: 20.33 mld. Kč"],"type":"scatter","mode":"lines","line":{"width":5.6692913385826778,"color":"rgba(9,97,106,1)","dash":"solid"},"hoveron":"points","name":"Dávky státní sociální podpory a dávky pěstounské péče","legendgroup":"Dávky státní sociální podpory a dávky pěstounské péče","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[2018,2019,2020,2021,2022,2023,2024],"y":[1.67657141526,2.7964227882900001,3.15998356346,3.61070579615,2.8862273526500002,1.69988693084,0.82015937066],"text":["Rok: 2018 <br>Odvětví: Komunální služby a územní rozvoj <br>Výdaje: 1.68 mld. Kč","Rok: 2019 <br>Odvětví: Komunální služby a územní rozvoj <br>Výdaje: 2.8 mld. Kč","Rok: 2020 <br>Odvětví: Komunální služby a územní rozvoj <br>Výdaje: 3.16 mld. Kč","Rok: 2021 <br>Odvětví: Komunální služby a územní rozvoj <br>Výdaje: 3.61 mld. Kč","Rok: 2022 <br>Odvětví: Komunální služby a územní rozvoj <br>Výdaje: 2.89 mld. Kč","Rok: 2023 <br>Odvětví: Komunální služby a územní rozvoj <br>Výdaje: 1.7 mld. Kč","Rok: 2024 <br>Odvětví: Komunální služby a územní rozvoj <br>Výdaje: 0.82 mld. Kč"],"type":"scatter","mode":"lines","line":{"width":5.6692913385826778,"color":"rgba(2,146,144,1)","dash":"solid"},"hoveron":"points","name":"Komunální služby a územní rozvoj","legendgroup":"Komunální služby a územní rozvoj","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[2015,2016,2017,2018,2019,2020,2021,2022,2023,2024],"y":[1.6773676289600001,2.2493404589599999,3.1941818671500002,1.2126270560799999,2.1132997519700001,1.31701081322,0.97534286400000003,0.94263653754999999,2.2349673913600001,0.69131391674999998],"text":["Rok: 2015 <br>Odvětví: Ochrana ovzduší a klimatu <br>Výdaje: 1.68 mld. Kč","Rok: 2016 <br>Odvětví: Ochrana ovzduší a klimatu <br>Výdaje: 2.25 mld. Kč","Rok: 2017 <br>Odvětví: Ochrana ovzduší a klimatu <br>Výdaje: 3.19 mld. Kč","Rok: 2018 <br>Odvětví: Ochrana ovzduší a klimatu <br>Výdaje: 1.21 mld. Kč","Rok: 2019 <br>Odvětví: Ochrana ovzduší a klimatu <br>Výdaje: 2.11 mld. Kč","Rok: 2020 <br>Odvětví: Ochrana ovzduší a klimatu <br>Výdaje: 1.32 mld. Kč","Rok: 2021 <br>Odvětví: Ochrana ovzduší a klimatu <br>Výdaje: 0.98 mld. Kč","Rok: 2022 <br>Odvětví: Ochrana ovzduší a klimatu <br>Výdaje: 0.94 mld. Kč","Rok: 2023 <br>Odvětví: Ochrana ovzduší a klimatu <br>Výdaje: 2.23 mld. Kč","Rok: 2024 <br>Odvětví: Ochrana ovzduší a klimatu <br>Výdaje: 0.69 mld. Kč"],"type":"scatter","mode":"lines","line":{"width":5.6692913385826778,"color":"rgba(70,163,112,1)","dash":"solid"},"hoveron":"points","name":"Ochrana ovzduší a klimatu","legendgroup":"Ochrana ovzduší a klimatu","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[2015,2016,2017,2018,2019,2020,2021],"y":[0.66460017107000002,0.46405308710000004,0.19152440866000001,0,0,0,0],"text":["Rok: 2015 <br>Odvětví: Ostatní činnost v oblasti bydlení, komunálních služeb a úz. rozv. <br>Výdaje: 0.66 mld. Kč","Rok: 2016 <br>Odvětví: Ostatní činnost v oblasti bydlení, komunálních služeb a úz. rozv. <br>Výdaje: 0.46 mld. Kč","Rok: 2017 <br>Odvětví: Ostatní činnost v oblasti bydlení, komunálních služeb a úz. rozv. <br>Výdaje: 0.19 mld. Kč","Rok: 2018 <br>Odvětví: Ostatní činnost v oblasti bydlení, komunálních služeb a úz. rozv. <br>Výdaje: 0 mld. Kč","Rok: 2019 <br>Odvětví: Ostatní činnost v oblasti bydlení, komunálních služeb a úz. rozv. <br>Výdaje: 0 mld. Kč","Rok: 2020 <br>Odvětví: Ostatní činnost v oblasti bydlení, komunálních služeb a úz. rozv. <br>Výdaje: 0 mld. Kč","Rok: 2021 <br>Odvětví: Ostatní činnost v oblasti bydlení, komunálních služeb a úz. rozv. <br>Výdaje: 0 mld. Kč"],"type":"scatter","mode":"lines","line":{"width":5.6692913385826778,"color":"rgba(176,165,44,1)","dash":"solid"},"hoveron":"points","name":"Ostatní činnost v oblasti bydlení, komunálních služeb a úz. rozv.","legendgroup":"Ostatní činnost v oblasti bydlení, komunálních služeb a úz. rozv.","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[2015,2016,2017,2018,2019,2020,2021,2022,2023,2024],"y":[6.7056526725900003,6.1896334054099995,5.5863772922199999,5.4263460964599997,6.1337736404399994,6.9382058991499997,7.2597953133500006,6.96437401758,5.8336510214300006,4.9146220281000002],"text":["Rok: 2015 <br>Odvětví: Rozvoj bydlení a bytové hospodářství <br>Výdaje: 6.71 mld. Kč","Rok: 2016 <br>Odvětví: Rozvoj bydlení a bytové hospodářství <br>Výdaje: 6.19 mld. Kč","Rok: 2017 <br>Odvětví: Rozvoj bydlení a bytové hospodářství <br>Výdaje: 5.59 mld. Kč","Rok: 2018 <br>Odvětví: Rozvoj bydlení a bytové hospodářství <br>Výdaje: 5.43 mld. Kč","Rok: 2019 <br>Odvětví: Rozvoj bydlení a bytové hospodářství <br>Výdaje: 6.13 mld. Kč","Rok: 2020 <br>Odvětví: Rozvoj bydlení a bytové hospodářství <br>Výdaje: 6.94 mld. Kč","Rok: 2021 <br>Odvětví: Rozvoj bydlení a bytové hospodářství <br>Výdaje: 7.26 mld. Kč","Rok: 2022 <br>Odvětví: Rozvoj bydlení a bytové hospodářství <br>Výdaje: 6.96 mld. Kč","Rok: 2023 <br>Odvětví: Rozvoj bydlení a bytové hospodářství <br>Výdaje: 5.83 mld. Kč","Rok: 2024 <br>Odvětví: Rozvoj bydlení a bytové hospodářství <br>Výdaje: 4.91 mld. Kč"],"type":"scatter","mode":"lines","line":{"width":5.6692913385826778,"color":"rgba(245,158,14,1)","dash":"solid"},"hoveron":"points","name":"Rozvoj bydlení a bytové hospodářství","legendgroup":"Rozvoj bydlení a bytové hospodářství","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[2015,2016,2017,2018,2019,2020,2021,2022,2023,2024],"y":[0.017831,0.032006400999999997,0.023600388999999999,0.0082595670000000006,0.01070192675,0.10281950179999999,0.090053458209999993,0.013486108,0.014732843000000001,0.018406545],"text":["Rok: 2015 <br>Odvětví: Služby sociální prevence <br>Výdaje: 0.02 mld. Kč","Rok: 2016 <br>Odvětví: Služby sociální prevence <br>Výdaje: 0.03 mld. Kč","Rok: 2017 <br>Odvětví: Služby sociální prevence <br>Výdaje: 0.02 mld. Kč","Rok: 2018 <br>Odvětví: Služby sociální prevence <br>Výdaje: 0.01 mld. Kč","Rok: 2019 <br>Odvětví: Služby sociální prevence <br>Výdaje: 0.01 mld. Kč","Rok: 2020 <br>Odvětví: Služby sociální prevence <br>Výdaje: 0.1 mld. Kč","Rok: 2021 <br>Odvětví: Služby sociální prevence <br>Výdaje: 0.09 mld. Kč","Rok: 2022 <br>Odvětví: Služby sociální prevence <br>Výdaje: 0.01 mld. Kč","Rok: 2023 <br>Odvětví: Služby sociální prevence <br>Výdaje: 0.01 mld. Kč","Rok: 2024 <br>Odvětví: Služby sociální prevence <br>Výdaje: 0.02 mld. Kč"],"type":"scatter","mode":"lines","line":{"width":5.6692913385826778,"color":"rgba(241,135,56,1)","dash":"solid"},"hoveron":"points","name":"Služby sociální prevence","legendgroup":"Služby sociální prevence","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[2015,2016,2017,2018,2019,2020,2021,2022,2023,2024],"y":[0.70368427190999994,1.02549356101,1.58813889794,1.6295806145999998,1.8447968074100001,2.2908671036199997,2.8772400939499998,7.7262381892299992,29.393578740099997,33.00478224247],"text":["Rok: 2015 <br>Odvětví: Záležitosti těžebního průmyslu a energetiky <br>Výdaje: 0.7 mld. Kč","Rok: 2016 <br>Odvětví: Záležitosti těžebního průmyslu a energetiky <br>Výdaje: 1.03 mld. Kč","Rok: 2017 <br>Odvětví: Záležitosti těžebního průmyslu a energetiky <br>Výdaje: 1.59 mld. Kč","Rok: 2018 <br>Odvětví: Záležitosti těžebního průmyslu a energetiky <br>Výdaje: 1.63 mld. Kč","Rok: 2019 <br>Odvětví: Záležitosti těžebního průmyslu a energetiky <br>Výdaje: 1.84 mld. Kč","Rok: 2020 <br>Odvětví: Záležitosti těžebního průmyslu a energetiky <br>Výdaje: 2.29 mld. Kč","Rok: 2021 <br>Odvětví: Záležitosti těžebního průmyslu a energetiky <br>Výdaje: 2.88 mld. Kč","Rok: 2022 <br>Odvětví: Záležitosti těžebního průmyslu a energetiky <br>Výdaje: 7.73 mld. Kč","Rok: 2023 <br>Odvětví: Záležitosti těžebního průmyslu a energetiky <br>Výdaje: 29.39 mld. Kč","Rok: 2024 <br>Odvětví: Záležitosti těžebního průmyslu a energetiky <br>Výdaje: 33 mld. Kč"],"type":"scatter","mode":"lines","line":{"width":5.6692913385826778,"color":"rgba(237,113,99,1)","dash":"solid"},"hoveron":"points","name":"Záležitosti těžebního průmyslu a energetiky","legendgroup":"Záležitosti těžebního průmyslu a energetiky","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null}],"layout":{"margin":{"t":23.305936073059364,"r":7.3059360730593621,"b":24.690038964857905,"l":37.260273972602747},"paper_bgcolor":"rgba(255,255,255,1)","font":{"color":"rgba(0,0,0,1)","family":"","size":14.611872146118724},"xaxis":{"domain":[0,1],"automargin":true,"type":"linear","autorange":false,"range":[2014.55,2024.45],"tickmode":"array","ticktext":["2015","2016","2017","2018","2019","2020","2021","2022","2023","2024"],"tickvals":[2015,2016,2017,2018,2019,2020,2021,2022,2023,2024],"categoryorder":"array","categoryarray":["2015","2016","2017","2018","2019","2020","2021","2022","2023","2024"],"nticks":null,"ticks":"","tickcolor":null,"ticklen":3.6529680365296811,"tickwidth":0,"showticklabels":true,"tickfont":{"color":"rgba(77,77,77,1)","family":"","size":11.68949771689498},"tickangle":-45,"showline":false,"linecolor":null,"linewidth":0,"showgrid":true,"gridcolor":"rgba(235,235,235,1)","gridwidth":0,"zeroline":false,"anchor":"y","title":{"text":"","font":{"color":null,"family":null,"size":0}},"hoverformat":".2f"},"yaxis":{"domain":[0,1],"automargin":true,"type":"linear","autorange":false,"range":[-1.6502391121235001,34.655021354593501],"tickmode":"array","ticktext":["0","10","20","30"],"tickvals":[0,10,20,29.999999999999996],"categoryorder":"array","categoryarray":["0","10","20","30"],"nticks":null,"ticks":"","tickcolor":null,"ticklen":3.6529680365296811,"tickwidth":0,"showticklabels":true,"tickfont":{"color":"rgba(77,77,77,1)","family":"","size":11.68949771689498},"tickangle":-0,"showline":false,"linecolor":null,"linewidth":0,"showgrid":true,"gridcolor":"rgba(235,235,235,1)","gridwidth":0,"zeroline":false,"anchor":"x","title":{"text":"Výdaje [mld. Kč]","font":{"color":"rgba(0,0,0,1)","family":"","size":14.611872146118724}},"hoverformat":".2f"},"shapes":[{"type":"rect","fillcolor":null,"line":{"color":null,"width":0,"linetype":[]},"yref":"paper","xref":"paper","layer":"below","x0":0,"x1":1,"y0":0,"y1":1}],"showlegend":true,"legend":{"bgcolor":null,"bordercolor":null,"borderwidth":0,"font":{"color":"rgba(0,0,0,1)","family":"","size":11.68949771689498},"title":{"text":"Odvětví","font":{"color":null,"family":null,"size":0}},"orientation":"h"},"hovermode":"closest","barmode":"relative"},"config":{"doubleClick":"reset","modeBarButtonsToAdd":["hoverclosest","hovercompare"],"showSendToCloud":false},"source":"A","attrs":{"e303348632b":{"x":{},"y":{},"text":{},"colour":{},"type":"scatter"}},"cur_data":"e303348632b","visdat":{"e303348632b":["function (y) ","x"]},"highlight":{"on":"plotly_click","persistent":false,"dynamic":false,"selectize":false,"opacityDim":0.20000000000000001,"selected":{"opacity":1},"debounce":0},"shinyEvents":["plotly_hover","plotly_click","plotly_selected","plotly_relayout","plotly_brushed","plotly_brushing","plotly_clickannotation","plotly_doubleclick","plotly_deselect","plotly_afterplot","plotly_sunburstclick"],"base_url":"https://plot.ly"},"evals":[],"jsHooks":{"render":[{"code":"function(el){\n el.setAttribute('role','img');\n el.setAttribute('aria-label','Liniový graf výdajů státního rozpočtu na bydlení (včetně výdajů s nepřímým dopadem) v Česku v miliardách Kč podle odvětví. Zobrazuje se výše a složení výdajů na bydlení v čase od roku 2015. Popis dostupný v textu nad grafem v části Výdaje s nepřímým dopadem na bydlení.');\n }","data":null}]}}
V NZÚ byly taky vyhlašovány výzvy na zateplení bytových domů (v období 14-23 za cca 1 mld. Kč), průměrné výdaje na jednu akci jsou výrazně vyšší než pro rodinné domy (cca 800 tis. Kč)
S1 Table
DOI: 10.1371/journal.pbio.3002720
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00034251
Supplementary Information
DOI: 10.1038/s41467-024-50973-y
Resource: (WB Cat# WBStrain00034068,RRID:WB-STRAIN:WBStrain00034068)
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00034068
Supplementary Information
DOI: 10.1038/s41467-024-50973-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00004114
Supplementary Information
DOI: 10.1038/s41467-024-50973-y
Resource: (WB Cat# WBStrain00000001,RRID:WB-STRAIN:WBStrain00000001)
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00000001
Supplementary Information
DOI: 10.1038/s41467-024-50973-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00026452
Supplementary Information
DOI: 10.1038/s41467-024-50973-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00022511
Supplementary Information
DOI: 10.1038/s41467-024-50973-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00034070
Supplementary Information
DOI: 10.1038/s41467-024-50973-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00007852
Supplementary Information
DOI: 10.1038/s41467-024-50973-y
Resource: (WB Cat# WBStrain00005096,RRID:WB-STRAIN:WBStrain00005096)
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00005096
Supplementary Information
DOI: 10.1038/s41467-024-50973-y
Resource: RRID:WB-STRAIN:WBStrain00004861
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00004861
Supplementary Information
DOI: 10.1038/s41467-024-50973-y
Resource: None
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00034066
RRID:SCR_002865
DOI: 10.1186/s13019-025-03628-y
Resource: SPSS (RRID:SCR_002865)
Curator: @scibot
SciCrunch record: RRID:SCR_002865
RRID:SCR_001362
DOI: 10.1038/s41598-025-24136-y
Resource: NiLearn (RRID:SCR_001362)
Curator: @scibot
SciCrunch record: RRID:SCR_001362
RRID:SCR_002502
DOI: 10.1038/s41598-025-24136-y
Resource: Nipype (RRID:SCR_002502)
Curator: @scibot
SciCrunch record: RRID:SCR_002502
Reviewer #2 (Public review):
Summary:
Wang et al. engineered an optimized ACE2 mutant by introducing two mutations (T92Q and H374N) and fused this ACE2 mutant to human IgG1-Fc (B5-D3). Experimental results suggest that B5-D3 exhibits broad-spectrum neutralization capacity and confers effective protection upon intranasal administration in SARS-CoV-2-infected K18-hACE2 mice. Transcriptomic analysis suggests that B5-D3 induces early immune activation in lung tissues of infected mice. Fluorescence-based bio-distribution assay further indicates rapid accumulation of B5-D3 in the respiratory tract, particularly in airway macrophages. Further investigation shows that B5-D3 promotes viral phagocytic clearance by macrophages via an Fc-mediated effector function, namely antibody-dependent cellular phagocytosis (ADCP), while simultaneously blocking ACE2-mediated viral infection in epithelial cells. These results provide insights into improving decoy treatments against SARS-CoV-2 and other potential respiratory viruses.
Strengths:
The protective effect of this ACE2-Fc fusion protein against SARS-CoV-2 infection has been evaluated in a quite comprehensive way.
Weaknesses:
(1) The paper lacks an explanation regarding the reason for the combination of mutations listed in Supplementary Figure 2b. For example, for the mutations that enhance spike protein binding, B2-B6 does not fully align with the mutations listed in Table S1 of Reference 4, yet no specific criteria are provided. Second, for the mutations that abolished enzymatic activity, while D1 and D2, D3, D4, and D5 are cited from References 12, 11, and 33, respectively, the reason for combining D3 and D4 into A2, and D1 and D2 into A3 remains unexplained. It is also unclear whether some of these other possible combinations have been tested. Furthermore, for the B5-derived mutations, only double-mutant combinations with D1-D5 are tested, with no attempt made to evaluate triple mutations involving A2 or A3.
(2) Figures 1b, 1d, and 1e lack statistical analyses, making it difficult to determine whether B5 and D3 exhibit significant advantages. For Wuhan-Hu-1 strain, B2 and B5 are similar, and for D614G strain, B2, B3, B4, B5, and B6 display comparable results. However, only the glycosylation-related single mutant B5 is chosen for further combinatorial constructs. Moreover, for VOC/VOI strains, B5 is superior to B5-D3; for the Alpha strain, B5-D4 and B5-D5 are superior to B5-D3; and for the Delta and Lambda strains, B5-D5 is superior to B5-D3. These observations further highlight the need for a clearer explanation of the selection strategy.
(3) Figure 1e does not specify the construct form of the control hIgG1, namely whether it is an hIgG1 Fc fragment or a full-length hIgG1 protein. If the full-length form is used, the design of its Fab region should be clarified to ensure the accuracy and comparability of the experimental control.
(4) In Figure 2a, all three PBS control mice died, whereas in Figure 2f, three out of five PBS control mice died, with the remaining showing gradual weight recovery. This discrepancy may reflect individual immune variations within the control groups, and it is necessary to clarify whether potential autoimmune factors could have affected the comparability of the results. Also, the mouse experiments suffer from insufficient sample sizes, which affects the statistical power and reliability of the results. In Figure 2a, each group contains only 4 replicates, one of which was used for lung tissue sampling. As a result, body weight monitoring data is derived from only 3 mice per group (the figure legend indicating n=4 should be corrected to n=3). Such a small sample size limits the robustness of the conclusions. Similarly, in Figure 2f, although each group has 5 replicates, body weight data are presented for only 4 mice, with no explanation provided for the exclusion of the fifth mouse. Furthermore, the lung tissue experiments in Figure 3a include only 3 replicates, which is also inadequate.
(5) Compared to 6 hours, intranasal administration of B5-D3 at 24 hours before viral infection results in reduced protective efficacy. However, only survival and body weight data are provided, with no supporting evidence from virological assays such as viral titer measurement. Therefore, the long-term effectiveness lacks sufficient experimental validation.
(6) In Figures 3b and 3c, viral spike (S) and nucleocapsid (N) RNA relative expression levels are quantified by qPCR. The results show significant individual variation within the B5-D3-LALA treatment group: one mouse exhibits high S and N expression, while the other two show low expression. Viral load levels are also inconsistent: two mice have high viral loads, and one has a low viral load. Due to this variability, the available data are insufficient to robustly support the conclusion.
(7) Figure 3e: "H&E staining indicated alveolar thickening in all groups," including the Mock group. Since the Mock group did not receive virus or active drug treatment, this observed change may result from local tissue reaction induced by the intranasal inoculation procedure itself, rather than specific immune activation. A control group (no manipulation) should be set to rule out potential confounding effects of the experimental procedure on tissue morphology, thereby allowing a more accurate assessment of the drug's effects.
(8) In Supplementary Figure 11b, a considerable number of alveolar macrophages (AMs) are observed in both the PBS and B5-D3 groups. This makes it difficult to determine whether the observed accumulation is specifically induced by B5-D3.
(9) In the flow cytometry experiment shown in Figure 5, the PBS control group is not labeled with AF750, which necessarily results in a value of zero for "B5-D3+ cells" on the y-axis. An appropriate control (e.g., hIgG1-Fc labeled with AF750) should be included.
(10) The Methods section: a more detailed description of the experimental procedures involving HIV p24 and SARS-CoV-2 should be included.
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public review):
Summary
This work provides important new evidence of the cognitive and neural mechanisms that give rise to feelings of shame and guilt, as well as their transformation into compensatory behavior. The authors use a well-designed interpersonal task to manipulate responsibility and harm, eliciting varying levels of shame and guilt in participants. The study combines behavioral, computational, and neuroimaging approaches to offer a comprehensive account of how these emotions are experienced and acted upon. Notably, the findings reveal distinct patterns in how harm and responsibility contribute to guilt and shame and how these factors are integrated into compensatory decision-making.
Strengths
(1) Investigating both guilt and shame in a single experimental framework allows for a direct comparison of their behavioral and neural effects while minimizing confounds.
(2) The study provides a novel contribution to the literature by exploring the neural bases underlying the conversion of shame into behavior.
(3) The task is creative and ecologically valid, simulating a realistic social situation while retaining experimental control.
(4) Computational modeling and fMRI analysis yield converging evidence for a quotient-based integration of harm and responsibility in guiding compensatory behavior.
We are grateful for your thoughtful summary of our work’s strengths and greatly appreciate these positive words.
We would like to note that, in accordance with the journal’s requirements, we have uploaded both a clean version of the revised manuscript and a version with all modifications highlighted in blue.
Weakness
(1) Post-experimental self-reports rely both on memory and on the understanding of the conceptual difference between the two emotions. Additionally, it is unclear whether the 16 scenarios were presented in random order; sequential presentation could have introduced contrast effects or demand characteristics.
Thank you for pointing out the two limitations of the experimental paradigm. We fully agree with your point. Participants recalled and reported their feelings of guilt and shame immediately after completing the task, which likely ensured reasonably accurate state reports. We acknowledge, however, that in-task assessments might provide greater precision. We opted against them to examine altruistic decision-making in a more natural context, as in-task assessments could have heightened participants’ awareness of guilt and shame and biased their altruistic decisions. Post-task assessments also reduced fMRI scanning time, minimizing discomfort from prolonged immobility and thereby preserving data quality.
In the present study, assessing guilt and shame required participants to distinguish conceptually between the two emotions. Most research with adult participants has adopted this approach, relying on direct self-reports of emotional intensity under the assumption that adults can differentiate between guilt and shame (Michl et al., 2014; Wagner et al., 2011; Zhu et al., 2019). However, we acknowledge that this approach may be less suitable for studies involving children, who may not yet have a clear understanding of the distinction between guilt and shame.
The limitations have been added into the Discussion section (Page 47): “This research has several limitations. First, post-task assessments of guilt and shame, unlike in-task assessments, rely on memory and may thus be less precise, although in-task assessments could have heightened participants’ awareness of these emotions and biased their decisions. Second, our measures of guilt and shame depend on participants’ conceptual understanding of the two emotions. While this is common practice in studies with adult participants (Michl et al., 2014; Wagner et al., 2011; Zhu et al., 2019), it may be less appropriate for research involving children.”
We apologize for the confusion. The 16 scenarios were presented in a random order. We have clarified this in the revised manuscript (Page 13): “After the interpersonal game, the outcomes of the experimental trials were re-presented in a random order.”
(2) In the neural analysis of emotion sensitivity, the authors identify brain regions correlated with responsibility-driven shame sensitivity and then use those brain regions as masks to test whether they were more involved in the responsibility-driven shame sensitivity than the other types of emotion sensitivity. I wonder if this is biasing the results. Would it be better to use a cross-validation approach? A similar issue might arise in "Activation analysis (neural basis of compensatory sensitivity)."
Thank you for this valuable comment. We replaced the original analyses with a leave-one-subject-out (LOSO) cross-validation approach, which minimizes bias in secondary tests due to non-independence (Esterman et al., 2010). The findings were largely consistent with the original results, except that two previously significant effects became marginally significant (one effect changed from P = 0.012 to P = 0.053; the other from P = 0.044 to P = 0.062). Although we believe the new results do not alter our main conclusions, marginally significant findings should be interpreted with caution. We have noted this point in the Discussion section (Page 48): “… marginally significant results should be viewed cautiously and warrant further examination in future studies with larger sample sizes.”
In the revised manuscript, we have described the cross-validation procedure in detail and reported the corresponding results. Please see the Method section, Page 23: “The results showed that the neural responses in the temporoparietal junction/superior temporal sulcus (TPJ/STS) and precentral cortex/postcentral cortex/supplementary motor area (PRC/POC/SMA) were negatively correlated with the responsibility-driven shame sensitivity. To test whether these regions were more involved in responsibilitydriven shame sensitivity than in other types of emotion sensitivity, we implemented a leave-one-subject-out (LOSO) cross-validation procedure (e.g., Esterman et al., 2010). In each fold, clusters in the TPJ/STS and PRC/POC/SMA showing significant correlations with responsibility-driven shame sensitivity were identified at the group level based on N-1 participants. These clusters, defined as regions of interest (ROI), were then applied to the left-out participant, from whom we extracted the mean parameter estimates (i.e., neural response values). If, in a given fold, no suprathreshold cluster was detected within the TPJ/STS or PRC/POC/SMA after correction, or if the two regions merged into a single cluster that could not be separated, the corresponding value was coded as missing. Repeating this procedure across all folds yielded an independent set of ROI-based estimates for each participant. In the LOSO crossvalidation procedure, the TPJ/STS and PRC/POC/SMA merged into a single inseparable cluster in two folds, and no suprathreshold cluster was detected within the TPJ/STS in one fold. These instances were coded as missing, resulting in valid data from 39 participants for the TPJ/STS and 40 participants for the PRC/POC/SMA. We then correlated these estimates with all four types of emotion sensitivities and compared the correlation with responsibility-driven shame sensitivity against those with the other sensitivities using Z tests (Pearson and Filon's Z).” and Page 24: “To directly test whether these regions were more involved in one of the two types of compensatory sensitivity, we applied the same LOSO cross-validation procedure described above. In this procedure, no suprathreshold cluster was detected within the LPFC in one fold and within the TP in 27 folds. These cases were coded as missing, resulting in valid data from 42 participants for the bilateral IPL, 41 participants for the LPFC, and 15 participants for the TP. The limited sample size for the TP likely reflects that its effect was only marginally above the correction threshold, such that the reduced power in cross-validation often rendered it nonsignificant. Because the sample size for the TP was too small and the results may therefore be unreliable, we did not pursue further analyses for this region. The independent ROI-based estimates were then correlated with both guilt-driven and shame-driven compensatory sensitivities, and the strength of the correlations was compared using Z tests (Pearson and Filon's Z).”
Please see the Results section, Pages 34 and 35: “To assess whether these brain regions were specifically involved in responsibility-driven shame sensitivity, we compared the Pearson correlations between their activity and all types of emotion sensitivities. The results demonstrated the domain specificity of these regions, by revealing that the TPJ/STS cluster had significantly stronger negative responses to responsibility-driven shame sensitivity than to responsibility-driven guilt sensitivity (Z = 2.44, P = 0.015) and harm-driven shame sensitivity (Z = 3.38, P < 0.001), and a marginally stronger negative response to harm-driven guilt sensitivity (Z = 1.87, P = 0.062) (Figure 4C; Supplementary Table 14). In addition, the sensorimotor areas (i.e., precentral cortex (PRC), postcentral cortex (POC), and supplementary motor area (SMA)) exhibited the similar activation pattern as the TPJ/STS (Figure 4B and 4C; Supplementary Tables 13 and 14).” and Page 35: “The results revealed that the left LPFC was more engaged in shame-driven compensatory sensitivity (Z = 1.93, P = 0.053), as its activity showed a marginally stronger positive correlation with shamedriven sensitivity than with guilt-driven sensitivity (Figure 5C). No significant difference was found in the Pearson correlations between the activity of the bilateral IPL and the two types of sensitivities (Supplementary Table 16). For the TP, the effective sample size was too small to yield reliable results (see Methods).”
(1) Regarding the traits of guilt and shame, I appreciate using the scores from the subscales (evaluations and action tendencies) separately for the analyses (instead of a composite score). An issue with using the actions subscales when measuring guilt and shame proneness is that the behavioral tendencies for each emotion get conflated with their definitions, risking circularity. It is reassuring that the behavior evaluation subscale was significantly correlated with compensatory behavior (not only the action tendencies subscale). However, the absence of significant neural correlates for the behavior evaluation subscale raises questions: Do the authors have thoughts on why this might be the case, and any implications?
We are grateful for this important comment. According to the Guilt and Shame Proneness Scale, trait guilt comprises two dimensions: negative behavior evaluations and repair action tendencies (Cohen et al., 2011). Behaviorally, both dimensions were significantly correlated with participants’ compensatory behavior (negative behavior evaluations: R = 0.39, P = 0.010; repair action tendencies: R = 0.33, P = 0.030). Neurally, while repair action tendencies were significantly associated with activity in the aMCC and other brain areas, negative behavior evaluations showed no significant neural correlates. The absence of significant neural correlates for negative behavior evaluations may be due to several factors. In addition to common explanations (e.g., limited sample size reducing the power to detect weak neural correlates or subtle effects obscured by fMRI noise), another possibility is that this dimension influences neural responses indirectly through intermediate processes not captured in our study (e.g., specific motivational states). We have added a discussion of the non-significant result to the revised manuscript (Page 47): “However, the neural correlates of negative behavior evaluations (another dimension of trait guilt) were absent. The reasons underlying the non-significant neural finding may be multifaceted. One possibility is that negative behavior evaluations influence neural responses indirectly through intermediate processes not captured in our study (e.g., specific motivational states).”
In addition, to avoid misunderstanding, the revised manuscript specifies at the appropriate places that the neural findings pertain to repair action tendencies rather than to trait guilt in general. For instance, see Pages 46 and 47: “Furthermore, we found neural responses in the aMCC mediated the relationship between repair action tendencies (one dimension of trait guilt) and compensation… Accordingly, our fMRI findings suggest that individuals with stronger tendency to engage in compensation across various moral violation scenarios (indicated by their repair action tendencies) are more sensitive to the severity of the violation and therefore engage in greater compensatory behavior.”
(2) Regarding the computational model finding that participants seem to disregard selfinterest, do the authors believe it may reflect the relatively small endowment at stake? Do the authors believe this behavior would persist if the stakes were higher?
Additionally, might the type of harm inflicted (e.g., electric shock vs. less stigmatized/less ethically charged harm like placing a hand in ice-cold water) influence the weight of self-interest in decision-making?
Taken together, the conclusions of the paper are well supported by the data. It would be valuable for future studies to validate these findings using alternative tasks or paradigms to ensure the robustness and generalizability of the observed behavioral and neural mechanisms.
Thank you for these important questions. As you suggested, we believe that the relatively small personal stakes in our task (a maximum loss of 5 Chinese yuan) likely explain why the computational model indicated that participants disregarded selfinterest. We also agree that when the harm to others is less morally charged, people may be more inclined to consider self-interest in compensatory decision-making. Overall, the more stigmatized the harm and the smaller the personal stakes, the more likely individuals are to disregard self-interest and focus solely on making appropriate compensation.
We have added the following passage to the Discussion section (Page 42): “Notably, in many computational models of social decision-making, self-interest plays a crucial role (e.g., Wu et al., 2024). However, our computational findings suggest that participants disregarded self-interest during compensatory decision-making. A possible explanation is that the personal stakes in our task were relatively small (a maximum loss of 5 Chinese yuan), whereas the harm inflicted on the receiver was highly stigmatized (i.e., an electric shock). Under conditions where the harm is highly salient and the cost of compensation is low, participants may be inclined to disregard selfinterest and focus solely on making appropriate compensation.”
Reviewer #2 (Public review):
Summary
The authors combined behavioral experiments, computational modeling, and functional magnetic resonance imaging (fMRI) to investigate the psychological and neural mechanisms underlying guilt, shame, and the altruistic behaviors driven by these emotions. The results revealed that guilt is more strongly associated with harm, whereas shame is more closely linked to responsibility. Compared to shame, guilt elicited a higher level of altruistic behavior. Computational modeling demonstrated how individuals integrate information about harm and responsibility. The fMRI findings identified a set of brain regions involved in representing harm and responsibility, transforming responsibility into feelings of shame, converting guilt and shame into altruistic actions, and mediating the effect of trait guilt on compensatory behavior.
Strengths
This study offers a significant contribution to the literature on social emotions by moving beyond prior research that typically focused on isolated aspects of guilt and shame. The study presents a comprehensive examination of these emotions, encompassing their cognitive antecedents, affective experiences, behavioral consequences, trait-level characteristics, and neural correlates. The authors have introduced a novel experimental task that enables such a systematic investigation and holds strong potential for future research applications. The computational modeling procedures were implemented in accordance with current field standards. The findings are rich and offer meaningful theoretical insights. The manuscript is well written, and the results are clearly and logically presented.
We are thankful for your considerate acknowledgment of our work’s strengths and truly value your positive comments.
We would like to note that, in accordance with the journal’s requirements, we have uploaded both a clean version of the revised manuscript and a version with all modifications highlighted in blue.
Weakness
In this study, participants' feelings of guilt and shame were assessed retrospectively, after they had completed all altruistic decision-making tasks. This reliance on memorybased self-reports may introduce recall bias, potentially compromising the accuracy of the emotion measurements.
Thank you for this crucial comment. We fully agree that measuring guilt and shame after the task may affect accuracy to some extent. However, because participants reported their emotions immediately after completing the task, we believe their recollections were reasonably accurate. In designing the experiment, we considered intask assessments, but this approach risked heightening participants’ awareness of guilt and shame and thereby interfering with compensatory decisions. After careful consideration, we ultimately chose post-task assessments of these emotions. A similar approach has been adopted in prior research on gratitude, where post-task assessments were also used (Yu et al., 2018).
In the revised manuscript, we have specified the limitations of both post-task and intask assessments of guilt and shame (Page 47): “… post-task assessments of guilt and shame, unlike in-task assessments, rely on memory and may thus be less precise, although in-task assessments could have heightened participants’ awareness of these emotions and biased their decisions.”.
In many behavioral economic models, self-interest plays a central role in shaping individual decision-making, including moral decisions. However, the model comparison results in this study suggest that models without a self-interest component (such as Model 1.3) outperform those that incorporate it (such as Model 1.1 and Model 1.2). The authors have not provided a satisfactory explanation for this counterintuitive finding.
Thank you for this important comment. In the revised manuscript, we have provided a possible explanation (Page 42): “Notably, in many computational models of social decision-making, self-interest plays a crucial role (e.g., Wu et al., 2024). However, our computational findings suggest that participants disregarded self-interest during compensatory decision-making. A possible explanation is that the personal stakes in our task were relatively small (a maximum loss of 5 Chinese yuan), whereas the harm inflicted on the receiver was highly stigmatized (i.e., an electric shock). Under conditions where the harm is highly salient and the cost of compensation is low, participants may be inclined to disregard self-interest and focus solely on making appropriate compensation.”
The phrases "individuals integrate harm and responsibility in the form of a quotient" and "harm and responsibility are integrated in the form of a quotient" appear in the Abstract and Discussion sections. However, based on the results of the computational modeling, it is more accurate to state that "harm and the number of wrongdoers are integrated in the form of a quotient." The current phrasing misleadingly suggests that participants represent information as harm divided by responsibility, which does not align with the modeling results. This potentially confusing expression should be revised for clarity and accuracy.
We sincerely thank you for this helpful suggestion and apologize for the confusion caused. We have removed expressions such as “harm and responsibility are integrated in the form of a quotient” from the manuscript. Instead, we now state more precisely that “harm and the number of wrongdoers are integrated in the form of a quotient.”
However, in certain contexts we continue to discuss harm and responsibility. Introducing “the number of wrongdoers” in these places would appear abrupt, so we have opted for alternative phrasing. For example, on Page 3, we now write:
“Computational modeling results indicated that the integration of harm and responsibility by individuals is consistent with the phenomenon of responsibility diffusion.” Similarly, on Page 49, we state: “Notably, harm and responsibility are integrated in a manner consistent with responsibility diffusion prior to influencing guilt-driven and shame-driven compensation.”
In the Discussion, the authors state: "Since no brain region associated with social cognition showed significant responses to harm or responsibility, it appears that the human brain encodes a unified measure integrating harm and responsibility (i.e., the quotient) rather than processing them as separate entities when both are relevant to subsequent emotional experience and decision-making." However, this interpretation overstates the implications of the null fMRI findings. The absence of significant activation in response to harm or responsibility does not necessarily imply that the brain does not represent these dimensions separately. Null results can arise from various factors, including limitations in the sensitivity of fMRI. It is possible that more finegrained techniques, such as intracranial electrophysiological recordings, could reveal distinct neural representations of harm and responsibility. The interpretation of these null findings should be made with greater caution.
Thank you for this reminder. In the revised manuscript, we have provided a more cautious interpretation of the results (Page 43): “Although the fMRI findings revealed that no brain region associated with social cognition showed significant responses to harm or responsibility, this does not suggest that the human brain encodes only a unified measure integrating harm and responsibility and does not process them as separate entities. Using more fine-grained techniques, such as intracranial electrophysiological recordings, it may still be possible to observe independent neural representations of harm and responsibility.”
Reviewer #3 (Public review):
Summary
Zhu et al. set out to elucidate how the moral emotions of guilt and shame emerge from specific cognitive antecedents - harm and responsibility - and how these emotions subsequently drive compensatory behavior. Consistent with their prediction derived from functionalist theories of emotion, their behavioral findings indicate that guilt is more influenced by harm, whereas shame is more influenced by responsibility. In line with previous research, their results also demonstrate that guilt has a stronger facilitating effect on compensatory behavior than shame. Furthermore, computational modeling and neuroimaging results suggest that individuals integrate harm and responsibility information into a composite representation of the individual's share of the harm caused. Brain areas such as the striatum, insula, temporoparietal junction, lateral prefrontal cortex, and cingulate cortex were implicated in distinct stages of the processing of guilt and/or shame. In general, this work makes an important contribution to the field of moral emotions. Its impact could be further enhanced by clarifying methodological details, offering a more nuanced interpretation of the findings, and discussing their potential practical implications in greater depth.
Strengths
First, this work conceptualizes guilt and shame as processes unfolding across distinct stages (cognitive appraisal, emotional experience, and behavioral response) and investigates the psychological and neural characteristics associated with their transitions from one stage to the next.
Second, the well-designed experiment effectively manipulates harm and responsibility - two critical antecedents of guilt and shame.
Third, the findings deepen our understanding of the mechanisms underlying guilt and shame beyond what has been established in previous research.
We truly appreciate your acknowledgment of our work’s strengths and your encouraging feedback.
We would like to note that, in accordance with the journal’s requirements, we have uploaded both a clean version of the revised manuscript and a version with all modifications highlighted in blue.
Weakness
Over the course of the task, participants may gradually become aware of their high error rate in the dot estimation task. This could lead them to discount their own judgments and become inclined to rely on the choices of other deciders. It is unclear whether participants in the experiment had the opportunity to observe or inquire about others' choices. This point is important, as the compensatory decision-making process may differ depending on whether choices are made independently or influenced by external input.
Thank you for pointing this out. We apologize for not making the experimental procedure sufficiently clear. Participants (as deciders) were informed that each decider performed the dot estimation independently and was unaware of the estimations made by the other deciders. We now have clarified this point in the revised manuscript (Pages 10 and 11): “Each decider indicated whether the number of dots was more than or less than 20 based on their own estimation by pressing a corresponding button (dots estimation period, < 2.5 s) and was unaware of the estimations made by other deciders”.
Given the inherent complexity of human decision-making, it is crucial to acknowledge that, although the authors compared eight candidate models, other plausible alternatives may exist. As such, caution is warranted when interpreting the computational modeling results.
Thank you for this comment. We fully agree with your opinion. Although we tried to build a conceptually comprehensive model space based on prior research and our own understanding, we did not include all plausible models, nor would it be feasible to do so. We acknowledge it as a limitation in the revised manuscript (Page 47): “... although we aimed to construct a conceptually comprehensive computational model space informed by prior research and our own understanding, it does not encompass all plausible models. Future research is encouraged to explore additional possibilities.”
I do not agree with the authors' claim that "computational modeling results indicated that individuals integrate harm and responsibility in the form of a quotient" (i.e., harm/responsibility). Rather, the findings appear to suggest that individuals may form a composite representation of the harm attributable to each individual (i.e., harm/the number of people involved). The explanation of the modeling results ought to be precise.
We appreciate your comment and apologize for the imprecise description. In the revised manuscript, we now use the expressions “… integrate harm and the number of wrongdoers in the form of a quotient.” and “… the integration of harm and responsibility by individuals is consistent with the phenomenon of responsibility diffusion.” For example, on Page 19, we state: “It assumes that individuals neglect their self-interest, have a compensatory baseline, and integrate harm and the number of wrongdoers in the form of a quotient.” On Page 3, we state: “Computational modeling results indicated that the integration of harm and responsibility by individuals is consistent with the phenomenon of responsibility diffusion.”
Many studies have reported positive associations between trait gratitude, social value orientation, and altruistic behavior. It would be helpful if the authors could provide an explanation about why this study failed to replicate these associations.
Thanks a lot for this important comment. We have now added an explanation into the revised manuscript (Page 47): “Although previous research has found that trait gratitude and SVO are significantly associated with altruistic behavior in contexts such as donation (Van Lange et al., 2007; Yost-Dubrow & Dunham, 2018) and reciprocity (Ma et al., 2017; Yost-Dubrow & Dunham, 2018), their associations with compensatory decisions in the present study were not significant. This suggests that the effects of trait gratitude and SVO on altruistic behavior are context-dependent and may not predict all forms of altruistic behavior.”
As the authors noted, guilt and shame are closely linked to various psychiatric disorders. It would be valuable to discuss whether this study has any implications for understanding or even informing the treatment of these disorders.
We are grateful for this advice. Although our study did not directly examine patients with psychological disorders, the findings offer insights into the regulation of guilt and shame. As these emotions are closely linked to various disorders, improving their regulation may help alleviate related symptoms. Accordingly, we have added a paragraph highlighting the potential clinical relevance (Pages 48 and 49): “Our study has potential practical implications. The behavioral findings may help counselors understand how cognitive interventions targeting perceptions of harm and responsibility could influence experiences of guilt and shame. The neural findings highlight specific brain regions (e.g., TPJ) as potential intervention targets for regulating these emotions. Given the close links between guilt, shame, and various psychological disorders (e.g., Kim et al., 2011; Lee et al., 2001; Schuster et al., 2021), strategies to regulate these emotions may contribute to symptom alleviation. Nevertheless, because this study was conducted with healthy adults, caution is warranted when considering applications to other populations.”
Reviewer #1 (Recommendations for the authors):
(1) Would it be interesting to explore other categories of behavior apart from compensatory behavior?
Thanks a lot for this insightful question. We focused on a classic form of altruistic behavior, compensation. Future studies are encouraged to adapt our paradigm to examine other behaviors associated with guilt and/or shame, such as donation (Xu, 2022), avoidance (Shen et al., 2023), or aggression (Velotti et al., 2014). Please see Page 48: “Future research could combine this paradigm with other cognitive neuroscience methods, such as electroencephalography (EEG) or magnetoencephalography (MEG), and adapt it to investigate additional behaviors linked to guilt and shame, including donation (Xu, 2022), avoidance (Shen et al., 2023), and aggression (Velotti et al., 2014).”
(2) Did the computational model account for the position of the block (slider) at the start of each decision-making response (when participants had to decide how to divide the endowment)? Or are anchoring effects not relevant/ not a concern?
Thank you for this interesting question. In our task, the initial position of the slider was randomized across trials, and participants were explicitly informed of this in the instructions. This design minimized stable anchoring effects across trials, as participants could not rely on a consistent starting point. Although anchoring might still have influenced individual trial responses, we believe it is unlikely that such effects systematically biased our results, since randomization would tend to cancel them out across trials. Additionally, prior research has shown that when multiple anchors are presented, anchoring effects are reduced if the anchors contradict each other (Switzer
III & Sniezek, 1991). Therefore, we did not attempt to model potential anchoring effects. Nevertheless, future research could systematically manipulate slider starting positions to directly examine possible anchoring influences. In the revised manuscript, we have added a brief clarification (Page 11): “The initial position of the block was randomized across trials, which helped minimize stable anchoring effects across trials.”
(3) Was there a real receiver who experienced the shocks and received compensation? I think it is not completely clear in the paper.
We are sorry for not making this clear enough. The receiver was fictitious and did not actually exist. We have supplemented the Methods section with the following description (Page 12): “We told the participant a cover story that the receiver was played by another college student who was not present in the laboratory at the time. … In fact, the receiver did not actually exist.”.
(4) What was the rationale behind not having participants meet the receiver?
Thank you for this question. Having participants meet the receiver (i.e., the victim), played by a confederate, might have intensified their guilt and shame and produced a ceiling effect. In addition, the current approach simplified the experimental procedure and removed the need to recruit an additional confederate. These reasons have been added to the Methods section (Page 12): “Not having participants meet the receiver helped prevent excessive guilt and shame that might produce a ceiling effect, while also eliminating the need to recruit an additional confederate.”
Minor edits:
(1) Line 49: "the cognitive assessment triggers them", I think a word is missing.
(2) Line 227: says 'Slide' instead of 'Slider'.
(3) Lines 867/868: "No brain response showed significant correlation with responsibility-driven guilt sensitivity, harm-driven shame sensitivity, or responsibilitydriven shame sensitivity." I think it should be harm-driven guilt sensitivity, responsibility-driven guilt sensitivity, and harm-driven shame sensitivity.
(4) Supplementary Information Line 12: I think there is a typo ( 'severs' instead of 'serves')
We sincerely thank you for patiently pointing out these typos. We have corrected them accordingly.
(1) “the cognitive assessment triggers them” has been revised to “the cognitive antecedents that trigger them” (Page 2).
(2) “SVO Slide Measure” has been revised to “SVO Slider Measure” (Page 8).
(3) “No brain response showed significant correlation with responsibility-driven guilt sensitivity, harm-driven shame sensitivity, or responsibility-driven shame sensitivity." has been revised to “No brain response showed significant correlation with harm-driven guilt sensitivity, responsibility-driven guilt sensitivity, and harm-driven shame sensitivity.” (Page 35).
(4) “severs” has been revised to “serves” (see Supplementary Information). In addition, we have carefully checked the entire manuscript to correct any remaining typographical errors.
Reviewer #2 (Recommendations for the authors):
The statement that trait gratitude and SVO were measured "for exploratory purposes" would benefit from further clarification regarding the specific questions being explored.
Thank you for this valuable suggestion. In the revised manuscript, we have illustrated the exploratory purposes (Page 9): “We measured trait gratitude and SVO for exploratory purposes. Previous research has shown that both are linked to altruistic behavior, particularly in donation contexts (Van Lange et al., 2007; Yost-Dubrow & Dunham, 2018) and reciprocity contexts (Ma et al., 2017; Yost-Dubrow & Dunham, 2018). Here, we explored whether they also exert significant effects in a compensatory context.”
In the Methods section, the authors state: "To confirm the relationships between κ and guilt-driven and shame-driven compensatory sensitivities, we calculated the Pearson correlations between them." However, the Results section reports linear regression results rather than Pearson correlation coefficients, suggesting a possible inconsistency. The authors are advised to carefully check and clarify the analysis approach used.
We thank you for the careful reviewing and apologize for this mistake. We used a linear mixed-effects regression instead of Pearson correlations for the analysis. The mistake has been revised (Page 25): “To confirm the relationships between κ and guiltdriven and shame-driven compensatory sensitivities, we conducted a linear mixedeffects regression. κ was regressed onto guilt-driven and shame-driven compensatory sensitivities, with participant-specific random intercepts and random slopes for each fixed effect included as random effects.”
A more detailed discussion of how the current findings inform the regulation of guilt and shame would further strengthen the contribution of this study.
Thank you for this suggestion. We have added a paragraph discussing the implications for the regulation of guilt and shame (Pages 48 and 49): “Our study has potential practical implications. The behavioral findings may help counselors understand how cognitive interventions targeting perceptions of harm and responsibility could influence experiences of guilt and shame. The neural findings highlight specific brain regions (e.g., TPJ) as potential intervention targets for regulating these emotions. Given the close links between guilt, shame, and various psychological disorders (e.g., Kim et al., 2011; Lee et al., 2001; Schuster et al., 2021), strategies to regulate these emotions may contribute to symptom alleviation. Nevertheless, because this study was conducted with healthy adults, caution is warranted when considering applications to other populations.”
As fMRI provides only correlational evidence, establishing a causal link between neural activity and guilt- or shame-related cognition and behavior would require brain stimulation or other intervention-based methods. This may represent a promising direction for future research.
Thank you for this advice. We also agree that it is important for future research to establish the causal relationships between the observed brain activity, psychological processes, and behavior. We have added a corresponding discussion in the revised manuscript (Pages 47 and 48): “… fMRI cannot establish causality. Future studies using brain stimulation techniques (e.g., transcranial magnetic stimulation) are needed to clarify the causal role of brain regions in guilt-driven and shame-driven altruistic behavior.”
Reviewer #3 (Recommendations for the authors):
It was mentioned that emotions beyond guilt and shame, such as indebtedness, may also drive compensation. Were any additional types of emotion measured in the study?
Thank you for this question. We did not explicitly measure emotions other than guilt and shame. However, the parameter κ from our winning computational model captures the combined influence of various psychological processes on compensation, which may reflect the impact of emotions beyond guilt and shame (e.g., indebtedness). We acknowledge that measuring other emotions similar to guilt and shame may help to better understand their distinct contributions. This point has been added into the revised manuscript (Page 48): “… we did not explicitly measure emotions similar to guilt and shame (e.g., indebtedness), which would have been helpful for understanding their distinct contributions.”
The experimental task is complicated, raising the question of whether participants fully understood the instructions. For instance, one participant's compensation amount was zero. Could this reflect a misunderstanding of the task instructions?
Thanks a lot for this question. In our study, after reading the instructions, participants were required to complete a comprehension test on the experimental rules. If they made any mistakes, the experimenter provided additional explanations. Only after participants fully understood the rules and correctly answered all comprehension questions did they proceed to the main experimental task. We have clarified this procedure in the revised manuscript (Page 13): “Participants did not proceed to the interpersonal game until they had fully understood the experimental rules and passed a comprehension test.”
Making identical choices across different trials does not necessarily indicate that participants misunderstood the rules. Similar patterns, where participants made the same choices across trials, have also been observed in previous studies (Zhong et al., 2016; Zhu et al., 2021).
Reference
Cohen, T. R., Wolf, S. T., Panter, A. T., & Insko, C. A. (2011). Introducing the GASP scale: a new measure of guilt and shame proneness. Journal of Personality and Social Psychology, 100(5), 947–966. https://doi.org/10.1037/a0022641
Esterman, M., Tamber-Rosenau, B. J., Chiu, Y. C., & Yantis, S. (2010). Avoiding nonindependence in fMRI data analysis: Leave one subject out. NeuroImage, 50(2), 572–576. https://doi.org/10.1016/j.neuroimage.2009.10.092
Kim, S., Thibodeau, R., & Jorgensen, R. S. (2011). Shame, guilt, and depressive symptoms: A meta-analytic review. Psychological Bulletin, 137(1), 68. https://doi.org/10.1037/a0021466
Lee, D. A., Scragg, P., & Turner, S. (2001). The role of shame and guilt in traumatic events: A clinical model of shame-based and guilt-based PTSD. British Journal of Medical Psychology, 74(4), 451–466. https://doi.org/10.1348/000711201161109
Ma, L. K., Tunney, R. J., & Ferguson, E. (2017). Does gratitude enhance prosociality?: A meta-analytic review. Psychological Bulletin, 143(6), 601–635. https://doi.org/10.1037/bul0000103
Michl, P., Meindl, T., Meister, F., Born, C., Engel, R. R., Reiser, M., & Hennig-Fast, K. (2014). Neurobiological underpinnings of shame and guilt: A pilot fMRI study. Social Cognitive and Affective Neuroscience, 9(2), 150–157.
Schuster, P., Beutel, M. E., Hoyer, J., Leibing, E., Nolting, B., Salzer, S., Strauss, B., Wiltink, J., Steinert, C., & Leichsenring, F. (2021). The role of shame and guilt in social anxiety disorder. Journal of Affective Disorders Reports, 6, 100208. https://doi.org/10.1016/j.jadr.2021.100208
Shen, B., Chen, Y., He, Z., Li, W., Yu, H., & Zhou, X. (2023). The competition dynamics of approach and avoidance motivations following interpersonal transgression. Proceedings of the National Academy of Sciences, 120(40), e2302484120. https://doi.org/10.1073/pnas.230248412
Switzer III, F. S., & Sniezek, J. A. (1991). Judgment processes in motivation: Anchoring and adjustment effects on judgment and behavior. Organizational Behavior and Human Decision Processes, 49(2), 208–229. https://doi.org/10.1016/0749-5978(91)90049-Y
Van Lange, P. A. M., Bekkers, R., Schuyt, T. N. M., & Van Vugt, M. (2007). From games to giving: Social value orientation predicts donations to noble causes. Basic and Applied Social Psychology, 29(4), 375–384. https://doi.org/10.1080/01973530701665223
Velotti, P., Elison, J., & Garofalo, C. (2014). Shame and aggression: Different trajectories and implications. Aggression and Violent Behavior, 19(4), 454–461. https://doi.org/10.1016/j.avb.2014.04.011
Wagner, U., N’Diaye, K., Ethofer, T., & Vuilleumier, P. (2011). Guilt-specific processing in the prefrontal cortex. Cerebral Cortex, 21(11), 2461–2470. https://doi.org/10.1093/cercor/bhr016
Wu, X., Ren, X., Liu, C., & Zhang, H. (2024). The motive cocktail in altruistic behaviors. Nature Computational Science, 4, 659–676. https://doi.org/10.1038/s43588-024-00685-6
Xu, J. (2022). The impact of guilt and shame in charity advertising: The role of self- construal. Journal of Philanthropy and Marketing, 27(1). https://doi.org/10.1002/nvsm.1709
Yost-Dubrow, R., & Dunham, Y. (2018). Evidence for a relationship between trait gratitude and prosocial behaviour. Cognition and Emotion, 32(2), 397–403. https://doi.org/10.1080/02699931.2017.1289153
Yu, H., Gao, X., Zhou, Y., & Zhou, X. (2018). Decomposing gratitude: Representation and integration of cognitive antecedents of gratitude in the brain. Journal of Neuroscience, 38(21), 4886–4898. https://doi.org/10.1523/JNEUROSCI.2944-17.2018
Zhong, S., Chark, R., Hsu, M., & Chew, S. H. (2016). Computational substrates of social norm enforcement by unaffected third parties. NeuroImage, 129, 95–104. https://doi.org/10.1016/j.neuroimage.2016.01.040
Zhu, R., Feng, C., Zhang, S., Mai, X., & Liu, C. (2019). Differentiating guilt and shame in an interpersonal context with univariate activation and multivariate pattern analyses. NeuroImage, 186, 476486. https://doi.org/10.1016/j.neuroimage.2018.11.012
Zhu, R., Xu, Z., Su, S., Feng, C., Luo, Y., Tang, H., Zhang, S., Wu, X., Mai, X., & Liu, C. (2021). From gratitude to injustice: Neurocomputational mechanisms of gratitude-induced injustice. NeuroImage, 245, 118730. https://doi.org/10.1016/j.neuroimage.2021.118730
DA2123
DOI: 10.1172/JCI165814
Resource: RRID:WB-STRAIN:WBStrain00005592
Curator: @Apiekniewska
SciCrunch record: RRID:WB-STRAIN:WBStrain00005592
RRID:SCR_023250
DOI: 10.1038/s42255-025-01390-y
Resource: Stanford Neuroscience Gene Vector and Virus Core Facility (RRID:SCR_023250)
Curator: @scibot
SciCrunch record: RRID:SCR_023250
RRID:Addgene_111661
DOI: 10.1038/s42255-025-01390-y
Resource: RRID:Addgene_111661
Curator: @scibot
SciCrunch record: RRID:Addgene_111661
RRID:Addgene_111811
DOI: 10.1038/s42255-025-01390-y
Resource: RRID:Addgene_111811
Curator: @scibot
SciCrunch record: RRID:Addgene_111811
RRID:Addgene_136470
DOI: 10.1038/s42255-025-01390-y
Resource: RRID:Addgene_136470
Curator: @scibot
SciCrunch record: RRID:Addgene_136470
RRID:Addgene_50458
DOI: 10.1038/s42255-025-01390-y
Resource: RRID:Addgene_50458
Curator: @scibot
SciCrunch record: RRID:Addgene_50458
RRID:Addgene_178582
DOI: 10.1038/s42255-025-01390-y
Resource: RRID:Addgene_178582
Curator: @scibot
SciCrunch record: RRID:Addgene_178582
RRID:SCR_022432
DOI: 10.1038/s42255-025-01390-y
Resource: University of Pennsylvania Perelman School of Medicine Vector Core Facility (RRID:SCR_022432)
Curator: @scibot
SciCrunch record: RRID:SCR_022432
eglamento del Código Territorial para el Estado y los Municipios de Guanajuato
se repite
ведь они могут существовать формально
Непонятно, что это значит. В структуре конститутивного правила X считается за Y, значит, есть причинная сила для тех, для кого считается
Author Response:
Reviewer #1 (Public Review):
The work by Wang et al. examined how task-irrelevant, high-order rhythmic context could rescue the attentional blink effect via reorganizing items into different temporal chunks, as well as the neural correlates. In a series of behavioral experiments with several controls, they demonstrated that the detection performance of T2 was higher when occurring in different chunks from T1, compared to when T1 and T2 were in the same chunk. In EEG recordings, they further revealed that the chunk-related entrainment was significantly correlated with the behavioral effect, and the alpha-band power for T2 and its coupling to the low-frequency oscillation were also related to behavioral effect. They propose that the rhythmic context implements a second-order temporal structure to the first-order regularities posited in dynamic attention theory.
Overall, I find the results interesting and convincing, particularly the behavioral part. The manuscript is clearly written and the methods are sound. My major concerns are about the neural part, i.e., whether the work provides new scientific insights to our understanding of dynamic attention and its neural underpinnings.
1) A general concern is whether the observed behavioral related neural index, e.g., alpha-band power, cross-frequency coupling, could be simply explained in terms of ERP response for T2. For example, when the ERP response for T2 is larger for between-chunk condition compared to within-chunk condition, the alpha-power for T2 would be also larger for between-chunk condition. Likewise, this might also explain the cross-frequency coupling results. The authors should do more control analyses to address the possibility, e.g., plotting the ERP response for the two conditions and regressing them out from the oscillatory index.
Many thanks for the comment. In short, the enhancement in alpha power and cross-frequency coupling results in the between-cycle condition compared with those in the within-cycle condition cannot be accounted for by the ERP responses for T2.
In general, the rhythmic stimulation in the AB paradigm prevents EEG signals from returning to the baseline. Therefore, we cannot observe typical ERP components purely related to individual items, except for the P1 and N1 components related to the stream onset, which reveals no difference between the two conditions and are trailed by steady-state responses (SSRs) resonating at the stimulus rate (Fig. R1).
Fig. R1. ERPs aligned to stream onset. EEG signals were filtered between 1–30 Hz, baseline-corrected (-200 to 0 ms before stream onset) and averaged across the electrodes in left parieto-occipital area where 10-Hz alpha power showed attentional modulation effect.
To further inspect the potential differences in the target-related ERP signals between the within- and between-cycle conditions, we plotted the target-aligned waveforms for these experimental conditions. As shown in Fig. R2, a drop of ERP amplitude occurred for both conditions around T2 onset, and the difference between these two conditions was not significant (paired t-test estimated on mean amplitude every 20 ms from 0 to 700 ms relative to T1 onset, p > .05, FDR-corrected).
Fig. R2. ERPs aligned to T1 onset. EEG signals were filtered between 1–30 Hz, and baseline-corrected using signals -100 to 0 ms before T1 onset. The two dash lines indicate the onset of T1 and T2, respectively.
Since there is a trend of enhanced ERP response for the between-cycle relative to the within-cycle condition during the period of 0 to 100 ms after T2 onset (paired t-test on mean amplitude, p =.065, uncorrected), we then directly examined whether such post-T2 responses contribute to the behavioral attentional modulation effect and behavior-related neural indices. Crucially, we did not find any significant correlation of such T2-related ERP enhancement with the behavioral modulation index (BMI), or with the reported effects of alpha power and cross-frequency coupling (PAC). Furthermore, after controlling for the T2-related ERP responses, there still remains a significant correlation between the delta-alpha PAC and the BMI (rpartial = .596, p = .019), which is not surprising given that the PAC is calculated based on an 800-ms time window covering more pre-T2 than post-T2 periods (see the response to point #4 for details) rather than around the T2 onset. Taken together, these results clearly suggest that the T2-related ERP responses cannot explain the attentional modulation effect and the observed behavior-related neural indices.
2) The alpha-band increase for T2 is indeed contradictory to the well known inhibitory function of alpha-band in attention. How could a target that is better discriminated elicit stronger inhibitory response? Related to the above point, the observed enhancement in alpha-band power and its coupling to low-frequency oscillation might derive from an enhanced ERP response for T2 target.
Many thanks for the comment. We have briefly discussed this point in the revised manuscript (page 18, line 477).
A widely accepted function of alpha activity in attention is that alpha oscillations suppress irrelevant visual information during spatial selection (Kelly et al., 2006; Thut et al., 2006; Worden et al., 2000). However, it becomes a controversial issue when there exists rhythmic sensory stimulation at alpha-band, just like the situation in the current study where both the visual stream and the contextual auditory rhythm were emitted at 10 Hz. In such a case, alpha-band neural responses at the stimulation frequency can be interpreted as either passively evoked steady-state responses (SSR) or actively synchronized intrinsic brain rhythms. From the former perspective (i.e., the SSR view), an increase in the amplitude or power at the stimulus frequency may indicate an enhanced attentional allocation to the stimulus stream that may result in better target detection (Janson et al., 2014; Keil et al., 2006; Müller & Hübner, 2002). Conversely, the latter view of the inhibitory function of intrinsic alpha oscillations would produce the opposite prediction. In a previous AB study, Janson and colleagues (2014) investigated this issue by separating the stimulus-evoked activity at 12 Hz (using the same power analysis method as ours) from the endogenous alpha oscillations ranging from 10.35 to 11.25 Hz (as indexed by individual alpha frequency, IAF). Interestingly, they found a dissociation between these two alpha-band neural responses, showing that the RSVP frequency power was higher in non-AB trials (T2 detected) than in AB trials (T2 undetected) while the IAF power exhibited the opposite pattern. According to these findings, the currently observed increase in alpha power for the between-cycle condition may reflect more of the stimulus-driven processes related to attentional enhancement. However, we don’t negate the effect of intrinsic alpha oscillations in our study, as the current design is not sufficient to distinguish between these two processes. We have discussed this point in the revised manuscript (page 18, line 477). Also, we have to admit that “alpha power” may not be the most precise term to describe our findings of the stimulus-related results. Thus, we have specified it as “neural responses to first-order rhythms at 10 Hz” and “10-Hz alpha power” in the revised manuscript (see page 12 in the Results section and page 18 in the Discussion section).
As for the contribution of T2-related ERP response to the observed effect of 10 Hz power and cross-frequency coupling, please refer to our response to point #1.
References:
Janson, J., De Vos, M., Thorne, J. D., & Kranczioch, C. (2014). Endogenous and Rapid Serial Visual Presentation-induced Alpha Band Oscillations in the Attentional Blink. Journal of Cognitive Neuroscience, 26(7), 1454–1468. https://doi.org/10.1162/jocn_a_00551
Keil, A., Ihssen, N., & Heim, S. (2006). Early cortical facilitation for emotionally arousing targets during the attentional blink. BMC Biology, 4(1), 23. https://doi.org/10.1186/1741-7007-4-23
Kelly, S. P., Lalor, E. C., Reilly, R. B., & Foxe, J. J. (2006). Increases in Alpha Oscillatory Power Reflect an Active Retinotopic Mechanism for Distracter Suppression During Sustained Visuospatial Attention. Journal of Neurophysiology, 95(6), 3844–3851. https://doi.org/10.1152/jn.01234.2005
Müller, M. M., & Hübner, R. (2002). Can the Spotlight of Attention Be Shaped Like a Doughnut? Evidence From Steady-State Visual Evoked Potentials. Psychological Science, 13(2), 119–124. https://doi.org/10.1111/1467-9280.00422
Thut, G., Nietzel, A., Brandt, S., & Pascual-Leone, A. (2006). Alpha-band electroencephalographic activity over occipital cortex indexes visuospatial attention bias and predicts visual target detection. The Journal of Neuroscience : The Official Journal of the Society for Neuroscience, 26(37), 9494–9502. https://doi.org/10.1523/JNEUROSCI.0875-06.2006
Worden, M. S., Foxe, J. J., Wang, N., & Simpson, G. V. (2000). Anticipatory Biasing of Visuospatial Attention Indexed by Retinotopically Specific α-Bank Electroencephalography Increases over Occipital Cortex. Journal of Neuroscience, 20(6), RC63–RC63. https://doi.org/10.1523/JNEUROSCI.20-06-j0002.2000
3) To support that it is the context-induced entrainment that leads to the modulation in AB effect, the authors could examine pre-T2 response, e.g., alpha-power, and cross-frequency coupling, as well as its relationship to behavioral performance. I think the pre-stimulus response might be more convincing to support the authors' claim.
Many thanks for the insightful suggestion. We have conducted additional analyses.
Following this suggestion, we have examined the 10-Hz alpha power within the time window of -100–0 ms before T2 onset and found stronger activity for the between-cycle condition than for the within-cycle condition. This pre-T2 response is similar to the post-T2 response except that it is more restricted to the left parieto-occipital cluster (CP3, CP5, P3, P5, PO3, PO5, POZ, O1, OZ, t(15) = 2.774, p = .007), which partially overlaps with the cluster that exhibits a delta-alpha coupling effect significantly correlated with the BMI. We have incorporated these findings into the main text (page 12, line 315) and the Fig. 5A of the revised manuscript.
As for the coupling results reported in our manuscript, the coupling index (PAC) was calculated based on the activity during the second and third cycles (i.e., 400 to 1200 ms from stream onset) of the contextual rhythm, most of which covers the pre-T2 period as T2 always appeared in the third cycle for both conditions. Together, these results on pre-T2 10-Hz alpha power and cross-frequency coupling, as well as its relationship to behavioral performance, jointly suggest that the observed modulation effect is caused by the context-induced entrainment rather than being a by-product of post-T2 processing.
4) About the entrainment to rhythmic context and its relation to behavioral modulation index. Previous studies (e.g., Ding et al) have demonstrated the hierarchical temporal structure in speech signals, e.g., emergence of word-level entrainment introduced by language experience. Therefore, it is well expected that imposing a second-order structure on a visual stream would elicit the corresponding steady-state response. I understand that the new part and main focus here are the AB effects. The authors should add more texts explaining how their findings contribute new understandings to the neural mechanism for the intriguing phenomena.
Many thanks for the suggestion. We have provided more discussion in the revised manuscript (page 17, line 447).
We have provided more discussion on this important issue in the revised manuscript (page 17, line 447). In brief, our study demonstrates how cortical tracking of feature-based hierarchical structure reframes the deployment of attentional resources over visual streams. This effect, distinct from the hierarchical entrainment to speech signals (Ding et al., 2016; Gross et al., 2013), does not rely on previously acquired knowledge about the structured information and can be established automatically even when the higher-order structure comes from a task-irrelevant and cross-modal contextual rhythm. On the other hand, our finding sheds fresh light on the adaptive value of the structure-based entrainment effect by expanding its role from rhythmic information (e.g., speech) perception to temporal attention deployment. To our knowledge, few studies have tackled this issue in visual or speech processing.
References:
Ding, N., Melloni, L., Zhang, H., Tian, X., & Poeppel, D. (2016). Cortical tracking of hierarchical linguistic structures in connected speech. Nature Neuroscience, 19(1), 158–164. https://doi.org/10.1038/nn.4186
Gross, J., Hoogenboom, N., Thut, G., Schyns, P., Panzeri, S., Belin, P., & Garrod, S. (2013). Speech Rhythms and Multiplexed Oscillatory Sensory Coding in the Human Brain. PLoS Biol, 11(12). https://doi.org/10.1371/journal.pbio.1001752
Reviewer #2 (Public Review):
In cognitive neuroscience, a large number of studies proposed that neural entrainment, i.e., synchronization of neural activity and low-frequency external rhythms, is a key mechanism for temporal attention. In psychology and especially in vision, attentional blink is the most established paradigm to study temporal attention. Nevertheless, as far as I know, few studies try to link neural entrainment in the cognitive neuroscience literature with attentional blink in the psychology literature. The current study, however, bridges this gap.
The study provides new evidence for the dynamic attending theory using the attentional blink paradigm. Furthermore, it is shown that neural entrainment to the sensory rhythm, measured by EEG, is related to the attentional blink effect. The authors also show that event/chunk boundaries are not enough to modulate the attentional blink effect, and suggest that strict rhythmicity is required to modulate attention in time.
In general, I enjoyed reading the manuscript and only have a few relatively minor concerns.
1) Details about EEG analysis.
. First, each epoch is from -600 ms before the stimulus onset to 1600 ms after the stimulus onset. Therefore, the epoch is 2200 s in duration. However, zero-padding is needed to make the epoch duration 2000 s (for 0.5-Hz resolution). This is confusing. Furthermore, for a more conservative analysis, I recommend to also analyze the response between 400 ms and 1600 ms, to avoid the onset response, and show the results in a supplementary figure. The short duration reduces the frequency resolution but still allows seeing a 2.5-Hz response.
Thanks for the comments. Each epoch was indeed segmented from -600 to 1600 ms relative to the stimulus onset, but in the spectrum analysis, we only used EEG signals from stream onset (i.e., time point 0) to 1600 ms (see the Materials and Methods section) to investigate the oscillatory characteristics of the neural responses purely elicited by rhythmic stimuli. The 1.6-s signals were zero-padded into a 2-s duration to achieve a frequency resolution of 0.5 Hz.
According to the reviewer’s suggestion, we analyzed the EEG signals from 400 ms to 1600 ms relative to stream onset to avoid potential influence of the onset response, and showed the results in Figure 4. Basically, we can still observe spectral peaks at the stimulus frequencies of 2.5, 5 (the harmonic of 2.5 Hz), and 10 Hz for both power and ITPC spectrum. However, the peak magnitudes were much weaker than those of 1.6-s signals especially for 2.5 Hz, and the 2.5-Hz power did not survive the multiple comparisons correction across frequencies (FDR threshold of p < .05), which might be due to the relatively low signal-to-noise ratio for the analysis based on the 1.2-s epochs (only three cycles to estimate the activity at 2.5 Hz). Importantly, we did identify a significant cluster for 2.5 Hz ITPC in the left parieto-occipital region showing a positive correlation with the individuals’ BMI (Fig. R3; CP5, TP7, P5, P7, PO5, PO7, O1; r = .538, p = .016), which is consistent with the findings based on the longer epochs.
Fig. R3. Neural entrainment to contextual rhythms during the period of 400–1600 ms from stream onset. (A) The spectrum for inter-trial phase coherence (ITPC) of EEG signals from 400 to 1600 ms after the stimulus onset. Shaded areas indicate standard errors of the mean. (B) The 2.5-Hz ITPC was significantly correlated with the behavioral modulation index (BMI) in a parieto-occipital cluster, as indicated by orange stars in the scalp topographic map.
Second, "The preprocessed EEG signals were first corrected by subtracting the average activity of the entire stream for each epoch, and then averaged across trials for each condition, each participant, and each electrode." I have several concerns about this procedure.
(A) What is the entire stream? It's the average over time?
Yes, as for the power spectrum analysis, EEG signals were first demeaned by subtracting the average signals of the entire stream over time from onset to offset (i.e., from 0 to 1600 ms) before further analysis. We performed this procedure following previous studies on the entrainment to visual rhythms (Spaak et al., 2014). We have clarified this point in the “Power analysis” part of the Materials and Methods section (page 25, line 677).
References:
Spaak, E., Lange, F. P. de, & Jensen, O. (2014). Local Entrainment of Alpha Oscillations by Visual Stimuli Causes Cyclic Modulation of Perception. The Journal of Neuroscience, 34(10), 3536–3544. https://doi.org/10.1523/JNEUROSCI.4385-13.2014
(B) I suggest to do the Fourier transform first and average the spectrum over participants and electrodes. Averaging the EEG waveforms require the assumption that all electrodes/participants have the same response phase, which is not necessarily true.
Thanks for the suggestion. In an AB paradigm, the evoked neural responses are sufficiently time-locked to the periodic stimulation, so it is reasonable to quantify power estimate with spectral decomposition performed on trial-averaged EEG signals (i.e., evoked power). Moreover, our results of inter-trial phase coherence (ITPC), which estimated the phase-locking value across trials based on single-trial decomposed phase values, also provided supporting evidence that the EEG waveforms were temporally locked across trials to the 2.5-Hz temporal structure in the context session.
Nevertheless, we also took the reviewer’s suggestion seriously and analyzed the power spectrum on the average of single-trial spectral transforms, i.e., the induced power, which puts emphasis on the intrinsic non-phase-locked activities. In line with the results of evoked power and ITPC, the induced power spectrum in context session also peaked at 2.5 Hz and was significantly stronger than that in baseline session at 2.5 Hz (t(15) = 4.186, p < .001, FDR-corrected with a p value threshold < .001). Importantly, Person correlation analysis also revealed a positive cluster in the left parieto-occipital region, indicating the induced power at 2.5 Hz also had strong relevance with the attentional modulation effect (P7, PO7, PO5, PO3; r = .606, p = .006). We have added these additional findings to the revised manuscript (page 11, line 288; see also Figure 4—figure supplement 1).
2) The sequences are short, only containing 16 items and 4 cycles. Furthermore, the targets are presented in the 2nd or 3rd cycle. I suspect that a stronger effect may be observed if the sequence are longer, since attention may not well entrain to the external stimulus until a few cycles. In the first trial of the experiment, they participant may not have a chance to realize that the task-irrelevant auditory/visual stimulus has a cyclic nature and it is not likely that their attention will entrain to such cycles. As the experiment precedes, they learns that the stimulus is cyclic and may allocate their attention rhythmically. Therefore, I feel that the participants do not just rely on the rhythmic information within a trial but also rely on the stimulus history. Please discuss why short sequences are used and whether it is possible to see buildup of the effect over trials or over cycles within a trial.
Thanks for the comments. Typically, to induce a classic pattern of AB effect, the RSVP stream should contain 3–7 distractors before the first target (T1), with varying lengths of distractors (0–7) between two targets and at least 2 items after the second target (T2). In our study, we created the RSVP streams following these rules, which allowed us to observe the typical AB effect that T2 performance was deteriorated at Lag 2 relative to that at Lag 8. Nevertheless, we agree with the reviewer that longer streams would be better for building up the attentional entrainment effect, as we did observe the attentional modulation effect ramped up as the stream proceeded over cycles, consistent with the reviewer’s speculation. In Experiments 1a (using auditory context) and 2a (using color-defined visual context), we adopted two sets of target positions—an early one where T2 appeared at the 6th or 8th position (in the 2nd cycle) of the visual stream, and a late one where T2 appeared at the 10th or 12th position (in the 3rd cycle) of the visual stream. In the manuscript, we reported T2 performance with all the target positions combined, as no significant interaction was found between the target positions and the experimental conditions (ps. > .1). However, additional analysis demonstrated a trend toward an increase of the attentional modulation effect over cycles, from the early to the late positions. As shown in Fig. R4, the modulation effect went stronger and reached significance for the late positions (for Experiment 1a, t(15) = 2.83, p = .013, Cohen’s d = 0.707; for Experiment 2a, t(15) = 3.656, p = .002, Cohen’s d = 0.914) but showed a weaker trend for the early positions (for Experiment 1a, t(15) = 1.049, p = .311, Cohen’s d = 0.262; for Experiment 2a, t(15) = .606, p = .553, Cohen’s d = 0.152).
Fig. R4. Attentional modulation effect built up over cycles in Experiments 1a & 2a. Error bars represent 1 SEM; * p<0.05, ** p<0.01.
However, we did not observe an obvious buildup effect across trials in our study. The modulation effect of contextual rhythms seems to be a quick process that the effect is evident in the first quarter of trials in Experiment 1a (for, t(15) = 2.703, p = .016, Cohen’s d = 0.676) and in the second quarter of trials in Experiment 2a (for, t(15) = 2.478, p = .026, Cohen’s d = 0.620.
3) The term "cycle" is used without definition in Results. Please define and mention that it's an abstract term and does not require the stimulus to have "cycles".
Thanks for the suggestion. By its definition, the term “cycle” refers to “an interval of time during which a sequence of a recurring succession of events or phenomena is completed” or “a course or series of events or operations that recur regularly and usually lead back to the starting point” (Merriam-Webster dictionary). In the current study, we stuck to the recurrent and regular nature of “cycle” in general while defined the specific meaning of “cycle” by feature-based periodic changes of the contextual stimuli in each experiment (page 5, line 101; also refer to Procedures in the Materials and Methods section for details). For example, in Experiment 1a, the background tone sequence changed its pitch value from high to low or vice versa isochronously at a rate of 2.5 Hz, thus forming a rhythmic context with structure-based cycles of 400 ms. Note that we did not use the more general term “chunk”, because arbitrary chunks without the regularity of cycles are insufficient to trigger the attentional modulation effect in the current study. Indeed, the effect was eliminated when we replaced the rhythmic cycles with irregular chunks (Experiments 1d & 1e).
4) Entrainment of attention is not necessarily related to neural entrainment to sensory stimulus, and there is considerable debate about whether neural entrainment to sensory stimulus should be called entrainment. Too much emphasis on terminology is of course counterproductive but a short discussion on these issues is probably necessary.
Thanks for the comments. As commonly accepted, entrainment is defined as the alignment of intrinsic neuronal activity to the temporal structure of external rhythmic inputs (Lakatos et al., 2019; Obleser & Kayser, 2019). Here, we are interested in the functional roles of cortical entrainment to the higher-order temporal structure imposed on first-order sensory stimulation, and used the term entrainment to describe the phase-locking neural responses to such hierarchical structure following literature on auditory and visual perception (Brookshire et al., 2017; Doelling & Poeppel, 2015). In our study, the consistent results of power and ITPC have provided strong evidence that neural entrainment at the structure level (2.5 Hz) is significantly correlated with the observed attentional modulation effect. However, this does not mean that the entrainment of attention is necessarily associated with neural entrainment to sensory stimulus in a broader context, as attention may also be guided by predictions based on non-isochronous temporal regularity without requiring stimulus-based oscillatory entrainment (Breska & Deouell, 2017; Morillon et al._2016).
On the other hand, there has been a debate about whether the neural alignment to rhythmic stimulation reflects active entrainment of endogenous oscillatory processes (i.e., induced activity) or a series of passively evoked steady-state responses (Keitel et al., 2019; Notbohm et al., 2016; Zoefel et al., 2018). The latter process is also referred to as “entrainment in a broad sense” by Obleser & Kayser (2019). Given that a presented rhythm always evokes event-related potentials, a better question might be whether the observed alignment reflects the entrainment of endogenous oscillations in addition to evoked steady-state responses. Here we attempted to tackle this issue by measuring the induced power, which emphasizes the intrinsic non-phase-locked activity, in addition to the phase-locked evoked power. Specifically, we quantified these two kinds of activities with the average of single-trial EEG power spectra and the power spectra of trial-averaged EEG signals, respectively, according to Keitel et al. (2019). In addition to the observation of evoked responses to the contextual structure, we also demonstrated an attention-related neural tracking of the higher-order temporal structure based on the induced power at 2.5 Hz (see Figure 4—figure supplement 1), suggesting that the observed attentional modulation effect is at least partially derived from the entrainment of intrinsic oscillatory brain activity. We have briefly discussed this point in the revised manuscript (page 17, line 460).
References:
Breska, A., & Deouell, L. Y. (2017). Neural mechanisms of rhythm-based temporal prediction: Delta phase-locking reflects temporal predictability but not rhythmic entrainment. PLOS Biology, 15(2), e2001665. https://doi.org/10.1371/journal.pbio.2001665
Brookshire, G., Lu, J., Nusbaum, H. C., Goldin-Meadow, S., & Casasanto, D. (2017). Visual cortex entrains to sign language. Proceedings of the National Academy of Sciences, 114(24), 6352–6357. https://doi.org/10.1073/pnas.1620350114
Doelling, K. B., & Poeppel, D. (2015). Cortical entrainment to music and its modulation by expertise. Proceedings of the National Academy of Sciences, 112(45), E6233–E6242. https://doi.org/10.1073/pnas.1508431112
Henry, M. J., Herrmann, B., & Obleser, J. (2014). Entrained neural oscillations in multiple frequency bands comodulate behavior. Proceedings of the National Academy of Sciences, 111(41), 14935–14940. https://doi.org/10.1073/pnas.1408741111
Keitel, C., Keitel, A., Benwell, C. S. Y., Daube, C., Thut, G., & Gross, J. (2019). Stimulus-Driven Brain Rhythms within the Alpha Band: The Attentional-Modulation Conundrum. The Journal of Neuroscience, 39(16), 3119–3129. https://doi.org/10.1523/JNEUROSCI.1633-18.2019
Lakatos, P., Gross, J., & Thut, G. (2019). A New Unifying Account of the Roles of Neuronal Entrainment. Current Biology, 29(18), R890–R905. https://doi.org/10.1016/j.cub.2019.07.075
Morillon, B., Schroeder, C. E., Wyart, V., & Arnal, L. H. (2016). Temporal Prediction in lieu of Periodic Stimulation. Journal of Neuroscience, 36(8), 2342–2347. https://doi.org/10.1523/JNEUROSCI.0836-15.2016
Notbohm, A., Kurths, J., & Herrmann, C. S. (2016). Modification of Brain Oscillations via Rhythmic Light Stimulation Provides Evidence for Entrainment but Not for Superposition of Event-Related Responses. Frontiers in Human Neuroscience, 10. https://doi.org/10.3389/fnhum.2016.00010
Obleser, J., & Kayser, C. (2019). Neural Entrainment and Attentional Selection in the Listening Brain. Trends in Cognitive Sciences, 23(11), 913–926. https://doi.org/10.1016/j.tics.2019.08.004
Zoefel, B., ten Oever, S., & Sack, A. T. (2018). The Involvement of Endogenous Neural Oscillations in the Processing of Rhythmic Input: More Than a Regular Repetition of Evoked Neural Responses. Frontiers in Neuroscience, 12. https://doi.org/10.3389/fnins.2018.00095
Reviewer #3 (Public Review):
The current experiment tests whether the attentional blink is affected by higher-order regularity based on rhythmic organization of contextual features (pitch, color, or motion). The results show that this is indeed the case: the AB effect is smaller when two targets appeared in two adjacent cycles (between-cycle condition) than within the same cycle defined by the background sounds. Experiment 2 shows that this also holds for temporal regularities in the visual domain and Experiment 3 for motion. Additional EEG analysis indicated that the findings obtained can be explained by cortical entrainment to the higher-order contextual structure. Critically feature-based structure of contextual rhythms at 2.5 Hz was correlated with the strength of the attentional modulation effect.
This is an intriguing and exciting finding. It is a clever and innovative approach to reduce the attention blink by presenting a rhythmic higher-order regularity. It is convincing that this pulling out of the AB is driven by cortical entrainment. Overall, the paper is clear, well written and provides adequate control conditions. There is a lot to like about this paper. Yet, there are particular concerns that need to be addressed. Below I outline these concerns:
1) The most pressing concern is the behavioral data. We have to ensure that we are dealing here with a attentional blink. The way the data is presented is not the typical way this is done. Typically in AB designs one see the T2 performance when T1 is ignored relative to when T1 has to be detected. This data is not provided. I am not sure whether this data is collected but if so the reader should see this.
Many thanks for the suggestion. We appreciate the reviewer for his/her thoughtful comments. To demonstrate the AB effect, we did include two T2 lag conditions in our study (Experiments 1a, 1b, 2a, and 2b)—a short-SOA condition where T2 was located at the second lag of T1 (i.e., SOA = 200 ms), and a long-SOA condition where T2 appeared at the 8th lag of T1 (i.e., SOA = 800 ms). In a typical AB effect, T2 performance at short lags is remarkably impaired compared with that at long lags. In our study, we consistently replicated this effect across the experiments, as reported in the Results section of Experiment 1 (page 5, line 106). Overall, the T2 detection accuracy conditioned on correct T1 response was significantly impaired in the short-SOA condition relative to that in the long-SOA condition (mean accuracy > 0.9 for all experiments), during both the context session and the baseline session. More crucially, when looking into the magnitude of the AB effect as measured by (ACClong-SOA - ACCshort-SOA)/ACClong-SOA, we still obtained a significant attentional modulation effect (for Experiment 1a, t(15) = -2.729, p = .016, Cohen’s d = 0.682; for Experiment 2a, t(15) = -4.143, p <.001, Cohen’s d = 1.036) similar to that reflected by the short-SOA condition alone, further confirming that cortical entrainment effectively influences the AB effect.
Although we included both the long- and short-SOA conditions in the current study, we focused on T2 performance in the short-SOA condition rather than along the whole AB curve for the following reasons. Firstly, for the long-SOA conditions, the T2 performance is at ceiling level, making it an inappropriate baseline to probe the attentional modulation effect. We focused on Lag 2 because previous research has identified a robust AB effect around the second lag (Raymond et al., 1992), which provides a reasonable and sensitive baseline to probe the potential modulation effect of the contextual auditory and visual rhythms. Note that instead of using multiple lags, we varied the length of the rhythmic cycles (i.e., a cycle of 300 ms, 400 ms, and 500 ms corresponding to a rhythm frequency of 3.3 Hz, 2.5 Hz, and 2 Hz, respectively, all within the delta band), and showed that the attentional modulation effect could be generalized to these different delta-band rhythmic contexts, regardless of the absolute positions of the targets within the rhythmic cycles.
As to the T1 performance, the overall accuracy was very high, ranging from 0.907 to 0.972, in all of our experiments. The corresponding results have been added to the Results section of the revised manuscript (page 5, line 103). Notably, we did not find T1-T2 trade-offs in most of our experiments, except in Experiment 2a where T1 performance showed a moderate decrease in the between-cycle condition relative to that in the within-cycle condition (mean ± SE: 0.888 ± 0.026 vs. 0.933 ± 0.016, respectively; t(15) = -2.217, p = .043). However, by examining the relationship between the modulation effects (i.e., the difference between the two experimental conditions) on T1 and T2, we did not find any significant correlation (p = .403), suggesting that the better performance for T2 was not simply due to the worse performance in detecting T1.
Finally, previous studies have shown that ignoring T1 would lead to ceiling-level T2 performance (Raymond et al., 1992). Therefore, we did not include such manipulation in the current study, as in that case, it would be almost impossible for us to detect any contextual modulation effect.
References:
Raymond, J. E., Shapiro, K. L., & Arnell, K. M. (1992). Temporary suppression of visual processing in an RSVP task: An attentional blink? Journal of Experimental Psychology: Human Perception and Performance, 18(3), 849–860. https://doi.org/10.1037/0096-1523.18.3.849
2) Also, there is only one lag tested. The ensure that we are dealing here with a true AB I would like to see that more than one lag is tested. In the ideal situation a full AB curve should be presented that includes several lags. This should be done for at least for one of the experiments. It would be informative as we can see how cortical entrainment affects the whole AB curve.
Many thanks for the suggestion. Please refer to our response to the point #1 for “Reviewer #3 (Public Review)”. In short, we did include two T2 lag conditions in our study (Experiments 1a, 1b, 2a and 2b), and the results replicated the typical AB effect. We have clarified this point in the revised manuscript (page 5, line 106).
3) Also, there is no data regarding T1 performance. It is important to show that this the better performance for T2 is not due to worse performance in detecting T1. So also please provide this data.
Many thanks for the suggestion. Please refer to our response to the point #1 or “Reviewer #3 (Public Review)”. We have reported the T1 performance in the revised manuscript (page 5, line 103), and the results didn’t show obvious T1-T2 trade-offs.
4) The authors identify the oscillatory characteristics of EEG signals in response to stimulus rhythms, by examined the FFT spectral peaks by subtracting the mean power of two nearest neighboring frequencies from the power at the stimulus frequency. I am not familiar with this procedure and would like to see some justification for using this technique.
According to previous studies (Nozaradan, 2011; Lenc e al., 2018), the procedure to subtract the average amplitude of neighboring frequency bins can remove unrelated background noise, like muscle activity or eye movement. If there were no EEG oscillatory responses characteristic of stimulus rhythms, the amplitude at a given frequency bin should be similar to the average of its neighbors, and thus no significant peaks could be observed in the subtracted spectrum.
References:
Lenc, T., Keller, P. E., Varlet, M., & Nozaradan, S. (2018). Neural tracking of the musical beat is enhanced by low-frequency sounds. Proceedings of the National Academy of Sciences, 115(32), 8221–8226. https://doi.org/10.1073/pnas.1801421115
Nozaradan, S., Peretz, I., Missal, M., & Mouraux, A. (2011). Tagging the Neuronal Entrainment to Beat and Meter. The Journal of Neuroscience, 31(28), 10234–10240. https://doi.org/10.1523/JNEUROSCI.0411-11.2011
Summary:
This work is of interest because it increases our understanding of the molecular mechanisms that distinguish subtypes of VIP interneurons in the cerebral cortex and because of the multiple ways in which the authors address the role of Prox1 in regulating synaptic function in these cells.
The authors would like to thank the reviewers for their constructive comments. In response, we would like to clarify a number of issues, as well as outline how we plan to resolve major concerns.
Reviewer #1:
Stachiak and colleagues examine the physiological effects of removing the homeobox TF Prox1 from two subtypes of VIP neurons, defined on the basis of their bipolar vs. multipolar morphology.
The results will be of interest to those in the field, since it is known from prior work that VIP interneurons are not a uniform class and that Prox1 is important for their development.
The authors first show that selective removal of a conditional Prox1 allele using a VIP cre driver line results in a change in paired pulse ratio of presumptive excitatory synaptic responses in multipolar but not bipolar VIP interneurons. The authors then use RNA-seq to identify differentially expressed genes that might contribute and highlight a roughly two-fold reduction in the expression of a transcript encoding a trans-synaptic protein Elfn1 known to contribute to reduced glutamate release in Sst+ interneurons. They then test the potential contribution of Elfn1 to the phenotype by examining whether loss of one allele of Elfn1 globally alters facilitation. They find that facilitation is reduced both by this genetic manipulation and by a pharmacological blockade of presynaptic mGluRs known to interact with Elfn1.
Although the results are interesting, and the authors have worked hard to make their case, the results are not definitive for several reasons:
1) The global reduction of Elfn1 may act cell autonomously, or may have other actions in other cell types. The pharmacological manipulation is less subject to this interpretation, but these results are not as convincing as they could be because the multipolar Prox1 KO cells (Fig. 3 J) still show substantial facilitation comparable, for example to the multipolar control cells in the Elfn1 Het experiment (controls in Fig. 3E). This raises a concern about control for multiple comparisons. Instead of comparing the 6 conditions in Fig 3 with individual t-tests, it may be more appropriate to use ANOVA with posthoc tests controlled for multiple comparisons.
The reviewer’s concerns regarding non-cell-autonomous actions of global Elfn1 KO are well founded. Significant phenotypic alterations have previously been reported, both in the physiology of SST neurons as well in the animals’ behavior (Stachniak, Sylwestrak, Scheiffele, Hall, & Ghosh, 2019; Tomioka et al., 2014). The homozygous Elfn1 KO mouse displays a hyperactive phenotype and epileptic activity after 3 months of age, suggesting generalcortical activity differences exist (Dolan & Mitchell, 2013; Tomioka et al., 2014). Nevertheless, we have not observed such changes in P17-21 Elfn1 heterozygous (Het) animals.
Comparing across different experimental animal lines, for example the multipolar Prox1 KO cells (Fig. 3 J) to the multipolar control cells in the Elfn1 Het experiment (controls in Fig. 3E), is in our view not advisable. There is a plethora of examples in the literature on the effect of mouse strain on even the most basic cellular functions and hence it is always expected that researchers use the correct control animals for their experiments, which in the best case scenario are littermate controls. For these reasons, we would argue that statistical comparisons across mouse lines is not ideal for our study. Elfn1 Het and MSOP data are presented side by side to illustrate that Elfn1 Hets (3C,E) phenocopy the effects of Prox1 deletion (3G,H,I,J). (See also point 3) MSOP effect sizes, however, do show significant differences by ANOVA with Bonferroni post-hoc (normalized change in EPSC amplitude; multipolar prox1 control: +12.1 ± 3.8%, KO: -8.4 ± 4.3%, bipolar prox1 control: -5.2 ± 4.3%, KO: -3.4 ± 4.7%, cell type x genotype interaction, p= 0.02, two way ANOVA).
2) The isolation of glutamatergic currents is not described. Were GABA antagonists present to block GABAergic currents? Especially with the Cs-based internal solutions used, chloride reversal potentials can be somewhat depolarized relative to the -65 mV holding potential. If IPSCs were included it would complicate the analysis.
No, in fact GABA antagonists were not present in these experiments. The holding voltage in our evoked synaptic experiments is -70 mV, which combined with low internal [Cl-] makes it highly unlikely that the excitatory synaptic responses we study are contaminated by GABA-mediated ones, even with a Cs MeSO4-based solution. Nevertheless, we have now performed additional experiments where glutamate receptor blockers were applied in bath and we observe a complete blockade of the synaptic events at -70mV proving that they are AMPA/NMDA receptor mediated. When holding the cell at 0mV with these blockers present, outward currents were clearly visible, suggesting intact GABA-mediated events.
3) The assumption that protein levels of Elfn1 are reduced to half in the het is untested. Synaptic proteins can be controlled at the level of translation and trafficking and WT may not have twice the level of this protein.
We thank reviewer for pointing this out. Our rationale for using the Elfn1 heterozygous animals is rather that transcript levels are reduced by half in heterozygous animals, to match the reduction we found in the mRNA levels of VIP Prox1 KO cells (Fig 2). The principle purpose of the Elfn1 KO experiment was to determine whether the change in Elfn1 transcript levels could be sufficient to explain the synaptic deficit observed in VIP Prox1 KO cells. As the reviewer notes, translational regulation and protein trafficking could ultimately result in even larger changes than 0.5x protein levels at the synapse. This may ultimately explain the observed multipolar/bipolar disparity, which cannot be explained by transcriptional regulation alone (Fig 4).
4) The authors are to be commended for checking whether Elfn1 is regulated by Prox1 only in the multipolar neurons, but unfortunately it is not. The authors speculate that the selective effects reflect a selective distribution of MgluR7, but without additional evidence it is hard to know how likely this explanation is.
Additional experiments are underway to better understand this mechanism.
Reviewer #2:
Stachniak et al., provide an interesting manuscript on the postnatal role of the critical transcription factor, Prox1, which has been shown to be important for many developmental aspects of CGE-derived interneurons. Using a combination of genetic mouse lines, electrophysiology, FACS + RNAseq and molecular imaging, the authors provide evidence that Prox1 is genetically upstream of Elfn1. Moreover, they go on to show that loss of Prox1 in VIP+ cells preferentially impacts those that are multipolar but not the bipolar subgroup characterized by the expression of calretinin. This latter finding is very interesting, as the field is still uncovering how these distinct subgroups emerge but are at a loss of good molecular tools to fully uncover these questions. Overall, this is a great combination of data that uses several different approaches to come to the conclusions presented. I have suggestions that I think would strengthen the manuscript:
1) Can the authors add a supplemental table showing the top 20-30 genes up and down regulated in their Prox1 KOS? This would make these, and additional, data more tenable to readers.
We would be happy to provide supplementary tables with candidate genes at both P8 and P12.
2) It is interesting that loss of Prox1 or Elfn1 leads to phenotypes in multipolar but are not present or mild in bipolar VIP+ cells. The authors test different hypotheses, which they are able to refute and discuss some ideas for how multipolar cells may be more affected by loss of Elfn1, even when the transcript is lost in both multipolar and bipolar after Prox1 deletion. If there is any way to expand upon these ideas experimentally, I believe it would greatly strengthen the manuscript. I understand there is no perfect experiment due to a lack of tools and reagents but if there is a way to develop one of the following ideas or something similar, it would be beneficial:
We thank the reviewer for the note.
a) Would it be possible to co-fill VIPCre labeled cells with biocytin and a retroviral tracer? Then, after the retroviral tracer had time to label a presynaptic cell, assess whether these were preferentially different between bipolar and multipolar cell types, the latter morphology determined by the biocytin fill? This would test whether each VIP+ subtype is differentially targeted.
Although this is a very elegant experiment and we would be excited to do it, we do feel that single-cell rabies virus tracing is technically very challenging and will take many months to troubleshoot before being able to acquire good data. Hence, we think it is beyond the scope of this study.
b) Another biocytin possibility would be to trace filled VIP+ cells and assess whether the dendrites of multipolar and bipolar cells differentially targeted distinct cortical lamina and whether these lamina, in the same section or parallel, were enriched for mGluR7+ afferents.
We thank the reviewer for their suggestion and we are planning on doing these kinds of experiments.
Reviewer #3:
In this work Stachiak and colleagues investigate the role of Prox1 on the development of VIP cells. Prox1 is expressed by the majority of GABAergic derived from the caudal ganglionic eminence (CGE), and as mentioned by the authors, Prox1 has been shown to be necessary for the differentiation, circuit integration, and maintenance of CGE-derived GABAergic cells. Here, Stachiak and colleagues show that removal of Prox1 in VIP cells leads to suppression of synaptic release probability onto cortical multipolar VIP cells in a mechanism dependent on Elfn1. This work is of interest for the field because it increases our understanding of differential synaptic maturation of VIP cells. The results are noteworthy, however the relevance of this manuscript would potentially be increased by addressing the following suggestions:
1) Include histology to show when exactly Prox1 is removed from multipolar and bipolar VIP-expressing cells by using the VIP-Cre mouse driver.
We can address this by performing an in-situ hybridization against Prox1 from P3 onwards (when Cre becomes active).
2) Clarify if the statistical analysis is done using n (number of cells) or N (number of animals). The analysis between control and mutants (both Prox1 and Elfn1) need to be done across animals and not cells.
Statistics for physiology were done across n (number of cells) while statistics for ISH are done across number of slices. We will clarify this point in the text and update the methods.
Regarding the statistics for the ISH, these have been done across n (number of slices) for control versus KO tissue (N = 3 and N = 2 animals, respectively). We will add more animals to this analysis to compare by animal instead, although we do not expect any change in the results.
Regarding the physiology, we would provide a two-pronged answer. We first of all feel that averaging synaptic responses for each animal would hide a good deal of the biological variability in PPR present in different cells (response Fig 1), the characterization of which is integral to the central findings of the paper. Secondly, to perform such analysis asked by the reviewer one would need to obtain recordings from ~10 animals or so per condition for each condition, which, to our knowledge, is something that is not standard when utilizing in vitro electrophysiological recordings from single cells. For example, in these very recent studies that have performed in vitro electrophysiological recordings all the statistics are performed using “n” number of cells and not the average of all the cells recorded per animal collapsed into a single data point. (Udakis, Pedrosa, Chamberlain, Clopath, & Mellor, 2020) https://www.nature.com/articles/s41467-020-18074-8
(Horvath, Piazza, Monteggia, & Kavalali, 2020) https://elifesciences.org/articles/52852
(Haas et al., 2018) https://elifesciences.org/articles/31755
Nevertheless, we have now re-run the analysis grouping the cells and averaging the values we get per animal, since we have obtained our data from many animals. The results are more or less indistinguishable from the ones presented in the original submission, except for on p value that rose to 0.07 from 0.03 due to the lack of the required number of animals. We hope that the new plots and statistics presented herein address the concern put forward by the reviewer.
Response Fig 1: A comparison of cell wise versus animal-wise analysis of synaptic physiology. Some cell to cell variability is hidden, and the reduction in numbers impacts the P values.
(A) PPR of multipolar Prox1 Control for 14 cells from 9 animals (n/N=14/9) under baseline conditions and with MSOP, cell-wise comparison p = 0.02 , t = 2.74 and (B) animal-wise comparisons (p = 0.04, t stat = 2.45). Statistics: paired t-test.
(C) PPR of multipolar Prox1 KO cells (n/N=9/8) under baseline conditions and with MSOP, cell-wise comparison p = 0.2, t = 1.33 and (D) animal-wise comparisons (p = 0.2, t stat = 1.56). Statistics: paired t-test. Comparisons for PPR of bipolar Prox1 Control (n/N=8/8) and KO cells (n/N=9/9) did not change.
(E) PPR for Prox1 control (n/N=18/11) and KO (n/N=13/11) bipolar VIP cells, cell-wise comparison p = 0.3, t = 1.1 and (F) animal-wise comparisons (p = 0.4, t stat = 0.93). Statistics: t-test.
(G) PPR of Elfn1 Control (n/N=12/4) and Het (n/N=12/4) bipolar VIP cells, cell-wise comparison p = 0.3, t = 1.06 and (H) animal-wise comparisons (p = 0.4, t stat = 0.93)
(I) PPR of Prox1 control (n/N=33/18) and KO (n/N=19/14) multipolar VIP cells, cell-wise comparison p = 0.03, t = 2.17. and (J) animal-wise comparisons (p = 0.07, t stat = 1.99).
(K) PPR of Elfn1 Control (n/N=14/6) and Het (n/N=20/8) multipolar VIP cells, cell-wise comparison p = 0.008, t = 2.84 and (L) animal-wise comparisons (p = 0.007, t stat = 3.23).
3) Clarify what are the parameters used to identify bipolar vs multipolar VIP cells. VIP cells comprise a wide variety of transcriptomic subtypes, and in the absence of using specific genetic markers for the different VIP subtypes, the authors should either include the reconstructions of all recorded cells or clarify if other methods were used.
We thank the reviewer for this comment. The cell parameter criteria will be amended in the methods: “Cell type was classified as bipolar vs. multipolar based on cell body morphology (ovoid vs. round) and number and orientation of dendritic processes emanating from it (2 or 3 dendrites perpendicular to pia (for bipolar) vs. 3 or more processes in diverse orientations (for multipolar). In addition, the laminar localization of the two populations differs, with multipolar cells found primarily in the upper layer 2, while bipolar cells are found throughout layers 2 and 3. Initial determination of cell classification was made prior to patching fluorescent-labelled cells, but whenever possible this initial assessment was confirmed with post-hoc verification of biocytin filled cells.”
Reference:
Dolan, J., & Mitchell, K. J. (2013). Mutation of Elfn1 in Mice Causes Seizures and Hyperactivity. PLOS ONE, 8(11), e80491. Retrieved from https://doi.org/10.1371/journal.pone.0080491
Haas, K. T., Compans, B., Letellier, M., Bartol, T. M., Grillo-Bosch, D., Sejnowski, T. J., … Hosy, E. (2018). Pre-post synaptic alignment through neuroligin-1 tunes synaptic transmission efficiency. ELife, 7, e31755. https://doi.org/10.7554/eLife.31755
Horvath, P. M., Piazza, M. K., Monteggia, L. M., & Kavalali, E. T. (2020). Spontaneous and evoked neurotransmission are partially segregated at inhibitory synapses. ELife, 9, e52852. https://doi.org/10.7554/eLife.52852
Stachniak, T. J., Sylwestrak, E. L., Scheiffele, P., Hall, B. J., & Ghosh, A. (2019). Elfn1-Induced Constitutive Activation of mGluR7 Determines Frequency-Dependent Recruitment of Somatostatin Interneurons. The Journal of Neuroscience, 39(23), 4461 LP – 4474. https://doi.org/10.1523/JNEUROSCI.2276-18.2019
Tomioka, N. H., Yasuda, H., Miyamoto, H., Hatayama, M., Morimura, N., Matsumoto, Y., … Aruga, J. (2014). Elfn1 recruits presynaptic mGluR7 in trans and its loss results in seizures. Nature Communications. https://doi.org/10.1038/ncomms5501
Udakis, M., Pedrosa, V., Chamberlain, S. E. L., Clopath, C., & Mellor, J. R. (2020). Interneuron-specific plasticity at parvalbumin and somatostatin inhibitory synapses onto CA1 pyramidal neurons shapes hippocampal output. Nature Communications, 11(1), 4395. https://doi.org/10.1038/s41467-020-18074-8
Author Response
1) Please comment on why many of the June samples failed to provide sufficient sequence information, especially since not all of them had low yields (supp table 2 and supp figure 5).
An extended paragraph about experimental intricacies of our study has been added to the Discussion. It has also been also slightly restructured to give a better and wider overview of how future freshwater monitoring studies using nanopore sequencing can be improved (page 18, lines 343-359).
We wish to highlight that all three MinION sequencing runs here analysed feature substantially higher data throughput than that of any other recent environmental 16S rRNA sequencing study with nanopore technology, as recently reviewed by Latorre-Pérez et al. (Biology Methods and Protocols 2020, doi:10.1093/biomethods/bpaa016). One of this work's sequencing runs has resulted in lower read numbers for water samples collected in June 2018 (~0.7 Million), in comparison to the ones collected in April and August 2018 (~2.1 and ~5.5 Million, respectively). While log-scale variabilities between MinION flow cell throughput have been widely reported for both 16S and shotgun metagenomics approaches (e.g. see Latorre-Pérez et al.), the count of barcode-specific 16S reads is nevertheless expected to be correlated with the barcode-specific amount of input DNA within a given sequencing run. As displayed in Supplementary Figure 7b, we see a positive, possibly logarithmic trend between the DNA concentration after 16S rDNA amplification and number of reads obtained. With few exceptions (April-6, April-9.1 and Apri-9.2), we find that sample pooling with original 16S rDNA concentrations of ≳4 ng/µl also results in the surpassing of the here-set (conservative) minimum read threshold of 37,000 for further analyses. Conversely, all June samples that failed to reach 37,000 reads did not pass the input concentration of 4 ng/µl, despite our attempt to balance their quantity during multiplexing.
We reason that such skews in the final barcode-specific read distribution would mainly arise from small concentration measurement errors, which undergo subsequent amplification during the upscaling with comparably large sample volume pipetting. While this can be compensated for by high overall flow cell throughput (e.g. see August-2, August-9.1, August-9.2), we think that future studies with much higher barcode numbers can circumvent this challenge by leveraging an exciting software solution: real-time selective sequencing via “Read Until”, as developed by Loose et al. (Nature Methods 2016, doi:10.1038/nmeth.3930). In the envisaged framework, incoming 16S read signals would be in situ screened for the sample-barcode which in our workflow is PCR-added to both the 5' and 3' end of each amplicon. Overrepresented barcodes would then be counterbalanced by targeted voltage inversion and pore "rejection" of such reads, until an even balance is reached. Lately, such methods have been computationally optimised, both through the usage of GPUs (Payne et al., bioRxiv 2020, https://doi.org/10.1101/2020.02.03.926956) and raw electrical signals (Kovaka et al., bioRxiv 2020, https://doi.org/10.1101/2020.02.03.931923).
2) It would be helpful if the authors could mention the amount (or proportion) of their sequenced 16S amplicons that provided species-level identification, since this is one of the advantages of nanopore sequencing.
We wish to emphasize that we intentionally refrained from reporting the proportion of 16S rRNA reads that could be classified at species level, since we are wary of any automated species level assignments even if the full-length 16S rRNA gene is being sequenced. While we list the reasons for this below, we appreciate the interest in the theoretical proportion of reads at species level assignment. We therefore re-analyzed our dataset, and now also provide the ratio of reads that could be classified at species level using Minimap2 (pages 16-17, lines 308-314).
To this end, we classified reads at species level if the species entry of the respective SILVA v.132 taxonomic ID was either not empty, or neither uncultured bacterium nor metagenome. Therefore, many unspecified classifications such as uncultured species of some bacterial genus are counted as species-level classifications, rendering our approach lenient towards a higher ratio of species level classifications. Still, the species level classification ratios remain low, on average at 16.2 % across all included river samples (genus-level: 65.6 %, family level: 76.6 %). The mock community, on the other hand, had a much higher species classification rate (>80 % in all three replicates), which is expected for a well-defined, well-referenced and divergent composition of only eight bacterial taxa, and thus re-validates our overall classification workflow.
On a theoretical level, we mainly refrain from automated across-the-board species level assignments because: (1) many species might differ by very few nucleotide differences within the 16S amplicon; distinguishing these from nanopore sequencing errors (here ~8 %) remains challenging (2) reference databases are incomplete and biased with respect to species level resolution, especially regarding certain environmental contexts; it is likely that species assignments would be guided by references available from more thoroughly studied niches than freshwater
Other recent studies have also shown that across-the-board species-level classification is not yet feasible with 16S nanopore sequencing, for example in comparison with Illumina data (Acharya et al., Scientific Reports 2019, doi:10.25405/data.ncl.9693533) which showed that “more reliable information can be obtained at genus and family level”, or in comparison with longer 16S-ITS-23S amplicons (Cusco et al., F1000Research 2019, doi: 10.12688/f1000research.16817.2), which “remarkably improved the taxonomy assignment at the species level”.
3) It is not entirely clear how the authors define their core microbiome. Are they reporting mainly the most abundant taxa (dominant core microbiome), and would this change if you look at a taxonomic rank below the family level? How does the core compare, for example, with other studies of this same river?
The here-presented core microbiome indeed represents the most abundant taxa, with relatively consistent profiles between samples. We used hierarchical clustering (Figure 4a, C2 and C4) on the bacterial family level, together with relative abundance to identify candidate taxa. Filtering these for median abundance > 0.1% across all samples resulted in 27 core microbiome families. To clarify this for the reader, we have added a new paragraph to the Material and Methods (section 2.7; page 29, lines 653-658).
We have also performed the same analysis on the bacterial genus level and now display the top 27 most abundant genera (median abundance > 0.2%), together with their corresponding families and hierarchical clustering analysis in a new Supplementary Figure 4. Overall, high robustness is observed with respect to the families of the core microbiome: out of the top 16 core families (Figure 4b), only the NS11-12 marine group family is not represented by the top 27 most abundant genera (Supplementary Figure 4b). We reason that this is likely because its corresponding genera are composed of relatively poorly resolved references of uncultured bacteria, which could thus not be further classified.
To the best of our knowledge, there are only two other reports that feature metagenomic data of the River Cam and its wastewater influx sources (Rowe et al., Water Science & Technology 2016, doi:10.2166/wst.2015.634; Rowe et al., Journal of Antimicrobial Chemotherapy 2017, doi:10.1093/jac/dkx017). While both of these primarily focus on the diversity and abundance of antimicrobial resistance genes using Illumina shotgun sequencing, they only provide limited taxonomic resolution on the river's core microbiome. Nonetheless, Rowe et al. (2016) specifically highlighted Sphingobium as the most abundant genus in a source location of the river (Ashwell, Hertfordshire). This genus belongs to the family of Sphingomonadaceae, which is also among the five most dominant families identified in our dataset. It thus forms part of what we define as the core microbiome of the River Cam (Figure 4b), and we have therefore highlighted this consistency in our manuscript's Discussion (page 17, lines 316-319).
4) Please consider revising the amount of information in some of the figures (such as figure 2 and figure 3). The resulting images are tiny, the legends become lengthy and the overall impact is reduced. Consider splitting these or moving some information to the supplements.
To follow this advice, we have split Figure 2 into two less compact figures. We have moved more detailed analyses of our classification tool benchmark to the supplement (now Supplementary Figure 1). Supplementary Figure 1 notably also contains a new summary of the systematic computational performance measurements of each classification tool (see minor suggestions).
Moreover, we here suggest that the original Figure 3 may be divided into two figures: one to visualise the sequencing output, data downsampling and distribution of the most abundant families (now Figure 3), and the other featuring the clustering of bacterial families and associated core microbiome (now Figure 4). We think that both the data summary and clustering/core microbiome analyses are of particular interest to the reader, and that they should be kept as part of the main analyses rather than the supplement – however, we are certainly happy to discuss alternative ideas with the reviewers and editors.
5) Given that the authors claim to provide a simple, fast and optimized workflow it would be good to mention how this workflow differs or provides faster and better analysis than previous work using amplicon sequencing with a MinION sequencer.
Data throughput, sequencing error rates and flow cell stability have seen rapid improvements since the commercial release of MinION in 2015. In consequence, bioinformatics community standards regarding raw data processing and integration steps are still lacking, as illustrated by a thorough recent benchmark of fast5 to fastq format "basecalling" methods (Wick et al., Genome Biology 2019, doi: 10.1186/s13059-019-1727-y).
Early on during our analyses, we noticed that a plethora of bespoke pipelines have been reported in recent 16S environmental surveys using MinION (e.g. Kerkhof et al., Microbiome 2017, 10.1186/s40168-017-0336-9; Cusco et al., F1000 Research 2018, 10.12688/f1000research.16817.2; Acharya et al., Scientific Reports 2019, 10.1038/s41598-019-51997-x; Nygaard et al., Scientific Reports 2020, doi: 10.1038/s41598-020-59771-0). This underlines a need for more unified bioinformatics standards of (full-length) 16S amplicon data treatment, while similar benchmarks exist for short-read 16S metagenomics approaches, as well as for nanopore shotgun sequencing (e.g. Ye et al., Cell 2019, doi: 10.1016/j.cell.2019.07.010; Latorre-Pérez et al., Scientific Reports 2020, doi:10.1038/s41598-020-70491-3).
By adding a thorough speed and memory usage summary (new Supplementary Figure 1b), in addition to our (mis)classification performance tests based on both mock and complex microbial community analyses, we provide the reader with a broad overview of existing options. While the widely used Kraken 2 and Centrifuge methods provide exceptional speed, we find that this comes with a noticeable tradeoff in taxonomic assignment accuracy. We reason that Minimap2 alignments provide a solid compromise between speed and classification performance, with the MAPseq software offering a viable alternative should memory usage limitation apply to users.
We intend to extend this benchmarking process to future tools, and to update it on our GitHub page (https://github.com/d-j-k/puntseq). This page notably also hosts a range of easy-to-use scripts for employing downstream 16S analysis and visualization approaches, including ordination, clustering and alignment tests.
The revised Discussion now emphasises the specific advancements of our study with respect to freshwater analysis and more general standardisation of nanopore 16S sequencing, also in contrast to previous amplicon nanopore sequencing approaches in which only one or two bioinformatics workflows were tested (page 16, lines 297-306).
They also mention that nanopore sequencing is an "inexpensive, easily adaptable and scalable framework" The term "inexpensive" doesn't seem appropriate since it is relative. In addition, they should also discuss that although it is technically convenient in some aspects compared to other sequencers, there are still protocol steps that need certain reagents and equipment that is similar or the same to those needed for other sequencing platforms. Common bottlenecks such as DNA extraction methods, sample preservation and the presence of inhibitory compounds should be mentioned.
We agree with the reviewers that “inexpensive” is indeed a relative term, which needs further clarification. We therefore now state that this approach is “cost-effective” and discuss future developments such as the 96-sample barcoding kits and Flongle flow cells for small-scale water diagnostics applications, which will arguably render lower per-sample analysis costs in the future (page 18, lines 361-365).
Other investigators (e.g. Boykin et al., Genes 2019, doi:10.3390/genes10090632; Acharya et al., Water Technology 2020, doi:10.1016/j.watres.2020.116112) have recently shown that the full application of DNA extraction and in-field nanopore sequencing can be achieved at comparably low expense: Boykin et al. studied cassava plant pathogens using barcoded nanopore shotgun sequencing, and estimated costs of ~45 USD per sample, while we calculate ~100 USD per sample in this study. Acharya et al. undertook in situ water monitoring between Birtley, UK and Addis Ababa, Ethiopia, estimated ~75-150 USD per sample and purchased all necessary equipment for ~10,000 GBP – again, we think that this lies roughly within a similar range as our (local) study's total cost of ~3,670 GBP (Supplementary Table 6).
The revised manuscript now mentions the possibility of increasing sequencing yield by improving DNA extraction methods, by taking sample storage and potential inhibitory compounds into account in the planning phase (page 18, lines 348-352).
Minor points:
-Please include a reference to the statement saying that the river Cam is notorious for the "infections such as leptospirosis".
There are indeed several media reports that link leptospirosis risk to the local River Cam (e.g. https://www.cambridge-news.co.uk/news/cambridge-news/weils-disease-river-cam-leptosirosis-14919008 or https://www.bbc.com/news/uk-england-cambridgeshire-29060018). As we, however, did not find a scientific source for this information, we have slightly adjusted the statement in our manuscript from referring to Cambridge to instead referring to the entire United Kingdom. Accordingly, we now cite two reports from Public Health England (PHE) about serial leptospirosis prevalence in the United Kingdom (page 13, lines 226-227).
-Please check figure 7 for consistency across panels, such as shading in violet and labels on the figures that do not seem to correspond with what is stated in the legend. Please mention what the numbers correspond to in outer ring. Check legend, where it says genes is probably genus.
Thank you for pointing this out. We have revised (now labelled) Figure 8 and removed all inconsistencies between the panels. The legend has also been updated, which now includes a description of the number labelling of the tree, and a clearer differentiation between the colour coding of the tree nodes and the background highlighting of individual nanopore reads.
-Page 6. There is a "data not shown" comment in the text: "Benchmarking of the classification tools on one aquatic sample further confirmed Minimap2's reliable performance in a complex bacterial community, although other tools such as SPINGO (Allard, Ryan, Jeffery, & Claesson, 2015), MAPseq (Matias Rodrigues, Schmidt, Tackmann, & von Mering, 2017), or IDTAXA (Murali et al., 2018) also produced highly concordant results despite variations in speed and memory usage (data not shown)." There appears to be no good reason that this data is not shown. In case the speed and memory usage was not recorded, is advisable to rerun the analysis and quantify these variables, rather than mentioning them and not reporting them. Otherwise, provide an explanation for not showing the data please.
This is a valid point, and we agree with the reviewers that it is worth properly following up on this initial observation. To this end, our revised manuscript now entails a systematic characterisation of the twelve tools' runtime and memory usage performance. This has been added as Supplementary Figure 1b and under the new Materials and Methods section 2.2.4 (page 26, lines 556-562), while the corresponding results and their implications are discussed on page 16, lines 301-306. Particularly with respect to the runtime measurements, it is worth noting that these can differ by several orders of magnitude between the classifiers, thus providing an additional clarification on our choice of the - relatively fast - Minimap2 alignments.
-In Figure 4, it would be important to calculate if the family PCA component contribution differences in time are differentially significant. In Panel B, depicted is the most evident variance difference but what about other taxa which might not be very abundant but differ in time? One can use the fitFeatureModel function from the metagenomeSeq R library and a P-adjusted threshold value of 0.05, to validate abundance differences in addition to your analysis.
To assess if the PC component contribution of Figure 5 (previously Figure 4) significantly differed between the three time points, we have applied non-parametric tests to all season-grouped samples except for the mock community controls. We first applied Kruskal-Wallis H-test for independent samples, followed by post-hoc comparisons using two-sided Mann-Whitney U rank tests.
The Kruskal-Wallis test established a significant difference in PC component contributions between the three time points (p = 0.0049), with most of the difference stemming from divergence between April and August samples according to the post-hoc tests (p = 0.0022). The June sampled seemed to be more similar to the August ones (p = 0.66) than to the ones from April (p = 0.11), recapitulating the results of our hierarchical clustering analysis (Figure 4a).
We have followed the reviewers' advice and applied a complementary approach, using the fitFeatureModel of metagenomeSeq to fit a zero-inflated log-normal mixture model of each bacterial taxon against the time points. As only three independent variables can be accounted for by the model (including the intercept), we have chosen to investigate the difference between the spring (April) and summer (June, August) months to capture the previously identified difference between these months. At a nominal P-value threshold of 0.05, this analysis identifies seven families to significantly differ in their relative composition between spring and summer, namely Cyanobiaceae, Armatimonadaceae, Listeriaceae, Carnobacteriaceae, Azospirillaceae, Cryomorphaceae, and Microbacteriaceae. Three out of these seven families were also detected by the PCA component analysis (Carnobacteriacaea, Azospirillaceae, Microbacteriaceae) and two more (Listeriacaea, Armatimonadaceae) occured in the top 15 % of that analysis (out of 357 families).
This approach represents a useful validation of our principal component analysis' capture of likely seasonal divergence, but moreover allows for a direct assessment of differential bacterial composition across time points. We have therefore integrated the analysis into our manuscript (page 10, lines 184-186; Materials and Methods section 2.6, page 29, lines 641-647) – thank you again for this suggestion.
-Page 12-13. In the paragraph: "Using multiple sequence alignments between nanopore reads and pathogenic species references, we further resolved the phylogenies of three common potentially pathogenic genera occurring in our river samples, Legionella, Salmonella and Pseudomonas (Figure 7a-c; Material and Methods). While Legionella and Salmonella diversities presented negligible levels of known harmful species, a cluster of reads in downstream sections indicated a low abundance of the opportunistic, environmental pathogen Pseudomonas aeruginosa (Figure 7c). We also found significant variations in relative abundances of the Leptospira genus, which was recently described to be enriched in wastewater effluents in Germany (Numberger et al., 2019) (Figure 7d)."
Here it is important to mention the relative abundance in the sample. While no further experiments are needed, the authors should mention and discuss that the presence of DNA from pathogens in the sample has to be confirmed by other microbiology methodologies, to validate if there are viable organisms. Definitively, it is a big warning finding pathogen's DNA but also, since it is characterized only at genus level, further investigation using whole metagenome shotgun sequencing or isolation, would be important.
We agree that further microbiological assays, particularly target-specific species isolation and culturing, would be essential to validate the presence of living pathogenic bacteria. Accordingly, our revised Discussion now contains a paragraph that encourages such experiments as part of the design of future studies (with a fully-equipped laboratory infrastructure); page 17, 338-341.
-Page 15: "This might help to establish this family as an indicator for bacterial community shifts along with water temperature fluctuations."
Temperature might not be the main factor for the shift. There could be other factors that were not measured that could contribute to this shift. There are several parameters that are not measured and are related to water quality (COD, organic matter, PO4, etc).
We agree that this was a simplified statement, given our currently limited number of samples, and have therefore slightly expanded on this point (page 17, lines 323-325). It is indeed possible that differential Carnobacteriaceae abundances between the time point measurements may have arisen not as a consequence of temperature fluctuations (alone), but instead as a consequence of the observed hydrochemical changes like e.g. Ca2+, Mg2+, HCO3- (Figure 6b-c) or possible even water flow speed reductions (Supplementary Figure 6d).
-"A number of experimental intricacies should be addressed towards future nanopore freshwater sequencing studies with our approach, mostly by scrutinising water DNA extraction yields, PCR biases and molar imbalances in barcode multiplexing (Figure 3a; Supplementary Figure 5)."
Here you could elaborate more on the challenges, as mentioned previously.
We realise that we had not discussed the challenges in enough detail, and the Discussion now contains a substantially more detailed description of these intricacies (page 18, lines 343-359).
Author Response:
Evaluation Summary:
Since DBS of the habenula is a new treatment, these are the first data of its kind and potentially of high interest to the field. Although the study mostly confirms findings from animal studies rather than bringing up completely new aspects of emotion processing, it certainly closes a knowledge gap. This paper is of interest to neuroscientists studying emotions and clinicians treating psychiatric disorders. Specifically the paper shows that the habenula is involved in processing of negative emotions and that it is synchronized to the prefrontal cortex in the theta band. These are important insights into the electrophysiology of emotion processing in the human brain.
The authors are very grateful for the reviewers’ positive comments on our study. We also thank all the reviewers for the comments which has helped to improve the manuscript.
Reviewer #1 (Public Review):
The study by Huang et al. report on direct recordings (using DBS electrodes) from the human habenula in conjunction with MEG recordings in 9 patients. Participants were shown emotional pictures. The key finding was a transient increase in theta/alpha activity with negative compared to positive stimuli. Furthermore, there was a later increase in oscillatory coupling in the same band. These are important data, as there are few reports of direct recordings from the habenula together with the MEG in humans performing cognitive tasks. The findings do provide novel insight into the network dynamics associated with the processing of emotional stimuli and particular the role of the habenula.
Recommendations:
How can we be sure that the recordings from the habenula are not contaminated by volume conduction; i.e. signals from neighbouring regions? I do understand that bipolar signals were considered for the DBS electrode leads. However, high-frequency power (gamma band and up) is often associated with spiking/MUA and considered less prone to volume conduction. I propose to also investigate that high-frequency gamma band activity recorded from the bipolar DBS electrodes and relate to the emotional faces. This will provide more certainty that the measured activity indeed stems from the habenula.
We thank the reviewer for the comment. As the reviewer pointed out, bipolar macroelectrode can detect locally generated potentials, as demonstrated in the case of recordings from subthalamic nucleus and especially when the macroelectrodes are inside the subthalamic nucleus (Marmor et al., 2017). However, considering the size of the habenula and the size of the DBS electrode contacts, we have to acknowledge that we cannot completely exclude the possibility that the recordings are contaminated by volume conduction of activities from neighbouring areas, as shown in Bertone-Cueto et al. 2019. We have now added extra information about the size of the habenula and acknowledged the potential contamination of activities from neighbouring areas through volume conduction in the ‘Limitation’:
"Another caveat we would like to acknowledge that the human habenula is a small region. Existing data from structural MRI scans reported combined habenula (the sum of the left and right hemispheres) volumes of ~ 30–36 mm3 (Savitz et al., 2011a; Savitz et al., 2011b) which means each habenula has the size of 2~3 mm in each dimension, which may be even smaller than the standard functional MRI voxel size (Lawson et al., 2013). The size of the habenula is also small relative to the standard DBS electrodes (as shown in Fig. 2A). The electrodes used in this study (Medtronic 3389) have electrode diameter of 1.27 mm with each contact length of 1.5 mm, and contact spacing of 0.5 mm. We have tried different ways to confirm the location of the electrode and to select the contacts that is within or closest to the habenula: 1.) the MRI was co-registered with a CT image (General Electric, Waukesha, WI, USA) with the Leksell stereotactic frame to obtain the coordinate values of the tip of the electrode; 2.) Post-operative CT was co-registered to pre-operative T1 MRI using a two-stage linear registration using Lead-DBS software. We used bipolar signals constructed from neighbouring macroelectrode recordings, which have been shown to detect locally generated potentials from subthalamic nucleus and especially when the macroelectrodes are inside the subthalamic nucleus (Marmor et al., 2017). Considering that not all contacts for bipolar LFP construction are in the habenula in this study, as shown in Fig. 2, we cannot exclude the possibility that the activities we measured are contaminated by activities from neighbouring areas through volume conduction. In particular, the human habenula is surrounded by thalamus and adjacent to the posterior end of the medial dorsal thalamus, so we may have captured activities from the medial dorsal thalamus. However, we also showed that those bipolar LFPs from contacts in the habenula tend to have a peak in the theta/alpha band in the power spectra density (PSD); whereas recordings from contacts outside the habenula tend to have extra peak in beta frequency band in the PSD. This supports the habenula origin of the emotional valence related changes in the theta/alpha activities reported here."
We have also looked at gamma band oscillations or high frequency activities in the recordings. However, we didn’t observe any peak in high frequency band in the average power spectral density, or any consistent difference in the high frequency activities induced by the emotional stimuli (Fig. S1). We suspect that high frequency activities related to MUA/spiking are very local and have very small amplitude, so they are not picked up by the bipolar LFPs measured from contacts with both the contact area for each contact and the between-contact space quite large comparative to the size of the habenula.
A
B
Figure S1. (A) Power spectral density of habenula LFPs across all time period when emotional stimuli were presented. The bold blue line and shadowed region indicates the mean ± SEM across all recorded hemispheres and the thin grey lines show measurements from individual hemispheres. (B) Time-frequency representations of the power response relative to pre-stimulus baseline for different conditions showing habenula gamma and high frequency activity are not modulated by emotional
References:
Savitz JB, Bonne O, Nugent AC, Vythilingam M, Bogers W, Charney DS, et al. Habenula volume in post-traumatic stress disorder measured with high-resolution MRI. Biology of Mood & Anxiety Disorders 2011a; 1(1): 7.
Savitz JB, Nugent AC, Bogers W, Roiser JP, Bain EE, Neumeister A, et al. Habenula volume in bipolar disorder and major depressive disorder: a high-resolution magnetic resonance imaging study. Biological Psychiatry 2011b; 69(4): 336-43.
Lawson RP, Drevets WC, Roiser JP. Defining the habenula in human neuroimaging studies. NeuroImage 2013; 64: 722-7.
Marmor O, Valsky D, Joshua M, Bick AS, Arkadir D, Tamir I, et al. Local vs. volume conductance activity of field potentials in the human subthalamic nucleus. Journal of Neurophysiology 2017; 117(6): 2140-51.
Bertone-Cueto NI, Makarova J, Mosqueira A, García-Violini D, Sánchez-Peña R, Herreras O, et al. Volume-Conducted Origin of the Field Potential at the Lateral Habenula. Frontiers in Systems Neuroscience 2019; 13:78.
Figure 3: the alpha/theta band activity is very transient and not band-limited. Why refer to this as oscillatory? Can you exclude that the TFRs of power reflect the spectral power of ERPs rather than modulations of oscillations? I propose to also calculate the ERPs and perform the TFR of power on those. This might result in a re-interpretation of the early effects in theta/alpha band.
We agree with the reviewer that the activity increase in the first time window with short latency after the stimuli onset is very transient and not band-limited. This raise the question that whether this is oscillatory or a transient evoked activity. We have now looked at this initial transient activity in different ways: 1.) We quantified the ERP in LFPs locked to the stimuli onset for each emotional valence condition and for each habenula. We investigated whether there was difference in the amplitude or latency of the ERP for different stimuli emotional valence conditions. As showing in the following figure, there is ERP with stimuli onset with a positive peak at 402 ± 27 ms (neutral stimuli), 407 ± 35 ms (positive stimuli), 399 ± 30 ms (negative stimuli). The flowing figure (Fig. 3–figure supplement 1) will be submitted as figure supplement related to Fig. 3. However, there was no significant difference in ERP latency or amplitude caused by different emotional valence stimuli. 2.) We have quantified the pure non-phase-locked (induced only) power spectra by calculating the time-frequency power spectrogram after subtracting the ERP (the time-domain trial average) from time-domain neural signal on each trial (Kalcher and Pfurtscheller, 1995; Cohen and Donner, 2013). This shows very similar results as we reported in the main manuscript, as shown in Fig. 3–figure supplement 2. These further analyses show that even though there were event related potential changes time locked around the stimuli onset, and this ERP did NOT contribute to the initial broad-band activity increase at the early time window shown in plot A-C in Figure 3. The figures of the new analyses and following have now been added in the main text:
"In addition, we tested whether stimuli-related habenula LFP modulations primarily reflect a modulation of oscillations, which is not phase-locked to stimulus onset, or, alternatively, if they are attributed to evoked event-related potential (ERP). We quantified the ERP for each emotional valence condition for each habenula. There was no significant difference in ERP latency or amplitude caused by different emotional valence stimuli (Fig. 3–figure supplement 1). In addition, when only considering the non phase-locked activity by removing the ERP from the time series before frequency-time decomposition, the emotional valence effect (presented in Fig. 3–figure supplement 2) is very similar to those shown in Fig.3. These additional analyses demonstrated that the emotional valence effect in the LFP signal is more likely to be driven by non-phase-locked (induced only) activity."
A
B
Fig. 3–figure supplement 1. Event-related potential (ERP) in habenula LFP signals in different emotional valence (neutral, positive and negative) conditions. (A) Averaged ERP waveforms across patients for different conditions. (B) Peak latency and amplitude (Mean ± SEM) of the ERP components for different conditions.
Fig. 3–figure supplement 2. Non-phase-locked activity in different emotional valence (neutral, positive and negative) conditions (N = 18). (A) Time-frequency representation of the power changes relative to pre-stimulus baseline for three conditions. Significant clusters (p < 0.05, non-parametric permutation test) are encircled with a solid black line. (B) Time-frequency representation of the power response difference between negative and positive valence stimuli, showing significant increased activity the theta/alpha band (5-10 Hz) at short latency (100-500 ms) and another increased theta activity (4-7 Hz) at long latencies (2700-3300 ms) with negative stimuli (p < 0.05, non-parametric permutation test). (C) Normalized power of the activities at theta/alpha (5-10 Hz) and theta (4-7 Hz) band over time. Significant difference between the negative and positive valence stimuli is marked by a shadowed bar (p < 0.05, corrected for multiple comparison).
References:
Kalcher J, Pfurtscheller G. Discrimination between phase-locked and non-phase-locked event-related EEG activity. Electroencephalography and Clinical Neurophysiology 1995; 94(5): 381-4.
Cohen MX, Donner TH. Midfrontal conflict-related theta-band power reflects neural oscillations that predict behavior. Journal of Neurophysiology 2013; 110(12): 2752-63.
Figure 4D: can you exclude that the frontal activity is not due to saccade artifacts? Only eye blink artifacts were reduced by the ICA approach. Trials with saccades should be identified in the MEG traces and rejected prior to further analysis.
We understand and appreciate the reviewer’s concern on the source of the activity modulations shown in Fig. 4D. We tried to minimise the eye movement or saccade in the recording by presenting all figures at the centre of the screen, scaling all presented figures to similar size, and presenting a white cross at the centre of the screen preparing the participants for the onset of the stimuli. Despite this, participants my still make eye movements and saccade in the recording. We used ICA to exclude the low frequency large amplitude artefacts which can be related to either eye blink or other large eye movements. However, this may not be able to exclude artefacts related to miniature saccades. As shown in Fig. 4D, on the sensor level, the sensors with significant difference between the negative vs. positive emotional valence condition clustered around frontal cortex, close to the eye area. However, we think this is not dominated by saccades because of the following two reasons:
1.) The power spectrum of the saccadic spike artifact in MEG is characterized by a broadband peak in the gamma band from roughly 30 to 120 Hz (Yuval-Greenberg et al., 2008; Keren et al., 2010). In this study the activity modulation we observed in the frontal sensors are limited to the theta/alpha frequency band, so it is different from the power spectra of the saccadic spike artefact.
2.) The source of the saccadic spike artefacts in MEG measurement tend to be localized to the region of the extraocular muscles of both eyes (Carl et al., 2012).We used beamforming source localisation to identify the source of the activity modulation reported in Fig. 4D. This beamforming analysis identified the source to be in the Broadmann area 9 and 10 (shown in Fig. 5). This excludes the possibility that the activity modulation in the sensor level reported in Fig. 4D is due to saccades. In addition, Broadman area 9 and 10, have previously been associated with emotional stimulus processing (Bermpohl et al., 2006), Broadman area 9 in the left hemisphere has also been used as the target for repetitive transcranial magnetic stimulation (rTMS) as a treatment for drug-resistant depression (Cash et al., 2020). The source localisation results, together with previous literature on the function of the identified source area suggest that the activity modulation we observed in the frontal cortex is very likely to be related to emotional stimuli processing.
References:
Yuval-Greenberg S, Tomer O, Keren AS, Nelken I, Deouell LY. Transient induced gamma-band response in EEG as a manifestation of miniature saccades. Neuron 2008; 58(3): 429-41.
Keren AS, Yuval-Greenberg S, Deouell LY. Saccadic spike potentials in gamma-band EEG: characterization, detection and suppression. NeuroImage 2010; 49(3): 2248-63.
Carl C, Acik A, Konig P, Engel AK, Hipp JF. The saccadic spike artifact in MEG. NeuroImage 2012; 59(2): 1657-67.
Bermpohl F, Pascual-Leone A, Amedi A, Merabet LB, Fregni F, Gaab N, et al. Attentional modulation of emotional stimulus processing: an fMRI study using emotional expectancy. Human Brain Mapping 2006; 27(8): 662-77.
Cash RFH, Weigand A, Zalesky A, Siddiqi SH, Downar J, Fitzgerald PB, et al. Using Brain Imaging to Improve Spatial Targeting of Transcranial Magnetic Stimulation for Depression. Biological Psychiatry 2020.
The coherence modulations in Fig 5 occur quite late in time compared to the power modulations in Fig 3 and 4. When discussing the results (in e.g. the abstract) it reads as if these findings are reflecting the same process. How can the two effect reflect the same process if the timing is so different?
As the reviewer pointed out correctly, the time window where we observed the coherence modulations happened quite late in time compared to the initial power modulations in the frontal cortex and the habenula (Fig. 4). And there was another increase in the theta band activities in the habenula area even later, at around 3 second after stimuli onset when the emotional figure has already disappeared. Emotional response is composed of a number of factors, two of which are the initial reactivity to an emotional stimulus and the subsequent recovery once the stimulus terminates or ceases to be relevant (Schuyler et al., 2014). We think these neural effects we observed in the three different time windows may reflect different underlying processes. We have discussed this in the ‘Discussion’:
"These activity changes at different time windows may reflect the different neuropsychological processes underlying emotion perception including identification and appraisal of emotional material, production of affective states, and autonomic response regulation and recovery (Phillips et al., 2003a). The later effects of increased theta activities in the habenula when the stimuli disappeared were also supported by other literature showing that, there can be prolonged effects of negative stimuli in the neural structure involved in emotional processing (Haas et al., 2008; Puccetti et al., 2021). In particular, greater sustained patterns of brain activity in the medial prefrontal cortex when responding to blocks of negative facial expressions was associated with higher scores of neuroticism across participants (Haas et al., 2008). Slower amygdala recovery from negative images also predicts greater trait neuroticism, lower levels of likability of a set of social stimuli (neutral faces), and declined day-to-day psychological wellbeing (Schuyler et al., 2014; Puccetti et al., 2021)."
References:
Schuyler BS, Kral TR, Jacquart J, Burghy CA, Weng HY, Perlman DM, et al. Temporal dynamics of emotional responding: amygdala recovery predicts emotional traits. Social Cognitive and Affective Neuroscience 2014; 9(2): 176-81.
Phillips ML, Drevets WC, Rauch SL, Lane R. Neurobiology of emotion perception I: The neural basis of normal emotion perception. Biological Psychiatry 2003a; 54(5): 504-14.
Haas BW, Constable RT, Canli T. Stop the sadness: Neuroticism is associated with sustained medial prefrontal cortex response to emotional facial expressions. NeuroImage 2008; 42(1): 385-92.
Puccetti NA, Schaefer SM, van Reekum CM, Ong AD, Almeida DM, Ryff CD, et al. Linking Amygdala Persistence to Real-World Emotional Experience and Psychological Well-Being. Journal of Neuroscience 2021: JN-RM-1637-20.
Be explicit on the degrees of freedom in the statistical tests given that one subject was excluded from some of the tests.
We thank the reviewers for the comment. The number of samples used for each statistics analysis are stated in the title of the figures. We have now also added the degree of freedom in the main text when parametric statistical tests such as t-test or ANOVAs have been used. When permutation tests (which do not have any degrees of freedom associated with it) are used, we have now added the number of samples for the permutation test.
Reviewer #2 (Public Review):
In this study, Huang and colleagues recorded local field potentials from the lateral habenula in patients with psychiatric disorders who recently underwent surgery for deep brain stimulation (DBS). The authors combined these invasive measurements with non-invasive whole-head MEG recordings to study functional connectivity between the habenula and cortical areas. Since the lateral habenula is believed to be involved in the processing of emotions, and negative emotions in particular, the authors investigated whether brain activity in this region is related to emotional valence. They presented pictures inducing negative and positive emotions to the patients and found that theta and alpha activity in the habenula and frontal cortex increases when patients experience negative emotions. Functional connectivity between the habenula and the cortex was likewise increased in this band. The authors conclude that theta/alpha oscillations in the habenula-cortex network are involved in the processing of negative emotions in humans.
Because DBS of the habenula is a new treatment tested in this cohort in the framework of a clinical trial, these are the first data of its kind. Accordingly, they are of high interest to the field. Although the study mostly confirms findings from animal studies rather than bringing up completely new aspects of emotion processing, it certainly closes a knowledge gap.
In terms of community impact, I see the strengths of this paper in basic science rather than the clinical field. The authors demonstrate the involvement of theta oscillations in the habenula-prefrontal cortex network in emotion processing in the human brain. The potential of theta oscillations to serve as a marker in closed-loop DBS, as put forward by the authors, appears less relevant to me at this stage, given that the clinical effects and side-effects of habenula DBS are not known yet.
We thank the reviewers for the favourable comments about the implication of our study in basic science and about the value of our study in closing a knowledge gap. We agree that further studies would be required to make conclusions about the clinical effects and side-effects of habenula DBS.
Detailed comments:
The group-average MEG power spectrum (Fig. 4B) suggests that negative emotions lead to a sustained theta power increase and a similar effect, though possibly masked by a visual ERP, can be seen in the habenula (Fig. 3C). Yet the statistics identify brief elevations of habenula theta power at around 3s (which is very late), a brief elevation of prefrontal power a time 0 or even before (Fig. 4C) and a brief elevation of Habenula-MEG theta coherence around 1 s. It seems possible that this lack of consistency arises from a low signal-to-noise ratio. The data contain only 27 trails per condition on average and are contaminated by artifacts caused by the extension wires.
With regard to the nature of the activity modulation with short latency after stimuli onset: whether this is an ERP or oscillation? We have now investigated this. In summary, by analysing the ERP and removing the influence of the ERP from the total power spectra, we didn’t observe stimulus emotional valence related modulation in the ERP, and the modulation related to emotional valence in the pure induced (non-phase-locked) power spectra was similar to what we have observed in the total power shown in Fig. 3. Therefore, we argue that the theta/alpha increase with negative emotional stimuli we observed in both habenula and prefrontal cortex 0-500 ms after stimuli onset are not dominated by visual or other ERP.
With regard to the signal-to-noise ratio from only 27 trials per condition on average per participant: We have tried to clean the data by removing the trials with obvious artefacts characterised by increased measurements in the time domain over 5 times the standard deviation and increased activities across all frequency bands in the frequency domain. After removing the trials with artefacts, we have 27 trials per condition per subject on average. We agree that 27 trials per condition on average is not a high number, and increasing the number of trials would further increase the signal-to-noise ratio. However, our studies with EEG recordings and LFP recordings from externalised patients have shown that 30 trials was enough to identify reduction in the amplitude of post-movement beta oscillations at the beginning of visuomotor adaption in the motor cortex and STN (Tan et al., 2014a; Tan et al., 2014b). These results of motor error related modulation in the post-movement beta have been repeated by other studies from other groups. In Tan et al. 2014b, with simultaneous EEG and STN LFP measurements and a similar number of trials (around 30), we also quantified the time-course of STN-motor cortex coherence during voluntary movements. This pattern has also been repeated in a separate study from another group with around 50 trials per participant (Talakoub et al., 2016). In addition, similar behavioural paradigm (passive figure viewing paradigm) has been used in two previous studies with LFP recordings from STN from different patient groups (Brucke et al., 2007; Huebl et al., 2014). In both studies, a similar number of trials per condition around 27 was used. The authors have identified meaningful activity modulation in the STN by emotional stimuli. Therefore, we think the number of trials per condition was sufficient to identify emotional valence induced difference in the LFPs in the paradigm.
We agree that the measurement of coherence can be more susceptible to noise and suffer from the reduced signal-to-noise ratio in MEG recording. In Hirschmann et al. 2013, 5 minutes of resting recording and 5 minutes of movement recording from 10 PD patients were used to quantify movement related changes in STN-cortical coherence and how this was modulated by levodopa (Hirschmann et al., 2013). Litvak et al. (2012) have identified movement-related changes in the coherence between STN LFP and motor cortex with recording with simultaneous STN LFP and MEG recordings from 17 PD patients and 20 trials in average per participant per condition (Litvak et al., 2012). With similar methods, van Wijk et al. (2017) used recordings from 9 patients and around on average in 29 trials per hand per condition, and they identified reduced cortico-pallidal coherence in the low-beta decreases during movement (van Wijk et al., 2017). So the trial number per condition participant we used in this study are comparable to previous studies.
The DBS extension wires do reduce signal-to-noise ratio in the MEG recording. therefore the spatiotemporal Signal Space Separation (tSSS) method (Taulu and Simola, 2006) implemented in the MaxFilter software (Elekta Oy, Helsinki, Finland) has been applied in this study to suppress strong magnetic artifacts caused by extension wires. This method has been proved to work well in de-noising the magnetic artifacts and movement artifacts in MEG data in our previous studies (Cao et al., 2019; Cao et al., 2020). In addition, the beamforming method proposed by several studies (Litvak et al., 2010; Hirschmann et al., 2011; Litvak et al., 2011) has been used in this study. In Litvak et al., 2010, the artifacts caused by DBS extension wires was detailed described and the beamforming was demonstrated to effectively suppress artifacts and thereby enable both localization of cortical sources coherent with the deep brain nucleus. We have now added more details and these references about the data cleaning and the beamforming method in the main text. With the beamforming method, we did observe the standard movement-related modulation in the beta frequency band in the motor cortex with 9 trials of figure pressing movements, shown in the following figure for one patient as an example (Figure 5–figure supplement 1). This suggests that the beamforming method did work well to suppress the artefacts and help to localise the source with a low number of trials. The figure on movement-related modulation in the motor cortex in the MEG signals have now been added as a supplementary figure to demonstrate the effect of the beamforming.
Figure 5–figure supplement 1. (A) Time-frequency maps of MEG activity for right hand button press at sensor level from one participant (Case 8). (B) DICS beamforming source reconstruction of the areas with movement-related oscillation changes in the range of 12-30 Hz. The peak power was located in the left M1 area, MNI coordinate [-37, -12, 43].
References:
Tan H, Jenkinson N, Brown P. Dynamic neural correlates of motor error monitoring and adaptation during trial-to-trial learning. Journal of Neuroscience 2014a; 34(16): 5678-88.
Tan H, Zavala B, Pogosyan A, Ashkan K, Zrinzo L, Foltynie T, et al. Human subthalamic nucleus in movement error detection and its evaluation during visuomotor adaptation. Journal of Neuroscience 2014b; 34(50): 16744-54.
Talakoub O, Neagu B, Udupa K, Tsang E, Chen R, Popovic MR, et al. Time-course of coherence in the human basal ganglia during voluntary movements. Scientific Reports 2016; 6: 34930.
Brucke C, Kupsch A, Schneider GH, Hariz MI, Nuttin B, Kopp U, et al. The subthalamic region is activated during valence-related emotional processing in patients with Parkinson's disease. European Journal of Neuroscience 2007; 26(3): 767-74.
Huebl J, Spitzer B, Brucke C, Schonecker T, Kupsch A, Alesch F, et al. Oscillatory subthalamic nucleus activity is modulated by dopamine during emotional processing in Parkinson's disease. Cortex 2014; 60: 69-81.
Hirschmann J, Ozkurt TE, Butz M, Homburger M, Elben S, Hartmann CJ, et al. Differential modulation of STN-cortical and cortico-muscular coherence by movement and levodopa in Parkinson's disease. NeuroImage 2013; 68: 203-13.
Litvak V, Eusebio A, Jha A, Oostenveld R, Barnes G, Foltynie T, et al. Movement-related changes in local and long-range synchronization in Parkinson's disease revealed by simultaneous magnetoencephalography and intracranial recordings. Journal of Neuroscience 2012; 32(31): 10541-53.
van Wijk BCM, Neumann WJ, Schneider GH, Sander TH, Litvak V, Kuhn AA. Low-beta cortico-pallidal coherence decreases during movement and correlates with overall reaction time. NeuroImage 2017; 159: 1-8.
Taulu S, Simola J. Spatiotemporal signal space separation method for rejecting nearby interference in MEG measurements. Physics in Medicine and Biology 2006; 51(7): 1759-68.
Cao C, Huang P, Wang T, Zhan S, Liu W, Pan Y, et al. Cortico-subthalamic Coherence in a Patient With Dystonia Induced by Chorea-Acanthocytosis: A Case Report. Frontiers in Human Neuroscience 2019; 13: 163.
Cao C, Li D, Zhan S, Zhang C, Sun B, Litvak V. L-dopa treatment increases oscillatory power in the motor cortex of Parkinson's disease patients. NeuroImage Clinical 2020; 26: 102255.
Litvak V, Eusebio A, Jha A, Oostenveld R, Barnes GR, Penny WD, et al. Optimized beamforming for simultaneous MEG and intracranial local field potential recordings in deep brain stimulation patients. NeuroImage 2010; 50(4): 1578-88.
Litvak V, Jha A, Eusebio A, Oostenveld R, Foltynie T, Limousin P, et al. Resting oscillatory cortico-subthalamic connectivity in patients with Parkinson's disease. Brain 2011; 134(Pt 2): 359-74.
Hirschmann J, Ozkurt TE, Butz M, Homburger M, Elben S, Hartmann CJ, et al. Distinct oscillatory STN-cortical loops revealed by simultaneous MEG and local field potential recordings in patients with Parkinson's disease. NeuroImage 2011; 55(3): 1159-68.
I doubt that the correlation between habenula power and habenula-MEG coherence (Fig. 6C) is informative of emotion processing. First, power and coherence in close-by time windows are likely to to be correlated irrespective of the task/stimuli. Second, if meaningful, one would expect the strongest correlation for the negative condition, as this is the only condition with an increase of theta coherence and a subsequent increase of theta power in the habenula. This, however, does not appear to be the case.
The authors included the factors valence and arousal in their linear model and found that only valence correlated with electrophysiological effects. I suspect that arousal and valence scores are highly correlated. When fed with informative yet highly correlated variables, the significance of individual input variables becomes difficult to assess in many statistical models. Hence, I am not convinced that valence matters but arousal not.
For the correlation shown in Fig. 6C, we used a linear mixed-effect modelling (‘fitlme’ in Matlab) with different recorded subjects as random effects to investigate the correlations between the habenula power and habenula-MEG coherence at an earlier window, while considering all trials together. Therefore the reported value in the main text and in the figure (k = 0.2434 ± 0.1031, p = 0.0226, R2 = 0.104) show the within subjects correlation that are consistent across all measured subjects. The correlation is likely to be mediated by emotional valence condition, as negative emotional stimuli tend to be associated with both high habenula-MEG coherence and high theta power in the later time window tend to happen in the trials with.
The arousal scores are significantly different for the three valence conditions as shown in Fig. 1B. However, the arousal scores and the valence scores are not monotonically correlated, as shown in the following figure (Fig. S2). The emotional neutral figures have the lowest arousal value, but have the valence value sitting between the negative figures and the positive figures. We have now added the following sentence in the main text:
"This nonlinear and non-monotonic relationship between arousal scores and the emotional valence scores allowed us to differentiate the effect of the valence from arousal."
Table 2 in the main text show the results of the linear mixed-effect modelling with the neural signal as the dependent variable and the valence and arousal scores as independent variables. Because of the non-linear and non-monotonic relationship between the valence and arousal scores, we think the significance of individual input variables is valid in this statistical model. We have now added a new figure (shown below, Fig. 7) with scatter plots showing the relationship between the electrophysiological signal and the arousal and emotional valence scores separately using Spearman’s partial correlation analysis. In each scatter plot, each dot indicates the average measurement from one participant in one emotional valence condition. As shown in the following figure, the electrophysiological measurements linearly correlated with the valence score, but not with the arousal scores. However, the statistics reported in this figure considered all the dots together. The linear mixed effect modelling taking into account the interdependency of the measurements from the same participant. So the results reported in the main text using linear mixed effect modelling are statistically more valid, but supplementary figure here below illustrate the relationship.
Figure S2. Averaged valence and arousal ratings (mean ± SD) for figures of the three emotional condition. (B) Scatter plots showing the relationship between arousal and valence scores for each emotional condition for each participant.
Figure 7. Scatter plots showing how early theta/alpha band power increase in the frontal cortex (A), theta/alpha band frontal cortex-habenula coherence (B) and theta band power increase in habenula stimuli (C) changed with emotional valence (left column) and arousal (right column). Each dot shows the average of one participant in each categorical valence condition, which are also the source data of the multilevel modelling results presented in Table 2. The R and p value in the figure are the results of partial correlation considering all data points together.
Page 8: "The time-varying coherence was calculated for each trial". This is confusing because coherence quantifies the stability of a phase difference over time, i.e. it is a temporal average, not defined for individual trials. It has also been used to describe the phase difference stability over trials rather than time, and I assume this is the method applied here. Typically, the greatest coherence values coincide with event-related power increases, which is why I am surprised to see maximum coherence at 1s rather than immediately post-stimulus.
We thank the reviewer for pointing out this incorrect description. As the reviewer pointed out correctly, the method we used describe the phase difference stability over trials rather than time. We have now clarified how coherence was calculated and added more details in the methods:
"The time-varying cross trial coherence between each MEG sensor and the habenula LFP was first calculated for each emotional valence condition. For this, time-frequency auto- and cross-spectral densities in the theta/alpha frequency band (5-10 Hz) between the habenula LFP and each MEG channel at sensor level were calculated using the wavelet transform-based approach from -2000 to 4000 ms for each trial with 1 Hz steps using the Morlet wavelet and cycle number of 6. Cross-trial coherence spectra for each LFP-MEG channel combination was calculated for each emotional valence condition for each habenula using the function ‘ft_connectivityanalysis’ in Fieldtrip (version 20170628). Stimulus-related changes in coherence were assessed by expressing the time-resolved coherence spectra as a percentage change compared to the average value in the -2000 to -200 ms (pre-stimulus) time window for each frequency."
In the Morlet wavelet analysis we used here, the cycle number (C) determines the temporal resolution and frequency resolution for each frequency (F). The spectral bandwidth at a given frequency F is equal to 2F/C while the wavelet duration is equal to C/F/pi. We used a cycle number of 6. For theta band activities around 5 Hz, we will have the spectral bandwidth of 25/6 = 1.7 Hz and the wavelet duration of 6/5/pi = 0.38s = 380ms.
As the reviewer noticed, we observed increased activities across a wide frequency band in both habenula and the prefrontal cortex within 500 ms after stimuli onset. But the increase of cross-trial coherence starts at around 300 ms. The increase of coherence in a time window without increase of power in either of the two structures indicates a phase difference stability across trials in the oscillatory activities from the two regions, and this phase difference stability across trials was not secondary to power increase.
Reviewer #3 (Public Review):
This paper describes the oscillatory activity of the habenula using local field potentials, both within the region and, through the use of MEG, in connection to the prefrontal cortex. The characteristics of this activity were found to vary with the emotional valence but not with arousal. Sheding light on this is relevant, because the habenula is a promising target for deep brain stimulation.
In general, because I am not much on top of the literature on the habenula, I find difficult to judge about the novelty and the impact of this study. What I can say is that I do find the paper is well-written and very clear; and the methods, although quite basic (which is not bad), are sound and rigourous.
We thank the reviewer for the positive comments about the potential implication of our study and on the methods we used.
On the less positive side, even though I am aware that in this type of studies it is difficult to have high N, the very low N in this case makes me worry about the robustness and replicability of the results. I'm sure I have missed it and it's specified somewhere, but why is N different for the different figures? Is it because only 8 people had MEG? The number of trials seems also a somewhat low. Therefore, I feel the authors perhaps need to make an effort to make up for the short number of subjects in order to add confidence to the results. I would strongly recommend to bootstrap the statistical analysis and extract non-parametric confidence intervals instead of showing parametric standard errors whenever is appropriate. When doing that, it must be taken into account that each two of the habenula belong to the same person; i.e. one bootstraps the subjects not the habenula.
We do understand and appreciate the concern of the reviewer on the low sample numbers due to the strict recruitment criteria for this very early stage clinical trial: 9 patients for bilateral habenula LFPs, and 8 patients with good quality MEGs. Some information to justify the number of trials per condition for each participant has been provided in the reply to the Detailed Comments 1 from Reviewer 2. The sample number used in each analysis was included in the figures and in the main text.
We have used non-parametric cluster-based permutation approach (Maris and Oostenveld, 2007) for all the main results as shown in Fig. 3-5. Once the clusters (time window and frequency band) with significant differences for different emotional valence conditions have been identified, parametric statistical test was applied to the average values of the clusters to show the direction of the difference. These parametric statistics are secondary to the main non-parametric permutation test.
In addition, the DICS beamforming method was applied to localize cortical sources exhibiting stimuli-related power changes and cortical sources coherent with deep brain LFPs for each subject for positive and negative emotional valence conditions respectively. After source analysis, source statistics over subjects was performed. Non-parametric permutation testing with or without cluster-based correction for multiple comparisons was applied to statistically quantify the differences in cortical power source or coherence source between negative and positive emotional stimuli.
References:
Maris E, Oostenveld R. Nonparametric statistical testing of EEG- and MEG-data. Journal of Neuroscience Methods 2007; 164(1): 177-90.
Related to this point, the results in Figure 6 seem quite noisy, because interactions (i.e. coherence) are harder to estimate and N is low. For example, I have to make an effort of optimism to believe that Fig 6A is not just noise, and the result in Fig 6C is also a bit weak and perhaps driven by the blue point at the bottom. My read is that the authors didn't do permutation testing here, and just a parametric linear-mixed effect testing. I believe the authors should embed this into permutation testing to make sure that the extremes are not driving the current p-value.
We have now quantified the coherence between frontal cortex-habenula and occipital cortex-habenula separately (please see more details in the reply to Reviewer 2 (Recommendations for the authors 6). The new analysis showed that the increase in the theta/alpha band coherence around 1 s after the negative stimuli was only observed between prefrontal cortex-habenula and not between occipital cortex-habenula. This supports the argument that Fig. 6A is not just noise.
Reviewer #1:
Hutchings et al. report an updated cryo-electron tomography study of the yeast COP-II coat assembled around model membranes. The improved overall resolution and additional compositional states enabled the authors to identify new domains and interfaces--including what the authors hypothesize is a previously overlooked structural role for the SEC31 C-Terminal Domain (CTD). By perturbing a subset of these new features with mutants, the authors uncover some functional consequences pertaining to the flexibility or stability of COP-II assemblies.
Overall, the structural and functional work appears reliable, but certain questions and comments should be addressed prior to publication. However, this reviewer failed to appreciate the conceptual advance that warrants publication in a general biology journal like eLIFE. Rather, this study provides a valuable refinement of our understanding of COP-II that I believe is better suited to a more specialized, structure-focused journal.
We agree that in our original submission our description of the experimental setup, indeed similar to previous work, did not fully capture the novel findings of this paper. Rather than being simply a higher resolution structure of the COPII coat, in fact we have discovered new interactions in the COPII assembly network, and we have probed their functional roles, significantly changing our understanding of the mechanisms of COPII-mediated membrane curvature. In the revised submission we have included additional genetic data that further illuminate this mechanism, and have rewritten the text to better communicate the novel aspects of our work.
Our combination of structural, functional and genetic analyses goes beyond refining our textbook understanding of the COPII coat as a simple ‘adaptor and cage’, but rather it provides a completely new picture of how dynamic regulation of assembly and disassembly of a complex network leads to membrane remodelling.
These new insights have important implications for how coat assembly provides structural force to bend a membrane but is still able to adapt to distinct morphologies. These questions are at the forefront of protein secretion, where there is debate about how different types of carriers might be generated that can accommodate cargoes of different size.
Major Comments: 1) The authors belabor what this reviewer thinks is an unimportant comparison between the yeast reconstruction of the outer coat vertex with prior work on the human outer coat vertex. Considering the modest resolution of both the yeast and human reconstructions, the transformative changes in cryo-EM camera technology since the publication of the human complex, and the differences in sample preparation (inclusion of the membrane, cylindrical versus spherical assemblies, presence of inner coat components), I did not find this comparison informative. The speculations about a changing interface over evolutionary time are unwarranted and would require a detailed comparison of co-evolutionary changes at this interface. The simpler explanation is that this is a flexible vertex, observed at low resolution in both studies, plus the samples are very different.
We do agree that our proposal that the vertex interface changes over evolutionary time is speculative and we have removed this discussion. We agree that a co-evolutionary analysis will be enlightening here, but is beyond the scope of the current work.
We respectfully disagree with the reviewer’s interpretation that the difference between the two vertices is due to low resolution. The interfaces are clearly different, and the resolutions of the reconstructions are sufficient to state this. The reviewer’s suggestion that the difference in vertex orientation might be simply attributable to differences in sample, such as inclusion of the membrane, cylindrical versus spherical morphology, or presence of inner coat components were ruled out in our original submission: we resolved yeast vertices on spherical vesicles (in addition to those on tubes) and on membrane-less cages. These analyses clearly showed that neither the presence of a membrane, nor the change in geometry (tubular vs. spherical) affect vertex interactions. These experiments are presented in Supplementary Fig 4 (Supplementary Fig. 3 in the original version). Similarly, we discount that differences might be due to the presence or absence of inner coat components, since membrane-less cages were previously solved in both conditions and are no different in terms of their vertex structure (Stagg et al. Nature 2006 and Cell 2008).
We believe it is important to report on the differences between the two vertex structures. Nevertheless, we have shifted our emphasis on the functional aspects of vertex formation and moved the comparison between the two vertices to the supplement.
2) As one of the major take home messages of the paper, the presentation and discussion of the modeling and assignment of the SEC31-CTD could be clarified. First, it isn't clear from the figures or the movies if the connectivity makes sense. Where is the C-terminal end of the alpha-solenoid compared to this new domain? Can the authors plausibly account for the connectivity in terms of primary sequence? Please also include a side-by-side comparison of the SRA1 structure and the CTD homology model, along with some explanation of the quality of the model as measured by Modeller. Finally, even if the new density is the CTD, it isn't clear from the structure how this sub-stoichiometric and apparently flexible interaction enhances stability. Hence, when the authors wrote "when the [CTD] truncated form was the sole copy of Sec31 in yeast, cells were not viable, indicating that the novel interaction we detect is essential for COPII coat function." Maybe, but could this statement be a leap to far? Is it the putative interaction essential, or is the CTD itself essential for reasons that remain to be fully determined?
The CTD is separated from the C-terminus of the alpha solenoid domain by an extended domain (~350 amino acids) that is predicted to be disordered, and contains the PPP motifs and catalytic fragment that contact the inner coat. This is depicted in cartoon form in Figures 3A and 7, and discussed at length in the text. This arrangement explains why no connectivity is seen, or expected. We could highlight the C-terminus of the alpha-solenoid domain to emphasize where the disordered region should emerge from the rod, but connectivity of the disordered domain to the CTD could arise from multiple positions, including from an adjacent rod.
The reviewer’s point about the essentiality of the CTD being independent of its interaction with the Sec31 rod, is an important one. The basis for our model that the CTD enhances stability or rigidity of the coat is the yeast phenotype of Sec31-deltaCTD, which resembles that of a sec13 null. Both mutants are lethal, but rescued by deletion of emp24, which leads to more easily deformable membranes (Čopič et al. Science 2012). We agree that even if this model is true, the interaction of the CTD with Sec31 that our new structure reveals is not proven to drive rigidity or essentiality. We have tempered this hypothesis and added alternative possibilities to the discussion.
We have included the SRA1 structure in Supplementary Fig 5, as requested, and the model z-score in the Methods. The Z-score, as calculated by the proSA-web server is -6.07 (see figure below, black dot), and falls in line with experimentally determined structures including that of the template (PDB 2mgx, z-score = -5.38).
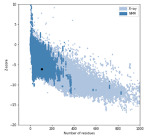
3) Are extra rods discussed in Fig. 4 are a curiosity of unclear functional significance? This reviewer is concerned that these extra rods could be an in vitro stoichiometry problem, rather than a functional property of COP-II.
This is an important point, that, as we state in the paper, cannot be answered at the moment: the resolution is too low to identify the residues involved in the interaction. Therefore we are hampered in our ability to assess the physiological importance of this interaction. We still believe the ‘extra’ rods are an important observation, as they clearly show that another mode of outer coat interaction, different from what was reported before, is possible.
The concern that interactions visualised in vitro might not be physiologically relevant is broadly applicable to structural biology approaches. However, our experimental approach uses samples that result from active membrane remodelling under near-physiological conditions, and we therefore expect these to be less prone to artefacts than most in vitro reconstitution approaches, where proteins are used at high concentrations and in high salt buffer conditions.
4) The clashsccore for the PDB is quite high--and I am dubious about the reliability of refining sidechain positions with maps at this resolution. In addition to the Ramchandran stats, I would like to see the Ramachandran plot as well as, for any residue-level claims, the density surrounding the modeled side chain (e.g. S742).
The clashscore is 13.2, which, according to molprobity, is in the 57th percentile for all structures and in the 97th for structures of similar resolutions. We would argue therefore that the clashscore is rather low. In fact, the model was refined from crystal structures previously obtained by other groups, which had worse clashscore (17), despite being at higher resolution. Our refinement has therefore improved the clashscore. During refinement we have chosen restraint levels appropriate to the resolution of our map (Afonine et al., Acta Cryst D 2018)
The Ramachandran plot is copied here and could be included in a supplemental figure if required. We make only one residue-level claim (S742), the density for which is indeed not visible at our resolution. We claim that S742 is close to the Sec23-23 interface, and do not propose any specific interactions. Nevertheless we have removed reference to S742 from the manuscript. We included this specific information because of the potential importance of this residue as a site of phosphorylation, thereby putting this interface in broader context for the general eLife reader.
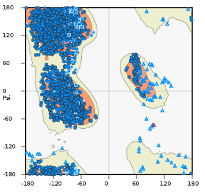
Minor Comments:
1) The authors wrote "To assess the relative positioning of the two coat layers, we analysed the localisation of inner coat subunits with respect to each outer coat vertex: for each aligned vertex particle, we superimposed the positions of all inner coat particles at close range, obtaining the average distribution of neighbouring inner coat subunits. From this 'neighbour plot' we did not detect any pattern, indicating random relative positions. This is consistent with a flexible linkage between the two layers that allows adaptation of the two lattices to different curvatures (Supplementary Fig 1E)." I do not understand this claim, since the pattern both looks far from random and the interactions depend on molecular interactions that are not random. Please clarify.
We apologize for the confusion: the pattern of each of the two coats are not random. Our sentence refers to the positions of inner and outer coats relative to each other. The two lattices have different parameters and the two layers are linked by flexible linkers (the 350 amino acids referred to above). We have now clarified the sentence.
2) Related to major point #1, the author wrote "We manually picked vertices and performed carefully controlled alignments." I do now know what it means to carefully control alignments, and fear this suggests human model bias.
We used different starting references for the alignments, with the precise aim to avoid model bias. For both vesicle and cage vertex datasets, we have aligned the subtomograms against either the vertex obtained from tubules, or the vertex from previously published membrane-less cages. In all cases, we retrieved a structure that resembles the one on tubules, suggesting that the vertex arrangement we observe isn’t simply the result of reference bias. This procedure is depicted in Supplementary Fig 4 (Supplementary Fig. 3 in the original manuscript), but we have now clarified it also in the methods section.
3) Why do some experiments use EDTA? I may be confused, but I was surprised to see the budding reaction employed 1mM GMPPNP, and 2.5mM EDTA (but no Magnesium?). Also, for the budding reaction, please replace or expand upon the "the 10% GUV (v/v)" with a mass or molar lipid-to-protein ratio.
We regret the confusion. As stated in the methods, all our budding reactions are performed in the presence of EDTA and Magnesium, which is present in the buffer (at 1.2 mM). The reason is to facilitate nucleotide exchange, as reported and validated in Bacia et al., Scientific Reports 2011.
Lipids in GUV preparations are difficult to quantify. We report the stock concentrations used, but in each preparation the amount of dry lipid that forms GUVs might be different, as is the concentration of GUVs after hydration. However since we analyse reactions where COPII proteins have bound and remodelled individual GUVs, we do not believe the protein/lipid ratio influences our structures.
4) Please cite the AnchorMap procedure.
We cite the SerialEM software, and are not aware of other citations specifically for the anchor map procedure.
5) Please edit for typos (focussing, functionl, others)
Done
Reviewer #2:
The manuscript describes new cryo-EM, biochemistry, and genetic data on the structure and function of the COPII coat. Several new discoveries are reported including the discovery of an extra density near the dimerization region of Sec13/31, and "extra rods" of Sec13/31 that also bind near the dimerization region. Additionally, they showed new interactions between the Sec31 C-terminal unstructured region and Sec23 that appear to bridge multiple Sec23 molecules. Finally, they increased the resolution of the Sec23/24 region of their structure compared to their previous studies and were able to resolve a previously unresolved L-loop in Sec23 that makes contact with Sar1. Most of their structural observations were nicely backed up with biochemical and genetic experiments which give confidence in their structural observations. Overall the paper is well-written and the conclusions justified.
However, this is the third iteration of structure determination of the COPII coat on membrane with essentially the same preparation and methods. Each time, there has been an incremental increase in resolution and new discoveries, but the impact of the present study is deemed to be modest. The science is good, but it may be more appropriate for a more specialized journal. Areas of specific concern are described below.
As described above, we respectfully disagree with this interpretation of the advance made by the current work. This work improves on previous work in many aspects. The resolution of the outer coat increases from over 40A to 10-12A, allowing visualisation of features that were not previously resolved, including a novel vertex arrangement, the Sec31 CTD, and the outer coat ‘extra rods’. An improved map of the inner coat also allows us to resolve the Sec23 ‘L-loop’. We would argue that these are not just extra details, but correspond to a suite of novel interactions that expand our understanding of the complex COPII assembly network. Moreover, we include biochemical and genetic experiments that not only back up our structural observations but bring new insights into COPII function. As pointed out in response to reviewer 1, we believe our work contributes a significant conceptual advance, and have modified the manuscript to convey this more effectively.
1) The abstract is vague and should be re-written with a better description of the work.
We have modified the abstract to specifically outline what we have done and the major new discoveries of this paper.
2) Line 166 - "Surprisingly, this mutant was capable of tubulating GUVs". This experiment gets to one of the fundamental unknown questions in COPII vesiculation. It is not clear what components are driving the membrane remodeling and at what stages during vesicle formation. Isn't it possible that the tubulation activity the authors observe in vitro is not being driven at all by Sec13/31 but rather Sec23/24-Sar1? Their Sec31ΔCTD data supports this idea because it lacks a clear ordered outer coat despite making tubules. An interesting experiment would be to see if tubules form in the absence of all of Sec13/31 except the disordered domain of Sec31 that the authors suggest crosslinks adjacent Sec23/24s.
This is an astute observation, and we agree with the reviewer that the source of membrane deformation is not fully understood. We favour the model that budding is driven significantly by the Sec23-24 array. To further support this, we have performed a new experiment, where we expressed Sec31ΔN in yeast cells lacking Emp24, which have more deformable membranes and are tolerant to the otherwise lethal deletion of Sec13. While Sec31ΔN in a wild type background did not support cell viability, this was rescued in a Δemp24 yeast strain, strongly supporting the hypothesis that a major contributor to membrane remodelling is the inner coat, with the outer coat becoming necessary to overcome membrane bending resistance that ensues from the presence of cargo. We now include these results in Figure 1.
However, we must also take into account the results presented in Fig. 6, where we show that weakening the Sec23-24 interface still leads to budding, but only if Sec13-31 is fully functional, and that in this case budding leads to connected pseudo-spherical vesicles rather than tubes. When Sec13-31 assembly is also impaired, tubes appear unstructured. We believe this strongly supports our conclusions that both inner and outer coat interactions are fundamental for membrane remodelling, and it is the interplay between the two that determines membrane morphology (i.e. tubes vs. spheres).
To dissect the roles of inner and outer coats even further, we have done the experiment that the reviewer suggests: we expressed Sec31768-1114, but the protein was not well-behaved and co-purified with chaperones. We believe the disordered domain aggregates when not scaffolded by the structured elements of the rod. Nonetheless, we used this fragment in a budding reaction, and could not see any budding. We did not include this experiment as it was inconclusive: the lack of functionality of the purified Sec31 fragment could be attributed to the inability of the disordered region to bind its inner coat partner in the absence of the scaffolding Sec13-31 rod. As an alternative approach, we have used a version of Sec31 that lacks the CTD, and harbours a His tag at the N-terminus (known from previous studies to partially disrupt vertex assembly). We think this construct is more likely to be near native, since both modifications on their own lead to functional protein. We could detect no tubulation with this construct by negative stain, while both control constructs (Sec31ΔCTD and Nhis-Sec31) gave tubulation. This suggests that the cross-linking function of Sec31 is not sufficient to tubulate GUV membranes, but some degree of functional outer coat organisation (either mediated by N- or C-terminal interactions) is needed. It is also possible that the lack of outer coat organisation might lead to less efficient recruitment to the inner coat and cross-linking activity. We have added this new observation to the manuscript.
3) Line 191 - "Inspecting cryo-tomograms of these tubules revealed no lozenge pattern for the outer 192 coat" - this phrasing is vague. The reviewer thinks that what they mean is that there is a lack of order for the Sec13/31 layer. Please clarify.
The reviewer is correct, we have changed the sentence.
4) Line 198 - "unambiguously confirming this density corresponds to 199 the CTD." This only confirms that it is the CTD if that were the only change and the Sec13/31 lattice still formed. Another possibility is that it is density from other Sec13/31 that only appears when the lattice is formed such as the "extra rods". One possibility is that the density is from the extra rods. The reviewer agrees that their interpretation is indeed the most likely, but it is not unambiguous. The authors should consider cross-linking mass spectrometry.
We have removed the word ‘unambiguously’, and changed to ‘confirming that this density most likely corresponds to the CTD’. Nonetheless, we believe that our interpretation is correct: the extra rods bind to a different position, and themselves also show the CTD appendage. In this experiment, the lack of the CTD was the only biochemical change.
5) In the Sec31ΔCTD section, the authors should comment on why ΔCTD is so deleterious to oligomer organization in yeast when cages form so abundantly in preparations of human Sec13/31 ΔC (Paraan et al 2018).
We have added a comment to address this. “Interestingly, human Sec31 proteins lacking the CTD assemble in cages, indicating that either the vertex is more stable for human proteins and sufficient for assembly, or that the CTD is important in the context of membrane budding but not for cage formation in high salt conditions.”
6) The data is good for the existence of the "extra rods", but significance and importance of them is not clear. How can these extra densities be distinguished from packing artifacts due to imperfections in the helical symmetry.
Please also see our response to point 3 from reviewer 1. Regarding the specific concern that artefacts might be a consequence of imperfection in the helical symmetry, we would argue such imperfections are indeed expected in physiological conditions, and to a much higher extent. For this reason interactions seen in the context of helical imperfections are likely to be relevant. In fact, in normal GTP hydrolysis conditions, we expect long tubes would not be able to form, and the outer coat to be present on a wide range of continuously changing membrane curvatures. We think that the ability of the coat to form many interactions when the symmetry is imperfect might be exactly what confers the coat its flexibility and adaptability.
7) Figure 5 is very hard to interpret and should be redone. Panels B and C are particularly hard to interpret.
We have made a new figure where we think clarity is improved.
8) The features present in Sec23/24 structure do not reflect the reported resolution of 4.7 Å. It seems that the resolution is overestimated.
We report an average resolution of 4.6 Å. In most of our map we can clearly distinguish beta strands, follow the twist of alpha helices and see bulky side chains. These features typically become visible at 4.5-5A resolution. We agree that some areas are worse than 4.6 Å, as typically expected for such a flexible assembly, but we believe that the average resolution value reported is accurate. We obtained the same resolution estimate using different software including relion, phenix and dynamo, so that is really the best value we can provide. To further convince ourselves that we have the resolution we claim, we sampled EM maps from the EMDB with the same stated resolution (we just took the 7 most recent ones which had an associated atomic model), and visualised their features at arbitrary positions. For both beta strands and alpha helices, we do not feel our map looks any worse than the others we have examined. We include a figure here.
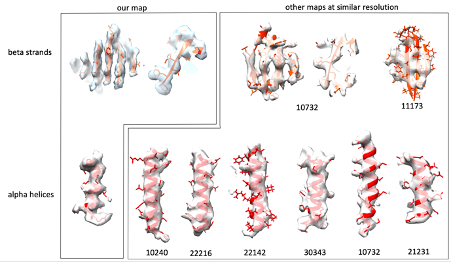
9) Lines 315/316 - "We have combined cryo-tomography with biochemical and genetic assays to obtain a complete picture of the assembled COPII coat at unprecedented resolution (Fig. 7)"
10) Figure 7. is a schematic model/picture the authors should reference a different figure or rephrase the sentence.
We now refer to Fig 7 in a more appropriate place.
Reviewer #3:
The manuscript by Hutchings et al. describes several previously uncharacterised molecular interactions in the coats of COP-II vesicles by using a reconstituted coats of yeast COPI-II. They have improved the resolution of the inner coat to 4.7A by tomography and subtomogram averaging, revealing detailed interactions, including those made by the so-called L-loop not observed before. Analysis of the outer layer also led to new interesting discoveries. The sec 31 CTD was assigned in the map by comparing the WT and deletion mutant STA-generated density maps. It seems to stabilise the COP-II coats and further evidence from yeast deletion mutants and microsome budding reconstitution experiments suggests that this stabilisation is required in vitro. Furthermore, COP-II rods that cover the membrane tubules in right-handed manner revealed sometimes an extra rod, which is not part of the canonical lattice, bound to them. The binding mode of these extra rods (which I refer to here a Y-shape) is different from the canonical two-fold symmetric vertex (X-shape). When the same binding mode is utilized on both sides of the extra rod (Y-Y) the rod seems to simply insert in the canonical lattice. However, when the Y-binding mode is utilized on one side of the rod and the X-binding mode on the other side, this leads to bridging different lattices together. This potentially contributes to increased flexibility in the outer coat, which maybe be required to adopt different membrane curvatures and shapes with different cargos. These observations build a picture where stabilising elements in both COP-II layers contribute to functional cargo transport. The paper makes significant novel findings that are described well. Technically the paper is excellent and the figures nicely support the text. I have only minor suggestions that I think would improve the text and figure.
We thank the reviewer for helpful suggestions which we agree improve the manuscript.
Minor Comments:
L 108: "We collected .... tomograms". While the meaning is clear to a specialist, this may sound somewhat odd to a generic reader. Perhaps you could say "We acquired cryo-EM data of COP-II induced tubules as tilt series that were subsequently used to reconstruct 3D tomograms of the tubules."
We have changed this as suggested
L 114: "we developed an unbiased, localisation-based approach". What is the part that was developed here? It seems that the inner layer particle coordinates where simply shifted to get starting points in the outer layer. Developing an approach sounds more substantial than this. Also, it's unclear what is unbiased about this approach. The whole point is that it's biased to certain regions (which is a good thing as it incorporates prior knowledge on the location of the structures).
We have modified the sentence to “To target the sparser outer coat lattice for STA, we used the refined coordinates of the inner coat to locate the outer coat tetrameric vertices”, and explain the approach in detail in the methods.
L 124: "The outer coat vertex was refined to a resolution of approximately ~12 A, revealing unprecedented detail of the molecular interactions between Sec31 molecules (Supplementary Fig 2A)". The map alone does not reveal molecular interactions; the main understanding comes from fitting of X-ray structures to the low-resolution map. Also "unprecedented detail" itself is somewhat problematic as the map of Noble et al (2013) of the Sec31 vertex is also at nominal resolution of 12 A. Furthermore, Supplementary Fig 2A does not reveal this "unprecedented detail", it shows the resolution estimation by FSC. To clarify, these points you could say: "Fitting of the Sec31 atomic model to our reconstruction vertex at 12-A resolution (Supplementary Fig 2A) revealed the molecular interactions between different copies of Sec31 in the membrane-assembled coat.
We have changed the sentence as suggested.
L 150: Can the authors exclude the possibility that the difference is due to differences in data processing? E.g. how the maps amplitudes have been adjusted?
Yes, we can exclude this scenario by measuring distances between vertices in the right and left handed direction. These measurements are only compatible with our vertex arrangement, and cannot be explained by the big deviation from 4-fold symmetry seen in the membrane-less cage vertices.
L 172: "that wrap tubules either in a left- or right-handed manner". Don't they do always both on each tubule? Now this sentence could be interpreted to mean that some tubules have a left-handed coat and some a right-handed coat.
We have changed this sentence to clarify. “Outer coat vertices are connected by Sec13-31 rods that wrap tubules both in a left- and right-handed manner.”
L276: "The difference map" hasn't been introduced earlier but is referred to here as if it has been.
We now introduce the difference map.
L299: Can "Secondary structure predictions" denote a protein region "highly prone to protein binding"?
Yes, this is done through DISOPRED3, a feature include in the PSIPRED server we used for our predictions. The reference is: Jones D.T., Cozzetto D. DISOPRED3: precise disordered region predictions with annotated protein-binding activity Bioinformatics. 2015; 31:857–863. We have now added this reference to the manuscript.
L316: It's true that the detail in the map of the inner coat is unprecedented and the model presented in Figure 7 is partially based on that. But here "unprecedented resolution" sounds strange as this sentence refers to a schematic model and not a map.
We have changed this by moving the reference to Fig 7 to a more appropriate place
L325: "have 'compacted' during evolution" -> remove. It's enough to say it's more compact in humans and less compact in yeast as there could have been different adaptations in different organisms at this interface.
We have changed as requested. See also our response to reviewer 1, point 1.
L327: What's exactly meant by "sequence diversity or variability at this density".
We have now clarified: “Since multiple charge clusters in yeast Sec31 may contribute to this interaction interface (Stancheva et al., 2020), the low resolution could be explained by the fact that the density is an average of different sequences.”
L606-607: The description of this custom data processing approach is difficult to follow. Why is in-plane flip needed and how is it used here?
Initially particles are picked ignoring tube directionality (as this cannot be assessed easily from the tomograms due to the pseudo-twofold symmetry of the Sec23/24/Sar1 trimer). So the in plane rotation of inner coat subunit could be near 0 or 180°. For each tube, both angles are sampled (in-plane flip). Most tubes result in the majority of particles being assigned one of the two orientations (which is then assumed as the tube directionality). Particles that do not conform are removed, and rare tubes where directionality cannot be determined are also removed. We have re-written the description to clarify these points: “Initial alignments were conducted on a tube-by-tube basis using the Dynamo in-plane flip setting to search in-plane rotation angles 180° apart. This allowed to assign directionality to each tube, and particles that were not conforming to it were discarded by using the Dynamo dtgrep_direction command in custom MATLAB scripts”
L627: "Z" here refers to the coordinate system of aligned particles not that of the original tomogram. Perhaps just say "shifted 8 pixels further away from the membrane".
Changed as requested.
L642-643: How can the "left-handed" and "right-handed" rods be separated here? These terms refer to the long-range organisation of the rods in the lattice it's not clear how they were separated in the early alignments.
They are separated by picking only one subset using the dynamo sub-boxing feature. This extracts boxes from the tomogram which are in set positions and orientation relative to the average of previously aligned subtomograms. From the average vertex structure, we sub-box rods at 4 different positions that correspond to the centre of the rods, and the 2-fold symmetric pairs are combined into the same dataset. We have clarified this in the text: “The refined positions of vertices were used to extract two distinct datasets of left and right-handed rods respectively using the dynamo sub-boxing feature.”
Figure 2B. It's difficult to see the difference between dark and light pink colours.
We have changed colours to enhance the difference.
Figure 3C. These panels report the relative frequency of neighbouring vertices at each position; "intensity" does not seem to be the right measure for this. You could say that the colour bar indicates the "relative frequency of neighbouring vertices at each position" and add detail how the values were scaled between 0 and 1. The same applies to SFigure 1E.
Changed as requested.
Figure 4. The COP-II rods themselves are relatively straight, and they are not left-handed or right-handed. Here, more accurate would be "architecture of COPII rods organised in a left-handed manner". (In the text the authors may of course define and then use this shorter expression if they so wish.) Panel 4B top panel could have the title "left-handed" and the lower panel should have the title "right-handed" (for consistency and clarity).
We have now defined left- and right-handed rods in the text, and have changed the figure and panel titles as requested.
We thank the reviewers for their comments, which will improve the quality of our manuscript.
Our study describes a novel approach to the identification of GTPase binding-partners. We recapitulated and augmented previous protein-protein interaction data for RAB18 and presented data validating some of our findings. In aggregate, our dataset suggested that RAB18 modulates the establishment of membrane contact sites and the transfer of lipid between closely apposed membranes.
In the original version of our manuscript, we stated that we were exploring the possibility that RAB18 contributes to cholesterol biosynthesis by mobilizing substrates or products of the Δ8-Δ7 sterol isomerase emopamil binding protein (EBP). While our manuscript was under review, we profiled sterols in wild-type and RAB18-null cells and assayed cholesterol biosynthesis in a panel of cell lines (Figure 1).
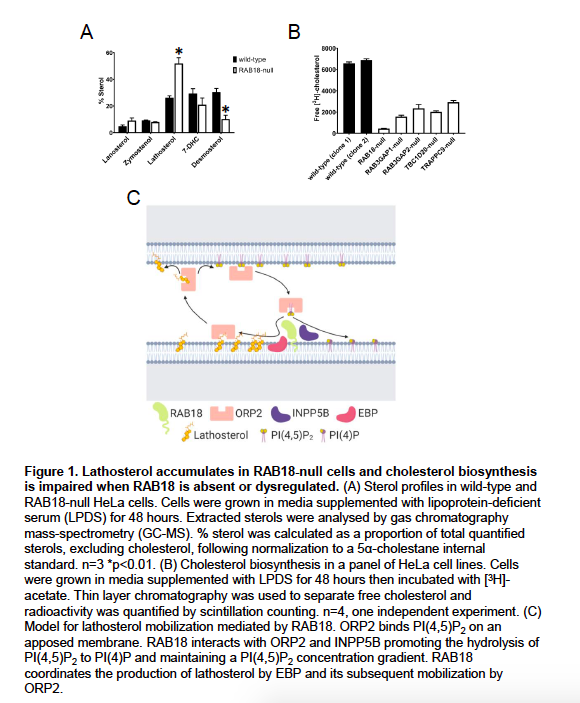
Our new data show that an EBP-product, lathosterol, accumulates in RAB18-null cells (p<0.01). Levels of a downstream cholesterol intermediate, desmosterol, are reduced in these cells (p<0.01) consistent with impaired delivery of substrates to post-EBP biosynthetic enzymes (Figure 1A). Further, our preliminary data suggests that cholesterol biosynthesis is substantially reduced when RAB18 is absent or dysregulated (4 technical replicates, one independent experiment) (Figure 1B).
Because of the clinical overlap between Micro syndrome and cholesterol biosynthesis disorders including Smith-Lemli-Opitz syndrome (SLOS; MIM 270400) and lathosterolosis (MIM 607330), our new findings suggest that impaired cholesterol biosynthesis may partly underlie Warburg Micro syndrome pathology. Therapeutic strategies have been developed for the treatment of SLOS and lathosterolosis, and so confirmation of our findings may spur development of similar strategies for Micro syndrome.
Our new findings provide further functional validation of our methodology and support our interpretation of our protein interaction data.
Reply to point 1)
Tetracycline-induced expression of wild-type and mutant BirA*-RAB18 fusion proteins in the stable HEK293 cell lines was quantified by densitometry (Figure 2).
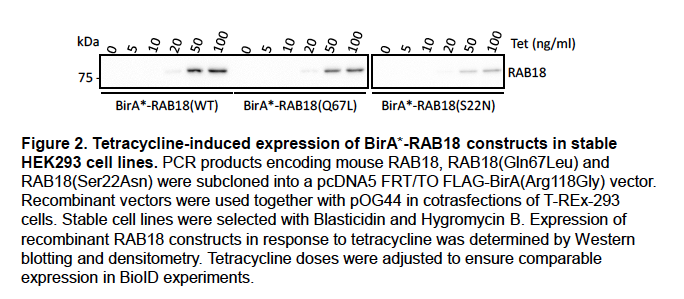
For the HEK293 BioID experiments, tetracycline dosage was adjusted to ensure comparable expression levels. We will include these data in supplemental material in an updated version of our manuscript.
The localization of wild-type and mutant forms of RAB18 in HEK293 cells is somewhat different consistent with previous reports (Ozeki et al. 2005)(Figure 3).
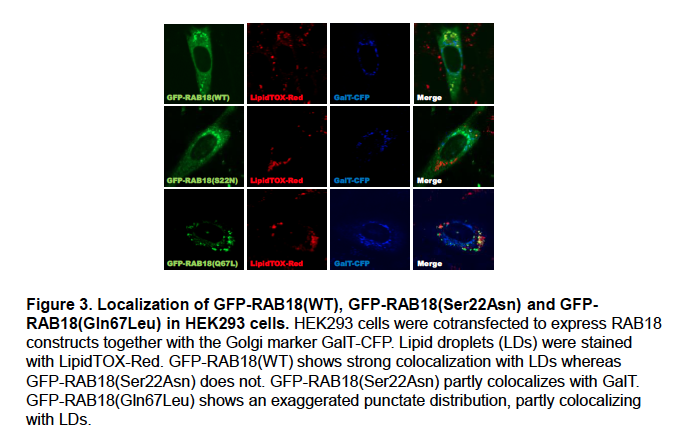
We feel that this may reflect the differential localization of ‘active’ and ‘inactive’ RAB18, with wild-type RAB18 corresponding to a mixture of the two. We will include these data in supplemental material in an updated version of our manuscript.
We acknowledge that the differential localization of wild-type and mutant BirA*-RAB18 might influence the compliment of proteins labeled by these constructs. Nevertheless, we feel that the RAB18(S22N):RAB18(WT) ratios are useful since they distinguish a number of previously-identified RAB18-interactors (manuscript, Figure 1B).
Reply to point 2)
For the HEK293 dataset, spectral counts are provided and for the HeLa dataset LFQ intensities were provided in the manuscript (manuscript, Tables S1 and S2 respectively). However, we did not find that these were useful classifiers for ranking functional interactions when used in isolation.
The extent of labelling produced in a BioID experiment is not wholly determined by the kinetics of protein-protein associations. It is also influenced by, for example, protein abundance, the number and location of exposed surface lysine residues, and protein stability over the timcourse of labelling. We feel that RAB18(S22N):RAB18(WT) and GEF-null:wild-type ratios were helpful in controlling for these factors. Further, that our comparative approach was effective in highlighting known RAB18-interactors and in identifying novel ones.
We acknowledge that our approach may omit some bona fide functional RAB18-interactions, but would argue that our aims were to augment existing functional RAB18-interaction data and avoid false-positives, rather than to emphasise completeness.
Reply to point 3)
We will include representative fluorescence images for the SEC22A, NBAS and ZW10 knockdown experiments in an updated version of our manuscript.
Unfortunately, a suitable antibody for determining knockdown efficiency of SEC22A at the protein level is not commercially available. We will determine SEC22A knockdown efficiency at the mRNA level using qPCR.
Reply to point 4)
Expression levels of wild-type and mutant RAB18 in the stable CHO cell lines generated for this study were determined by Western blotting and found to be comparable (Figure 4).
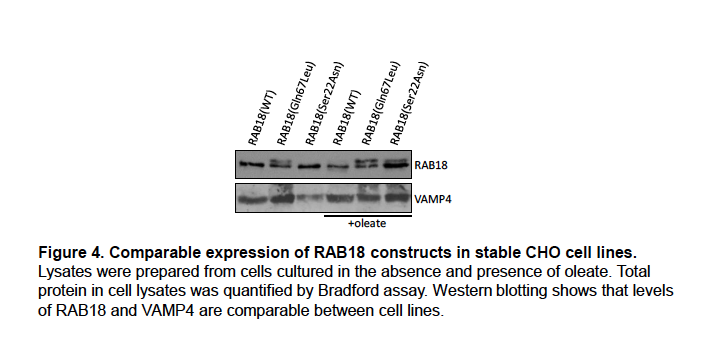
We will include these data in supplemental material in an updated version of our manuscript.
The levels of [14C]-CE were higher in RAB18(Gln67Leu) cells than in the other cell lines following loading with [14C]-oleate for 24 hours. We will amend the text to make this explicit. Our interpretation of the data is that ‘active’ RAB18 facilitates the mobilization of cholesterol. When cells are loaded with oleate, this promotes generation and storage of CE. Conversely, when cells are treated with HDL, it promotes more rapid efflux.
Our new data implicating RAB18 in the mobilization of lathosterol supports our interpretation of our loading and efflux experiments. In the light of our new data showing that de novo cholesterol biosynthesis is impaired when RAB18 is absent or dysregulated, it will be interesting to determine whether cholesterol synthesis is increased in the RAB18(Gln67Leu) cells.
Reply to point 1)
We anticipate that the approach of comparative proximity biotinylation in GEF-null and wild-type cell lines will be broadly useful in small GTPase research.
While RAB18 has previously been implicated in regulating membrane contacts, the identification of SEC22A as a RAB18-interactor adds to the previous model for their assembly.
While ORP2 and INPP5B have previously been shown to mediate cholesterol mobilization, the novel finding that they both interact with RAB18 adds to this work. We argue that RAB18-ORP2-INPP5B functions in an analogous manner to ARF1-OSBP-SAC1 in mediating sterol exchange. The broad Rab-binding specificity of multiple OSBP-homologs, and that of multiple phosphoinositide phosphatase enzymes, suggests that this may be a common conserved relationship.
Our new data indicating that RAB18 coordinates generation of sterol intermediates by EBP and their delivery to post-EBP biosynthetic enzymes reveals a new role for Rab proteins in lipid biogenesis. Most importantly, our new findings that RAB18 deficiency is associated with impaired cholesterol biogenesis suggest that Warburg Micro syndrome is a cholesterol biogenesis disorder. Further, that it may be amenable to therapeutic intervention.
Reply to point 2)
Recognising that the effect of RAB18 on cholesterol esterification and efflux could arise from various causes, we previously carried out Western blotting of the CHO cell lines for ABCA1 to determine whether this protein was involved (Figure 5).
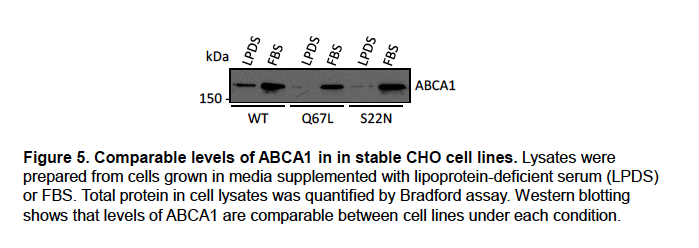
Similar levels of ABCA1 expression in these lines suggests it is not. We will include these data in supplemental material in an updated version of our manuscript.
We feel that our new data implicating RAB18 in lathosterol mobilization provides important insight into its role in cholesterol biogenesis. Further, it supports our previous suggestion that RAB18 mediates cholesterol mobilization.
Reply to point 3)
We agree that the established roles for ORP2, TMEM24/C2CD2L and PIP2 at the plasma membrane make this an extremely interesting area for future research; it is one we are actively investigating. However, we respectfully feel that to comprehensively explore the subcellular locations of RAB18-mediated sterol/PIP2 exchange requires another study and is beyond the scope of the present report.
Reply to point 1)
The RAB18-SPG20 interaction has already been validated with a co-immunoprecipitation experiment (Gillingham et al. 2014). We will update the text of our manuscript to make this more explicit, but do not feel it is necessary to recapitulate this work.
We argue in the manuscript that RAB18 may coordinate the assembly of a non-canonical SNARE complex incorporating SEC22A, STX18, BNIP1 and USE1. However, this role may be mediated through prior interaction with the NBAS-RINT1-ZW10 (NRZ) tethering complex and the SM-protein SCFD2 rather than through a direct interaction. We therefore feel that a RAB18-SEC22A interaction may be difficult to validate by conventional means.
The reciprocal experiments with BioID2(Gly40S)-SEC22A did provide tentative confirmation of the interaction together with evidence that a subset of SEC22A-interactions are attenuated when RAB18 is absent or dysregulated. In the light of our new findings reinforcing a role for RAB18 in sterol mobilization at membrane contact sites, it is interesting that one of these is DHRS7, an enzyme with steroids among its putative substrates.
Reply to point 2)
We previously analysed the localization of the BirA*-RAB18 fusion protein in HeLa cells (Figure 6).
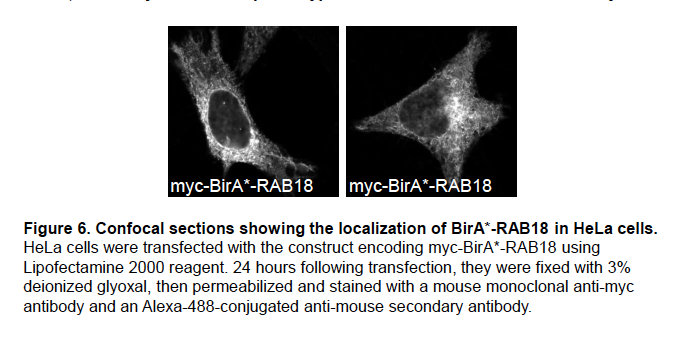
It shows a reticular staining pattern consistent with the reported localization of RAB18 to the ER (Gerondopoulos et al. 2014; Ozeki et al. 2005). We will include these data in supplemental material in an updated version of our manuscript.
Heterologous expression of the BirA*-RAB18 fusion protein in HeLa cells identified the interactions between RAB18 and EBP, ORP2 and INPP5B, for which we now have supportive functional evidence. Since the evidence for impaired lathosterol mobilization and cholesterol biosynthesis was derived from experiments on null-cells, in which endogenous protein expression is absent, we feel that rescue experiments are not necessary in the present study. However, such experiments could be highly useful in future studies.
Reply to point 3)
Our screening approach did use both a RAB3GAP-null:wild-type comparison (manuscript, Figure 2, Table S2) and also a RAB18(S22N):RAB18(WT) comparison (manuscript, Figure 1, Table S1). Differences should be expected between these datasets, since they used different cell lines and slightly different methodologies. Nevertheless, proteins identified in both datasets included the known RAB18 effectors NBAS, RINT1, ZW10 and SCFD2, and the novel potential effectors CAMSAP1 and FAM134B.
There is prior evidence for 12 of the 25 RAB3GAP-dependent RAB18 interactions we identified (manuscript, Figure 2D). Among the 6 lipid modifying/mobilizing proteins found exclusively in our HeLa dataset, we previously presented direct evidence for the interaction of RAB18 with TMCO4. We now also have strong functional evidence for its interaction with EBP, ORP2 and INPP5B.
Reply to point 4)
It has been reported that knockdown of SEC22B does not affect the size distribution of lipid droplets (Xu et al. 2018) Figure 8H). Nevertheless, we will carry out qPCR experiments to determine whether the SEC22A siRNAs used in our study affect SEC22B expression. We have found that exogenous expression of SEC22A can cause cellular toxicity. Rescue experiments would therefore be difficult to perform.
Reply to point 5)
The background fluorescence measured in SPG20-null cells and presented in Figure 4B in the manuscript does not imply that the SPG20 antibody shows significant cross-reactivity. Rather, it reflects the fact that fluorescence intensity is recorded by our Operetta microscope in arbitrary units.
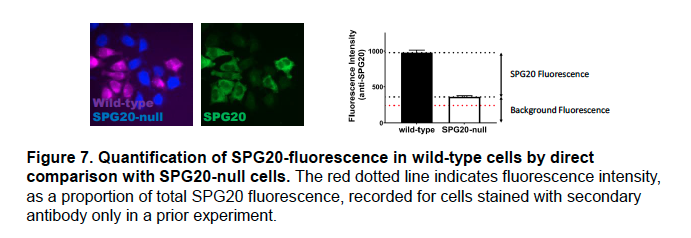
Above (Figure 7), is a version of the panel in which fluorescence from staining cells with only the secondary antibody is included (recorded in a previous experiment and expressed as a proportion of total SPG20 fluorescence in this experiment).
We have found that comparative fluorescence microscopy is more sensitive than immunoblotting. The SPG20 antibody we used to stain the HeLa cells has previously been used in quantitative fluorescence microscopy (Nicholson et al. 2015).
Furthermore, we showed corresponding, significantly reduced, expression of SPG20 in RAB18- and TBC1D20-null RPE1 cells, using quantitative proteomics (manuscript, Table S3).
We acknowledge that quantification of SPG20 transcript levels would clarify the level at which it is downregulated and will carry out qPCR experiments accordingly.
Reply to point 6)
We interpret both the enhanced CE-synthesis following oleate-loading and the rapid efflux upon incubation with HDL (manuscript, Figure 7A) as resulting from increased cholesterol mobilization. Our new data implicating RAB18 in the mobilization of lathosterol support this interpretation.
In the [3H]-cholesterol efflux assay (manuscript, Figure 7B) total [3H]-cholesterol loading at t=0 was 156392±8271 for RAB18(WT) cells, 168425±9103 for RAB18(Gln67Leu) cells and 148867±7609 (cpm determined through scintillation counting). Normalizing to total cellular radioactivity assured that differences in loading between replicates did not skew the results.
The candidate effector likely to directly mediate cholesterol mobilization is ORP2. It has been shown that ORP2 overexpression drives cholesterol to the plasma membrane (Wang et al. 2019). Further, there is evidence for reduced plasma membrane cholesterol in ORP2-null cells (Wang et al. 2019).
We previously carried out Western blotting of the CHO cell lines for ABCA1 to determine whether this protein was involved in altered efflux (Figure 5, above). Similar levels of ABCA1 expression in these lines suggests it is not. We will include these data in supplemental material in an updated version of our manuscript.
References
Gerondopoulos, A., R. N. Bastos, S. Yoshimura, R. Anderson, S. Carpanini, I. Aligianis, M. T. Handley, and F. A. Barr. 2014. 'Rab18 and a Rab18 GEF complex are required for normal ER structure', J Cell Biol, 205: 707-20.
Gillingham, A. K., R. Sinka, I. L. Torres, K. S. Lilley, and S. Munro. 2014. 'Toward a comprehensive map of the effectors of rab GTPases', Dev Cell, 31: 358-73.
Nicholson, J. M., J. C. Macedo, A. J. Mattingly, D. Wangsa, J. Camps, V. Lima, A. M. Gomes, S. Doria, T. Ried, E. Logarinho, and D. Cimini. 2015. 'Chromosome mis-segregation and cytokinesis failure in trisomic human cells', eLife, 4.
Ozeki, S., J. Cheng, K. Tauchi-Sato, N. Hatano, H. Taniguchi, and T. Fujimoto. 2005. 'Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane', J Cell Sci, 118: 2601-11.
Wang, H., Q. Ma, Y. Qi, J. Dong, X. Du, J. Rae, J. Wang, W. F. Wu, A. J. Brown, R. G. Parton, J. W. Wu, and H. Yang. 2019. 'ORP2 Delivers Cholesterol to the Plasma Membrane in Exchange for Phosphatidylinositol 4, 5-Bisphosphate (PI(4,5)P2)', Mol Cell, 73: 458-73 e7.
Xu, D., Y. Li, L. Wu, Y. Li, D. Zhao, J. Yu, T. Huang, C. Ferguson, R. G. Parton, H. Yang, and P. Li. 2018. 'Rab18 promotes lipid droplet (LD) growth by tethering the ER to LDs through SNARE and NRZ interactions', J Cell Biol, 217: 975-95.
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public Review):
Summary:
The study conducted by the Schuldiner's group advances the understanding of mitochondrial biology through the utilization of their bi-genomic (BiG) split-GFP assay, which they had previously developed and reported. This research endeavors to consolidate the catalog of matrix and inner membrane mitochondrial proteins. In their approach, a genetic framework was employed wherein a GFP fragment (GFP1-10) is encoded within the mitochondrial genome. Subsequently, a collection of strains was created, with each strain expressing a distinct protein tagged with the GFP11 fragment. The reconstitution of GFP fluorescence occurs upon the import of the protein under examination into the mitochondria.
We are grateful for the positive evaluation. We would like to clarify that the bi-genomic (BiG) split-GFP assay was developed by the labs of H. Becker and Roza Kucharzyk by highly laborious construction of the strain with mtDNA-encoded GFP<sub>1-10</sub> (Bader et al, 2020).
Strengths:
Notably, this assay was executed under six distinct conditions, facilitating the visualization of approximately 400 mitochondrial proteins. Remarkably, 50 proteins were conclusively assigned to mitochondria for the first time through this methodology. The strains developed and the extensive dataset generated in this study serve as a valuable resource for the comprehensive study of mitochondrial biology. Specifically, it provides a list of 50 "eclipsed" proteins whose role in mitochondria remains to be characterized.
Weaknesses:
The work could include some functional studies of at least one of the newly identified 50 proteins.
In response to this we have expanded the characterization of phenotypic effects resulting from changing the targeting signal and expression levels of the dually localized Gpp1 protein and expanded the data in Fig. 3, panels H and I.
Reviewer #2 (Public Review):
The authors addressed the question of how mitochondrial proteins that are dually localized or only to a minor fraction localized to mitochondria can be visualized on the whole genome scale. For this, they used an established and previously published method called BiG split-GFP, in which GFP strands 1-10 are encoded in the mitochondrial DNA and fused the GFP11 strand C-terminally to the yeast ORFs using the C-SWAT library. The generated library was imaged under different growth and stress conditions and yielded positive mitochondrial localization for approximately 400 proteins. The strength of this method is the detection of proteins that are dually localized with only a minor fraction within mitochondria, which so far has hampered their visualization due to strong fluorescent signals from other cellular localizations. The weakness of this method is that due to the localization of the GFP1-10 in the mitochondrial matrix, only matrix proteins and IM proteins with their C-termini facing the matrix can be detected. Also, proteins that are assembled into multimeric complexes (which will be the case for probably a high number of matrix and inner membrane-localized proteins) resulting in the C-terminal GFP11 being buried are likely not detected as positive hits in this approach. Taking these limitations into consideration, the authors provide a new library that can help in the identification of eclipsed protein distribution within mitochondria, thus further increasing our knowledge of the complete mitochondrial proteome. The approach of global tagging of the yeast genome is the logical consequence after the successful establishment of the BiG split-GFP for mitochondria. The authors also propose that their approach can be applied to investigate the topology of inner membrane proteins, however, for this, the inherent issue remains that it cannot be excluded that even the small GFP11 tag can impact on protein biogenesis and topology. Thus, the approach will not overcome the need to assess protein topology analysis via biochemical approaches on endogenous untagged proteins.
Reviewer #3 (Public Review):
Summary:
Here, Bykov et al move the bi-genomic split-GFP system they previously established to the genomewide level in order to obtain a more comprehensive list of mitochondrial matrix and inner membrane proteins. In this very elegant split-GFP system, the longer GFP fragment, GFP1-10, is encoded in the mitochondrial genome and the shorter one, GFP11, is C-terminally attached to every protein encoded in the genome of yeast Saccharomyces cerevisiae. GFP fluorescence can therefore only be reconstituted if the C-terminus of the protein is present in the mitochondrial matrix, either as part of a soluble protein, a peripheral membrane protein, or an integral inner membrane protein. The system, combined with high-throughput fluorescence microscopy of yeast cells grown under six different conditions, enabled the authors to visualize ca. 400 mitochondrial proteins, 50 of which were not visualised before and 8 of which were not shown to be mitochondrial before. The system appears to be particularly well suited for analysis of dually localized proteins and could potentially be used to study sorting pathways of mitochondrial inner membrane proteins.
Strengths:
Many fluorescence-based genome-wide screens were previously performed in yeast and were central to revealing the subcellular location of a large fraction of yeast proteome. Nonetheless, these screens also showed that tagging with full-length fluorescent proteins (FP) can affect both the function and targeting of proteins. The strength of the system used in the current manuscript is that the shorter tag is beneficial for the detection of a number of proteins whose targeting and/or function is affected by tagging with full-length FPs.
Furthermore, the system used here can nicely detect mitochondrial pools of dually localized proteins. It is especially useful when these pools are minor and their signals are therefore easily masked by the strong signals coming from the major, nonmitochondrial pools of the proteins.
Weaknesses:
My only concern is that the biological significance of the screen performed appears limited. The dataset obtained is largely in agreement with several previous proteomic screens but it is, unfortunately, not more comprehensive than them, rather the opposite. For proteins that were identified inside mitochondria for the first time here or were identified in an unexpected location within the organelle, it remains unclear whether these localizations represent some minor, missorted pools of proteins or are indeed functionally important fractions and/or productive translocation intermediates. The authors also allude to several potential applications of the system but do little to explore any of these directions.
We agree with the reviewer that a single method may not be used for the construction of the complete protein inventory of an organelle or its sub-compartment. We suggest that the value of our assay is in providing a complementary view to the existing data and approaches. For example, we confirm the matrix localization of several proteins that were only found in the two proteomic data and never verified before (Vögtle et al, 2017; Morgenstern et al, 2017). Given that proteomics is a very sensitive technique and false positives are hard to completely exclude, our complementary verification is valuable.
Reviewer #1 (Recommendations for the authors):
In my opinion, the manuscript can be published as it is, and I would expect that future work will advance the functional properties of the newly found mitochondrial proteins.
We thank the reviewer for their positive evaluation
Reviewer #2 (Recommendations for the authors)
(1) Due to the localization of the GFP1-10 in the matrix, only matrix and IM proteins with C-termini facing the matrix can be detected, this should be added e.g. in the heading of the first results part and discussed earlier in the manuscript. In addition, the limitation that assembly into protein complexes will likely preclude detection of matrix and IM proteins needs to be discussed.
To address the first point, we edited the title of the first section to only mention the visualization of the matrix-facing proteome and remove the words “inner membrane”. We also clarified early in the Results section that we only consider the matrix-facing C-termini by extending the sentence early in the results section “To compare our findings with published data, we created a unified list of 395 proteins that are observed with high confidence using our assay indicating that their C-terminus is positioned in the matrix (Fig. 2 – figure supplement 1B-D, Table S1).” (P. 6 Lines 1-3). Concluding the comparison with the earlier proteomic studies we also added the sentence “Many proteins are missing because their C-termini are facing the IMS” (P.8 Line 2).
To address the second point concerning the possible interference of the complex assembly and protein detection by our assay, we conducted an additional analysis. The analysis takes advantage of the protein complexes with known structures where we could estimate if the C-terminus with the GFP<sub>11</sub> tag would be available for GFP1-10 binding. We added the additional figure (Figure 3 – figure supplement 2) and following text in the Results section (P.7 Lines 22-34):
“To examine the influence of protein complex assembly on the performance of the BiG Mito-Split assay we analyzed the published structures of the mitoribosome and ATP synthase (Desai et al, 2017; Srivastava et al, 2018; Guo et al, 2017) and classified all proteins as either having C-termini in, or out of, the complex. There was no difference between the “in” and “out” groups in the percentage observed in the BiG Mito-Split collection (Fig. 3 – figure supplement 2A) suggesting that the majority of the GFP11tagged proteins have a chance to interact with GFP1-10 before (or instead of) assembling into the complex. PCR and western blot verification of eight strains with the tagged complex subunits for which we observed no signal showed that mitoribosomal proteins were incorrectly tagged or not expressed, and the ATP synthase subunits Atp7, Atp19, and Atp20 were expressed (Fig. 3 – Supplement 2B). Atp19 and Atp20 have their C-termini most likely oriented towards the IMS (Guo et al, 2017) while Atp7 is completely in the matrix and may be the one example of a subunit whose assembly into a complex prevents its detection by the BiG Mito-Split assay.”
We also consider related points on the interference of the tag and the influence of protein essentiality in the replies to points 3) and 12) of these reviews.
(2) The imaging data is of high quality, but the manuscript would greatly benefit from additional analysis to support the claims or hypothesis brought forward by the authors. The idea that the nonmitochondrial proteins are imported due to their high sequence similarity to MTS could be easily addressed at least for some of these proteins via import studies, as also suggested by the authors.
The idea that non-mitochondrial proteins may be imported into mitochondria due to occasional sequence similarity was recently demonstrated experimentally by (Oborská-Oplová et al, 2025). We incorporate this information in the Discussion section as follows (P. 14 Lines 10-16):
“It was also recently shown that the r-protein uS5 (encoded by RPS2 in yeast) has a latent MTS that is masked by a special mitochondrial avoidance segment (MAS) preceding it (Oborská-Oplová et al, 2025). The removal of the MAS leads to import of uS5 into mitochondria killing the cells. The case of uS5 is an example of occasional similarity between an r-protein and an MTS caused by similar requirements of positive charges for rRNA binding and mitochondrial import. It remains unclear if other r-proteins have a MAS and if there are other mechanisms that protect mitochondria from translocation of cytosolic proteins.”
We also conducted additional analysis to substantiate the claim that ribosomal (r)-proteins are similar in their physico-chemical properties to MTS-containing mitochondrial proteins. For this we chose not to use prediction algorithms like TartgetP and MitoFates that were already trained on the same dataset of yeast proteins to discriminate cytosolic and mitochondrial localization. Instead, we extended the analysis earlier made by (Woellhaf et al, 2014) and calculated several different properties such as charge, hydrophobicity, hydrophobic moment and amino acid content for mitochondrial MTS-containing proteins, cytosolic non-ribosomal proteins, and r-proteins. The analysis showed striking similarity of r-proteins and mitochondrial proteins. We incorporate a new Figure 3 – figure supplement 3 and the following text in the Results section (P. 8 Lines14-22):
“Five out of eight proteins are components of the cytosolic ribosome (r-proteins). In agreement with previous reports (Woellhaf et al, 2014) we find that their unique properties, such as charge, hydrophobicity and amino acid content, are indeed more similar to mitochondrial proteins than to cytosolic ones (Fig. 3 – figure supplement 3). Additional experiments with heterologous protein expression and in vitro import will be required to confirm the mitochondrial import and targeting mechanisms of these eight non-mitochondrial proteins. The data highlights that out of hundreds of very abundant proteins with high prediction scores only few are actually imported and highlights the importance of the mechanisms that help to avoid translocation of wrong proteins (Oborská-Oplová et al, 2025).”
To further prove the possibility of r-protein import into mitochondria we aimed to clone the r-proteins identified in this work for cell-free expression and import into purified mitochondria. Despite the large effort, we have succeeded in cloning and efficiently expressing only Rpl23a (Author response image 1 A). Rpl23a indeed forms proteinase-protected fractions in a membrane potential-dependent manner when incubated with mitochondria. The inverse import dynamics of Rpl23a could be either indicative of quick degradation inside mitochondria or of background signal during the import experiments (Author response image 1.A). To address the r-protein degradation possibility, we measured how does GFP signal change in the BiG Mito-Split diploid collection strains after blocking cytosolic translation with cycloheximide (CHX). For this we selected Mrpl12a, that had one of the highest signals. We did not detect any drop in fluorescence signal for Rpl12a and the control protein Mrpl6 (Author response image 1 B). This might indicate the lack of degradation, or the degradation of the whole protein except GFP<sub>11</sub> that remains connected to GFP<sub>1-10</sub>. Due to time constrains we could not perform all experiments for the whole set of potentially imported r-proteins. Since more experiments are required to clearly show the mechanisms of mitochondrial r-protein import, degradation, and toxicity, or possible moonlighting functions (such as import into mitochondria derived from pim1∆ strain, degradation assays, fractionations, and analyses with antibodies for native proteins) we decided not to include this new data into the manuscript itself.
Author response image 1.
The import of r-proteins into mitochondria and their stability. (A) Rpl23 was synthesized in vitro (Input), radiolabeled, and imported into mitochondria isolated from BY4741 strain as described before (Peleh et al, 2015); the import was performed for 5,10, or 15 minutes and mitochondria were treated with proteinase K (PK) to degrade nonimported proteins; some reactions were treated with the mix of valinomycin, antimycin, and oligomycin (VAO) to dissipate mitochondrial membrane potential; the proteins were visualized by SDS-PAGE and autoradiography (B) The strains from the diploid BiG Mito-Split collection were grown in YPD to mid-logarithmic growth phase, then CHX was added to block translation and cell aliquots were taken from the culture and analyzed by fluorescence microscopy at the indicated time points. Scale bar is 5 µm.
(3) The claim that the approach can be used to assess the topology of inner membrane proteins is problematic as the C-terminal tag can alter the biogenesis pathway of the protein or impact on the translocation dynamics (in particular as the imaging method applied here does not allow for analysis of dynamics). The hypothesis that the biogenesis route can be monitored is therefore far-reaching. To strengthen the hypothesis the authors should assess if the C-terminal GFP11 influences protein solubility by assessing protein aggregation of e.g. Rip1.
We agree with the reviewer that the tag and assembly of GFP<sub>1-10/11</sub> can further complicate the assessment of topology of the IM proteins that already have complex biogenesis routes (lateral transfer, conservative, and a Rip1-specific Bcs1 pathway). To emphasize that the assessment of the steady state topology needs to be backed up by additional biochemical approaches, we edited the beginning of the corresponding Results sections as follows (P. 11 Lines 2-6):
“Studying membrane protein biogenesis requires an accurate way to determine topology in vivo. The mitochondrial IM is one of the most protein-rich membranes in the cell supporting a wide variety of TMD topologies with complex biogenesis pathways. We aimed to find out if our BiG Mito-Split collection can accurately visualize the steady-state localization of membrane protein C-termini protruding into the matrix or trap protein transport intermediates” (inserted text is underlined).
The collection that we studied by microscopy is diploid and contains one WT copy of each 3xGFP<sub>11</sub>tagged gene. To assess the influence of the tag on the protein function we performed growth assays with haploid strains which have one 3xGFP<sub>11</sub>-tagged gene copy and no GFP<sub>1-10</sub>. We find that Rip13xGFP<sub>11</sub> displays slower growth on glycerol at 30˚C and even slower at 37˚C while tagged Qcr8, Qcr9, and Qcr10 grow normally (Author response image 2 A). Based on the growth assays and microscopy it is not possible to conclude whether the “Qcr” proteins’ biogenesis is affected by the tag. It may be that laterally sorted proteins are functional with the tag and constitute the majority while only a small portion is translocated into the matrix, trapped and visualized with GFP<sub>1-10</sub>. In case of Rip1 it was shown that C-terminal tag can affect its interaction with the chaperone Mzm1 and promote Rip1 aggregation (Cui et al, 2012). The extent of Rip1 function disruption can be different and depends on the tag. We hypothesize that our split-assay may trap the pre-translocation intermediate of Rip1 and can be helpful to study its interactors. To test this, we performed anti-GFP immune-precipitation (IP) using GFP-Trap beads (Author response image 2 B).
Author response image 2.
The influence of 3x-GFP11 on the function and processing of the inner membrane proteins. (A) Drop dilution assays with haploid strains from C-SWAT 3xGFP<Sub>11</sub> library on fermentative (YPD) and respiratory (YPGlycerol) media at different temperatures. (B) Immuno-precipitation with GFP-Trap agarose was performed on haploid strain that has only Rip1-3xGFP<sub>11</sub> and on the diploid strain derived from this haploid mated with BiG Mito-Split strain containing mtGFP<sub>1-10</sub> and WT untagged Rip1 using the lysis (1% TX-100) and washing protocols provided by the manufacturer; the total (T) and eluted with the Laemmli buffer (IP) samples were analyzed by immunoblotting with polyclonal rabbit antibodies against GFP (only visualizes GFP<Sub>11</sub> in these samples) and Rip1 (visualizes both tagged and WT Rip1). Polyclonal home-made rabbit antisera for GFP and Rip1 were kindly provided by Johannes Herrmann (Kaiserslautern) and Thomas Becker (Bonn); the antisera were diluted 1:500 for decorating the membranes.
We find that the haploid strain with Rip1-3xGFP<sub>11</sub> contains not only mature (m) and intermediate (i) forms but also an additional higher Mw band that we interpreted as precursor that was not cleaved by MPP. WT Rip1 in the diploid added two more lower Mw bands: (m) and (i) forms of the untagged Rip1. IP successfully enriched GFP<sub>1-10</sub> fragment as visualized by anti-GFP staining. Interestingly only the highest Mw Rip1-3xGFP<sub>11</sub> band was also enriched when anti-Rip1 antibodies were used to analyze the samples. This suggests that Rip1 precursor gets completely imported and interacts with GFP<sub>1-10</sub> and can be pulled down. It is however not processed. Processed Rip1 is not interacting with GFP<sub>1-10</sub>. Based on the literature we expect all Rip1 in the matrix to be cleaved by MPP including the one interacting with GFP. Due to this discrepancy, we did not include this data in the manuscript. This is however clear that the assay may be useful to analyze biogenesis intermediates of the IM and matrix proteins. To emphasize this, we added information on the C-terminal tagging of Rip1 in the Results section (P. 11 Lines 18-20):
“It was shown that a C-terminal tag on Rip1 can prevent its interaction with the chaperone Mzm1 and promote aggregation in the matrix (Cui et al, 2012). It is also possible that our assay visualizes this trapped biogenesis intermediate.”
We also added a note on biogenesis intermediates in the Discussion (P. 14 Line 36 onwards):
“It is possible that the proteins with C-termini that are translocated into the IMS from the matrix side can be trapped by the interaction with GFP<sub>1-10</sub>. In that case, our assay can be a useful tool to study these pre-translocation intermediates.”
(4) The hypothesis that the method can reveal new substrates for Bcs1 is interesting, and it would strongly increase the relevance for the scientific community if this would be directly tested, e.g. by deleting BCS1 and testing if more IM proteins are then detected by interaction with the matrix GFP110.
we attempted to move the BiG Mito-Split assay into haploid strains where BCS1 and other factors can be deleted, however, this was not successful. Since this was a big effort (We cloned 10 potential substrate proteins but none of them were expressed) we decided not to pursue this further.
(5) The screening of six different growth conditions reflects the strength of the high-throughput imaging readout. However, the interpretation of the data and additional follow-up on this is rather short and would be a nice addition to the present manuscript. In addition, one wonders, what was the rationale behind these six conditions (e.g. DTT treatment)? The direct metabolic shift from fermentation to respiration to boost mitochondrial biogenesis would be a highly interesting condition and the authors should consider adding this in the present manuscript.
we agree with the reviewer that the analysis of different conditions is a strength of this work. However, we did not reveal any clear protein groups with strong conditional import and thus it was hard to select a follow-up candidate. The selection of conditions was partially driven by the technical possibilities: the media change is challenging on the robotic system; heat shock conditions make microscope autofocus unstable; library strain growth on synthetic respiratory media is very slow and the media cannot be substituted with rich media due to its autofluorescence. However, the usage of the spinning disc confocal microscope allowed us to screen directly in synthetic oleate media which has a lot of background on widefield systems due to oil micelles. We extended the explanation of condition choice as follows (P. 4 Line 34 onwards):
“The diploid BiG Mito-Split collection was imaged in six conditions representing various carbon sources and a diversity of stressors the cells can adapt to: logarithmic growth on glucose as a control carbon source and oleic acid as a poorly studied carbon source; post-diauxic (stationary) phase after growth on glucose where mitochondria, are more active and inorganic phosphate (Pi) depletion that was recently described to enhance mitochondrial membrane potential (Ouyang et al, 2024); as stress conditions we chose growth on glucose in the presence of 1 mM dithiothreitol (DTT) that might interfere with the disulfide relay system in the IMS, and nitrogen starvation as a condition that may boost biosynthetic functions of mitochondria. DTT and nitrogen starvation were earlier used for a screen with the regular C’-GFP collection (Breker et al, 2013). Another important consideration for selecting the conditions was the technical feasibility to implement them on automated screening setups.”
Reviewer #3 (Recommendations for the authors )
(6) This is a very elegant and clearly written study. As mentioned above, my only concern is that the biological significance of the obtained data, at this stage, is rather limited. It would have been nice if the authors explored one of the potential applications of the system they propose. For example, it should be relatively easy to analyze whether Cox26, Qcr8, Qcr9, or Qcr10 are new substrates of Bsc1, as the authors speculate.
we thank the reviewer for their positive feedback. We addressed the biological application of the screen by including new data on metabolite concentrations in the strains where Gpp1 N-terminus was mutated leading to loss of the mitochondrial form. We added panels H and I to Figure 4, the new Supplementary Table S2 and appended the description of these results at the end of the third Results subsection (P. 10 Lines 19-35). Our data now show a role for the mitochondrial fraction of Gpp1 which adds mechanistic insight into this dually localized protein.
We also were interested in the applications of our system to the study of mitochondrial import. However, the study of Cox26, Qcr8, Qcr9, and Qcr10 was not successful (also related to point 4, Reviewer #2). We thus decided to investigate the import mechanisms of the poorly studied dually localized proteins Arc1, Fol3, and Hom6 (related to Figure 4 of the original manuscript). To this end, we expressed these proteins in vitro, radiolabeled, and performed import assays with purified mitochondria. Arc1 was not imported, Fol3 and Hom6 gave inconclusive results (Author response image 3). Since it is known that even some genuine fully or dually localized mitochondrial proteins such as Fum1 cannot be imported in vitro post-translationally (Knox et al, 1998), we cannot draw conclusions from these experiments and left them out of the revised manuscript. Additional investigation is required to clarify if there exist special cytosolic mechanisms for the import of these proteins that were not reconstituted in vitro such as co-translational import.
Author response image 3.
In vitro import of poorly studies dually localized proteins. Arc1, Fol3, and Hom6 were cloned into pGEM4 plasmid, synthesized in vitro (Input), radiolabeled, and imported into mitochondria isolated from BY4741 strain as described before (Peleh et al, 2015); the import was performed for 5,10, or 15 minutes and mitochondria were treated with proteinase K (PK) to degrade non-imported proteins; some reactions were treated with the mix of valinomycin, antimycin, and oligomycin (VAO) to dissipate mitochondrial membrane potential. The proteins were separated by SDS-PAGE and visualized by autoradiography.
Minor comments:
(7) It is unclear why the authors used the six growth conditions they used, and why for example a nonfermentable medium was not included at all.
we address this shortcoming in the reply to the previous point 5 (Reviewer #2).
(8) Page 2, line 17 - "Its" should be corrected to "its".
Changed
(9) Page 2, line 25 to the end of the paragraph - the authors refer to the TIM complex when actually the TIM23 complex is probably meant. Also, it would be clearer if the TIM22 complex was introduced as well, especially in the context of the sentence stating that "the IM is a major protein delivery destination in mitochondria".
This was corrected.
(10) Page 5, line 35 - "who´s" should be corrected to "whose".
This was corrected.
(11) Page 9, line 5 - "," after Gpp1 should probably be "and".
This was corrected.
(12) Page 11 - the authors discuss in several places the possible effects of tags and how they may interfere with "expression, stability and targeting of proteins". Protein function may also be dramatically affected by tags - a quick look into the dataset shows that several mitochondrial matrix and inner membrane proteins that are essential for cell viability were not identified in the screen, likely because their function is impaired.
we agree with the reviewer that the influence of tags needs to be carefully evaluated. This is not always possible in the context of whole genomic screens. Sometimes, yeast collections (and proteomic datasets) can miss well-known mitochondrial residents without a clear reason. To address this important point we conducted an additional analysis to look specifically at the essential proteins. We indeed found that several of the mitochondrial proteins that are essential for viability were absent from the collection at the start, but for those present, their essentiality did not impact the likelihood to be detected in our assay. To describe the analysis we added the following text and a Fig. 3 – figure supplement 2. Results now read (P.7 Lines 8-21):
“Next, we checked the two categories of proteins likely to give biased results in high-throughput screens of tagged collections: proteins essential for viability, and molecular complex subunits. To look at the first category we split the proteomic dataset of soluble matrix proteins (Vögtle et al. 2017) into essential and non-essential ones according to the annotations in the Saccharomyces Genome Database (SGD) (Wong et al, 2023). We found that there was no significant difference in the proportion of detected proteins in both groups (17 and 20 % accordingly), despite essential proteins being less represented in the initial library (Fig. 3 – figure supplement 2A). From the three essential proteins of the (Vögtle et al. 2017) dataset for which the strains present in our library but showed no signal, two were nucleoporins Nup57 and Nup116, and one was a genuine mitochondrial protein Ssc1. Polymerase chain reaction (PCR) and western blot verification showed that the Ssc1 strain was incorrect (Fig. 3 – figure supplement 2B). We conclude that essential proteins are more likely to be absent or improperly tagged in the original C’-SWAT collection, but the essentiality does not affect the results of the BiG Mito-Split assay.”
Discussion (P. 13 Lines 23-26):
“We did not find that protein complex components or essential proteins are more likely to be falsenegatives. However, some essential proteins were absent from the collection to start with (Fig. 3 – figure supplement 2A). Thus, a small tag allows visualization of even complex proteins.”
From our data it is difficult to estimate the effect of tagging on protein function. We also addressed the effect of tagging Rip1 as well as performed growth assays on the tagged small “Qcr proteins” in the reply to point 3 (Reviewer #2). It is also difficult to estimate the effect of GFP<sub>1-10</sub> and <sub>11</sub> complex assembly on protein function since the presence of functional, unassembled GFP<sub>11</sub> tagged pool cannot be ruled out in our assay.
Other changes
Figure and table numbers changed after new data additions.
A sentence added in the abstract to highlight the additional experiments on Gpp1 function: “We use structure-function analysis to characterize the dually localized protein Gpp1, revealing an upstream start codon that generates a mitochondrial targeting signal and explore its unique function.”
The reference to the PCR verification (Fig. 3 – Supplement 2B) of correct tagging of Ycr102c was added to the Results section (P.8 Line 6), western blot verification added on.
Added the Key Resources Table at the beginning of the Methods section.
Small grammar edits, see tracked changes.
References:
Bader G, Enkler L, Araiso Y, Hemmerle M, Binko K, Baranowska E, De Craene J-O, Ruer-Laventie J, Pieters J, Tribouillard-Tanvier D, et al (2020) Assigning mitochondrial localization of dual localized proteins using a yeast Bi-Genomic Mitochondrial-Split-GFP. eLife 9: e56649
Cui T-Z, Smith PM, Fox JL, Khalimonchuk O & Winge DR (2012) Late-Stage Maturation of the Rieske Fe/S Protein: Mzm1 Stabilizes Rip1 but Does Not Facilitate Its Translocation by the AAA ATPase Bcs1. Mol Cell Biol 32: 4400–4409
Desai N, Brown A, Amunts A & Ramakrishnan V (2017) The structure of the yeast mitochondrial ribosome. Science 355: 528–531
Guo H, Bueler SA & Rubinstein JL (2017) Atomic model for the dimeric FO region of mitochondrial ATP synthase. Science 358: 936–940
Knox C, Sass E, Neupert W & Pines O (1998) Import into Mitochondria, Folding and Retrograde Movement of Fumarase in Yeast. J Biol Chem 273: 25587–25593
Morgenstern M, Stiller SB, Lübbert P, Peikert CD, Dannenmaier S, Drepper F, Weill U, Höß P, Feuerstein R, Gebert M, et al (2017) Definition of a High-Confidence Mitochondrial Proteome at Quantitative Scale. Cell Rep 19: 2836–2852
Oborská-Oplová M, Geiger AG, Michel E, Klingauf-Nerurkar P, Dennerlein S, Bykov YS, Amodeo S, Schneider A, Schuldiner M, Rehling P, et al (2025) An avoidance segment resolves a lethal nuclear–mitochondrial targeting conflict during ribosome assembly. Nat Cell Biol 27: 336–346
Peleh V, Ramesh A & Herrmann JM (2015) Import of Proteins into Isolated Yeast Mitochondria. In Membrane Trafficking: Second Edition, Tang BL (ed) pp 37–50. New York, NY: Springer
Srivastava AP, Luo M, Zhou W, Symersky J, Bai D, Chambers MG, Faraldo-Gómez JD, Liao M & Mueller DM (2018) High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane. Science 360: eaas9699
Vögtle F-N, Burkhart JM, Gonczarowska-Jorge H, Kücükköse C, Taskin AA, Kopczynski D, Ahrends R, Mossmann D, Sickmann A, Zahedi RP, et al (2017) Landscape of submitochondrial protein distribution. Nat Commun 8: 290
Woellhaf MW, Hansen KG, Garth C & Herrmann JM (2014) Import of ribosomal proteins into yeast mitochondria. Biochem Cell Biol 92: 489–498
Author Response:
Reviewer #1:
This is a very interesting study that examines the neural processes underlying age-related changes in the ability to prioritize memory for value information. The behavioral results show that older subjects are better able to learn which information is valuable (i.e., more frequently presented) and are better at using value to prioritize memory. Importantly, prioritizing memory for high-value items is accompanied by stronger neural responses in the lateral PFC, and these responses mediate the effects of age on memory.
Strengths of this paper are the large sample size and the clever learning tasks. The results provide interesting insights into potential neurodevelopmental changes underlying the prioritization of memory.
There are also a few weaknesses:
First, the effects of age on repetition suppression in the parahippocampal cortex are relatively modest. It is not clear why repetition suppression effects should only be estimated using the first and last but not all presentations. The consideration of linear and quadratic effects of repetition number could provide a more reliable estimate and provide insights into age-related differences in the dynamics of frequency learning across multiple repetitions.
Thank you for this helpful suggestion. As recommended, we have now computed neural activation within our parahippocampal region of interest not just for the first and last appearance of each item during frequency learning, but for all appearances. Specifically we extended our repetition suppression analysis described in the manuscript to include all image repetitions (p. 36 - 37). Our new methods description reads:
“For each stimulus in the high-frequency condition, we examined repetition suppression by measuring activation within a parahippocampal ROI during the presentation of each item during frequency-learning. We defined our ROI by taking the peak voxel (x = 30, y = -39, z = -15) from the group-level first > last item appearance contrast for high-frequency items during frequency-learning and drawing a 5 mm sphere around it. This voxel was located in the right parahippocampal cortex, though we observed widespread and largely symmetric activation in bilateral parahippocampal cortex. To encompass both left and right parahippocampal cortex within our ROI, we mirrored the peak voxel sphere. For each participant, we modeled the neural response to each appearance of each item using the Least Squares-Separate approach (Mumford et al., 2014). Each first-level model included a regressor for the trial of interest, as well as separate regressors for the onsets of all other items, grouped by repetition number (e.g., a regressor for item onsets on their first appearance, a regressor for item onsets on their second appearance, etc.). Values that fell outside five standard deviations from the mean level of neural activation across all subjects and repetitions were excluded from subsequent analyses (18 out of 10,320 values; .01% of observations). In addition to examining neural activation as a function of stimulus repetition, we also computed an index of repetition suppression for each high-frequency item by computing the difference in mean beta values within our ROI on its first and last appearance.”
As suggested, we ran a mixed effects model examining the influence of linear and quadratic age and linear and quadratic repetition number on neural activation. In line with our whole-brain analysis, we observed a robust effect of linear and quadratic repetition number, suggesting that neural activation decreased non-linearly across stimulus repetitions. In addition, we observed significant interactions between our age and repetition number terms, suggesting that repetition suppression increased into early adulthood. Thus, although the relation we observed between age and repetition suppression is modest, the results from our new analyses suggest it is robust. Because these results largely aligned with the pattern of age-related change we observed in our analysis of repetition suppression indices, we continued to use that compressed metric in subsequent analyses looking at relations with behavior. However, we have updated our results section to include the full analysis taking into account all item repetitions, as suggested. Our updated manuscript now reads (p. 9):
“We next examined whether repetition suppression in the parahippocampal cortex changed with age. We defined a parahippocampal region of interest (ROI) by drawing a 5mm sphere around the peak voxel from the group-level first > last appearance contrast (x = 30, y = -39, z = -15), and mirrored it to encompass both right and left parahippocampal cortex (Figure 2C). For each participant, we modeled the neural response to each appearance of each high-frequency item. We then examined how neural activation changed as a function of repetition number and age. To account for non-linear effects of repetition number, we included linear and quadratic repetition number terms. In line with our whole-brain analysis, we observed a main effect of repetition number, F(1, 5016.0) = 30.64, p < .001, indicating that neural activation within the parahippocampal ROI decreased across repetitions. Further, we observed a main effect of quadratic repetition number, F(1, 9881.0) = 7.47, p = .006, indicating that the reduction in neural activity was greatest across earlier repetitions (Fig 3A). Importantly, the influence of repetition number on neural activation varied with both linear age, F(1, 7267.5) = 7.2, p = .007 and quadratic age , F(1, 7260.8) = 6.9, p = .009. Finally, we also observed interactions between quadratic repetition number and both linear and quadratic age (ps < .026). These age-related differences suggest that repetition suppression was greatest in adulthood, with the steepest increases occurring from late adolescence to early adulthood (Figure 3).”
"For each participant for each item, we also computed a “repetition suppression index” by taking the difference in mean beta values within our ROI on each item’s first and last appearance (Ward et al., 2013). These indices demonstrated a similar pattern of age- related variance — we found that the reduction of neural activity from the first to last appearance of the items varied positively with linear age, F(1, 78.32) = 3.97, p = .05, and negatively with quadratic age, F(1, 77.55) = 4.8, p = .031 (Figure 3B). Taken together, our behavioral and neural results suggest that sensitivity to the repetition of items in the environment was prevalent from childhood to adulthood but increased with age.”
In addition, in the main text on p. 10, we have now included the suggested scatter plot (see new Fig. 3B, below) as well as a modified version of our previous figure S2 to show neural activation across all repetitions in the parahippocampal cortex (see new Fig 3A). We thank the reviewer for this helpful suggestion, as we believe these new figures much more clearly illustrate the repetition suppression effects we observed during frequency learning.
Fig 3. (A) Neural activation within a bilateral parahippocampal cortex ROI decreased across stimulus repetitions both linearly, F(1, 5015.9) = 30.64, p < .001, and quadratically, F(1, 9881.0) = 7.47, p = .006. Repetition suppression increased with linear age, F(1, 7267.5) = 7.2, p = .007, and quadratic age F(1, 7260.8) = 6.9, p = .009. The horizontal black lines indicate median neural activation values. The lower and upper edges of the boxes indicate the first and third quartiles of the grouped data, and the vertical lines extend to the smallest value no further than 1.5 times the interquartile range. Grey dots indicate data points outside those values. (B) The decrease in neural activation in the bilateral PHC ROI from the first to fifth repetition of each item also increased with both linear age, F(1, 78.32) = 3.97, p = .05, and quadratic age, F(1, 77.55) = 4.8, p = .031.
Second, the behavioral data show effects of age on both initial frequency learning and the effects of item frequency on memory. It is not clear whether the behavioral findings reflect the effects of age on the ability to use value information to prioritize memory or simply better initial learning of value-related information on older subjects.
Thank you for raising this important point. Indeed, one of our main findings is that older participants are better both at learning the structure of their environments and also at using structured knowledge to strategically prioritize memory. In our original manuscript, we described results of a model that included participants’ explicit frequency reports as a predictor of memory. Model comparison revealed that participants’ frequency reports — which we interpret as reflecting their beliefs about the structure of the environment — predicted memory more strongly than the item’s true frequency. In other words, participants’ beliefs about the structure of the environment (even if incorrect) more strongly influenced their memory encoding than the true structure of the environment. Critically, however, frequency reports interacted with age to predict memory (Fig 8). Even when we accounted for age-related differences in knowledge of the structure of the environment, older participants demonstrated a stronger influence of frequency on memory, suggesting they were better able to use their beliefs to control subsequent associative encoding. We have now clarified our interpretation of this model in our discussion on p. 23:
“Importantly, though we observed age-related differences in participants’ learning of the structure of their environment, the strengthening of the relation between frequency reports and associative memory with increasing age suggests that age differences in learning cannot fully account for age differences in value-guided memory. Even when accounting for individual differences in participants’ explicit knowledge of the structure of the environment, older participants demonstrated a stronger relation between their beliefs about item frequency and associative memory, suggesting that they used their beliefs to guide memory to a greater degree than younger participants.”
As noted by the reviewer, however, our initial memory analysis did not account for age-related differences in participants’ initial, online learning of item frequency, and our neural analyses further did not account for age differences in explicit frequency reports. We have now run additional control analyses to account for the potential influence of individual differences in frequency learning on associative memory. Specifically, for each participant, we computed three metrics: 1.) their overall accuracy during frequency-learning, 2.) their overall accuracy for the last presentation of each item during frequency-learning (as suggested by Reviewer 2), and 3.) the mean magnitude of the error in their frequency reports. We then included these metrics as covariates in our memory analyses.
When we include these control variables in our model, we continue to observe a robust effect of frequency condition (p < .001) as well as robust interactions between frequency condition and linear and quadratic age (ps < .003) on associative memory accuracy. We also observed a main effect of frequency error magnitude on memory accuracy (p < .001). Here, however, we no longer observe main effects of age or quadratic age on overall memory accuracy. Given the relation we observed between frequency error magnitudes and age, the results from this model suggests that there may be age-related improvements in overall memory that influence both memory for associations as well as learning of and memory for item frequencies. The fact that age no longer relates to overall memory when controlling for frequency error magnitudes suggest that age-related variance in memory for item frequencies and memory for associations are strongly related within individuals. Importantly, however, age-related variance in memory for item frequencies did not explain age-related variance in the influence of frequency condition on associative memory, suggesting that there are developmental differences in the use of knowledge of environmental structure to prioritize valuable information in memory that persist even when controlling for age-related differences in initial learning of environmental regularities. Given the importance of this analysis in elucidating the relation between the learning of environmental structure and value-guided memory, we have now updated the results in the main text of our manuscript to include them. Specifically, on p. 13, we now write:
“Because we observed age-related differences in participants’ online learning of item frequencies and in their explicit frequency reports, we further examined whether these age differences in initial learning could account for the age differences we observed in associative memory. To do so, we ran an additional model in which we included each participant’s mean frequency learning accuracy, mean frequency learning accuracy on the last repetition of each item, and explicit report error magnitude as covariates. Here, explicit report error magnitude predicted overall memory performance, χ2(1) =13.05, p < .001, and we did not observe main effects of age or quadratic age on memory performance (ps > .20). However, we continued to observe a main effect of frequency condition, χ2(1) = 19.65 p < .001, as well as significant interactions between frequency condition and both linear age χ2(1) = 10.59, p = .001, and quadratic age χ2(1) = 9.15, p = .002. Thus, while age differences in initial learning related to overall memory performance, they did not account for age differences in the use of environmental regularities to strategically prioritize memory for valuable information.”
In addition, as suggested by the reviewer, we also included the three covariates as control variables in our mediation analysis. When controlling for online frequency learning and explicit frequency report errors, PFC activity continued to mediate the relation between age and memory difference scores. We have now included these results on p. 16 - 17 of the main text:
“Further, when we included quadratic age, WASI scores, online frequency learning accuracy, online frequency learning accuracy on the final repetition of each item, and mean explicit frequency report error magnitudes as control variables in the mediation analysis, PFC activation continued to mediate the relation between linear age and memory difference scores (standardized indirect effect: .56, 95% confidence interval: [.06, 1.35], p = .023; standardized direct effect; 1.75, 95% confidence interval: [.12, .3.38], p = .034).”
We also refer to these analyses when we interpret our findings in our discussion. On p. 23, we write:
“In addition, we continued to observe a robust interaction between age and frequency condition on associative memory, even when controlling for age-related change in the accuracy of both online frequency learning and explicit frequency reports. Thus, though we observed age differences in the learning of environmental regularities and in their influence on subsequent associative memory encoding, our developmental memory effects cannot be fully explained by differences in initial learning.”
We thank the reviewer for this constructive suggestion, as we believe these control analyses strengthen our interpretation of age differences in both the learning and use of environmental regularities to prioritize memory.
Reviewer #2:
Nussenbaum and Hartley provide novel neurobehavioral evidence of how individuals differentially use incrementally acquired information to guide goal-relevant memory encoding, highlighting roles for the medial temporal lobe during frequency learning, and the lateral prefrontal cortex for value-guided encoding/retrieval. This provides a novel behavioral phenomenology that gives great insight into the processes guiding adaptive memory formation based on prior experience. However, there were a few weaknesses throughout the paper that undermined an overall mechanistic understanding of the processes.
First, there was a lack of anatomical specificity in the discussion and interpretation of both prefrontal and striatal targets, as there is great heterogeneity across these regions that would infer very different behavioral processes.
We agree with the reviewer that our introduction and discussion would benefit from more anatomical granularity, and we did indeed have a priori predictions about more specific neural regions that might be involved in our task.
First, we expected that both the ventral and dorsal striatum might be responsive to stimulus value across our age range. Prior work has suggested that activity in the ventral striatum often correlates with the intrinsic value of a stimulus, whereas activity in the dorsal striatum may reflect goal-directed action values (Liljeholm & O’Doherty, 2012). In our task, we expected that high-frequency items may acquire intrinsic value during frequency-learning that is then reflected in the striatal response to these items during encoding. However, because participants were not rewarded when they encountered these images, but rather incentivized to encode associations involving them, we hypothesized that the dorsal striatum may represent the value of the ‘action’ of remembering each pair. In line with this prediction, the dorsal striatum, and the caudate in particular, have also been shown to be engaged during value-guided cognitive control (Hikosaka et al., 2014; Insel et al., 2017).
We have now revised our introduction to include greater specificity in our anatomical predictions on p. 3:
“When individuals need to remember information associated with previously encountered stimuli (e.g., the grocery store aisle where an ingredient is located), frequency knowledge may be instantiated as value signals, engaging regions along the mesolimbic dopamine pathway that have been implicated in reward anticipation and the encoding of stimulus and action values. These areas include the ventral tegmental area (VTA) and the ventral and dorsal striatum (Adcock et al., 2006; Liljeholm & O’Doherty, 2012; Shigemune et al., 2014).”
Though we initially predicted that encoding of high-value information would be associated with increased activation in both the ventral and dorsal striatum, the activation we observed was largely within the dorsal striatum, and specifically, the caudate. We have now revised our discussion accordingly on p. 26:
“Though we initially hypothesized that both the ventral and dorsal striatum may be involved in encoding of high-value information, the activation we observed was largely within the dorsal striatum, a region that may reflect the value of goal-directed actions (Liljeholm & O’Doherty, 2012). In our task, rather than each stimulus acquiring intrinsic value during frequency-learning, participants may have represented the value of the ‘action’ of remembering each pair during encoding.”
Second, while the ventromedial PFC often reflects value, given the control demands of our task, we expected to see greater activity in the dorsolateral PFC, which is often engaged in tasks that require the implementation of cognitive control (Botvinick & Braver, 2015). Thus, we hypothesized that individuals would show increased activation in the dlPFC during encoding of high- vs. low-value information, and that this activation would vary as a function of age. We have now clarified this hypothesis on p. 3:
“Value responses in the striatum may signal the need for increased engagement of the dorsolateral prefrontal cortex (dlPFC) (Botvinick & Braver, 2015), which supports the implementation of strategic control.”
In our discussion, we review disparate findings in the developmental literature and discuss factors that may contribute to these differences across studies. For example, in our discussion of Davidow et al. (2016), we highlight differences between their task design and the present study, focusing on how their task involved immediate receipt of reward at the time of encoding, while our task incentivized memory accuracy. We further note that studies that involve reward delivery at the time of encoding may engage different neural pathways than those that promote goal-directed encoding. Beyond Davidow et al. (2016), there are no other neuroimaging studies that examine the influence of reward on memory across development. Thus, we cannot relate our present neural findings to prior work on the development of value-guided memory. As we note in our discussion (p. 28), “Further work is needed to characterize both the influence of different types of reward signals on memory across development, as well as the development of the neural pathways that underlie age-related change in behavior.”
Second, age-related differences in neural activation emerged both during the initial frequency learning as well as during memory-guided adaptive encoding. While data from this initial phase was used to unpack the behavioral relationships on adaptive memory, a major weakness of the paper was not connecting these measures to neural activity during memory encoding/retrieval. This would be especially relevant given that both implicit and explicit measures of frequency predicted subsequent performance, but it is unclear which of these measures was guiding lateral PFC and caudate responses.
Thank you for this valuable suggestion. We agree that it would be interesting to link frequency- learning behavior to neural activity at encoding. As such, we have now conducted additional analyses to explore these relations.
In the original version of our manuscript, we examined behavior at the item level through mixed- effects models, and neural activation during encoding at the participant level. Thus, to examine the relation between frequency-learning metrics and neural activation at encoding, we created two additional participant-level metrics. For each participant we computed their average repetition suppression index, and a measure of frequency distance. The average repetition suppression index reflects the overall extent to which the participant demonstrated repetition suppression in response to the fifth presentation of the high-frequency items, and is computed by averaging each participant’s repetition suppression indices across items. We hypothesized that participants who demonstrated the greatest degree of repetition suppression might be the most sensitive to the difference between the 1- and 5-frequency items, and therefore, show the greatest differences in striatal and PFC activation during encoding of high- vs. low-value information. The frequency distance metric reflects the average distance between participants’ explicit frequency reports for items that appeared once and items that appeared five times, and is computed by averaging their explicit frequency reports for items in each frequency condition, and then subtracting the average reports in the low-frequency condition from those in the high- frequency condition. We hypothesized that participants with the largest frequency distances might similarly be the most sensitive to the difference between the 1- and 5-frequency items, and therefore, show the greatest differences in striatal and PFC activation during encoding of high- vs. low-value information.
We first wanted to confirm that the relations we observed between repetition suppression, frequency reports, and age, could also be observed at the participant level. In line with our prior, behavioral analyses, we found that age related to both mean repetition suppression indices (marginally; linear age: p = .067; quadratic age: p = .042); and frequency distances (linear and quadratic age: ps < .001).
In addition, we further tested whether these two metrics related to memory performance. In contrast to our item-level findings, we did not observe a significant relation between repetition suppression indices and memory (p = .83). We did observe an effect of frequency distance on memory performance. Specifically, we observed significant interactions between frequency distance and age (p = .014) and frequency distance and quadratic age (p = .021) on memory difference scores, such that the influence of frequency distance on memory difference scores increased with increasing age from childhood to adolescence.
We next examined how mean repetition suppression indices and frequency distances related to differential neural activation during encoding of high- and low-value pairs. In line with our memory findings, we did not observe any significant relations between mean repetition suppression indices and neural activation in the caudate or prefrontal cortex during encoding (ps > .15).
Frequency distance did not relate to caudate activation during encoding nor did we observe a frequency distance x age interaction effect (ps > .16). Frequency distance did, however, relate to differential PFC activation during encoding of high- vs. low-value pairs. Specifically, we observed a main effect of frequency distance on PFC activation (p = .0012), such that participants whose explicit reports of item frequency, were on average, more distinct across frequency conditions, demonstrated increased PFC activation during encoding of pairs involving high- vs. low-frequency items. Interestingly, when we included frequency distance in our model, we no longer observed a significant effect of age on differential PFC activation, nor did we observe a significant frequency distance x age interaction (ps > .13). These findings suggest that PFC activation during encoding may have, in part, reflected participants’ beliefs about the structure of the environment, with participants demonstrating stronger differential engagement of control processes across conditions when their representations of the conditions themselves were more distinct.
Finally, we examined how age, frequency distance, and PFC activation related to memory difference scores. Here, even when controlling for both frequency distance and PFC activation, we continued to observe main effects of age and quadratic age on memory difference scores (linear age: p = .006; quadratic age: p = .001). In line with our analysis of the relation between frequency reports and memory, these results suggest that age-related variance in value-guided memory may depend on both knowledge of the structure of the environment and use of that knowledge to effectively control encoding.
We have now added these results to our manuscript on p. 13 - 14. We write:
“Given the relations we observed between memory and both repetition suppression and frequency reports, we examined whether they related to neural activation in both our caudate and PFC ROI during encoding. To do so, we computed each participant’s average repetition suppression index, and their “frequency distance” — or the average difference in their explicit reports for items in the high- and low-frequency conditions. We expected that participants with greater average repetition suppression indices and greater frequency distances represented the high- and low-frequency items as more distinct from one another and therefore would show greater differences in neural activation at encoding across frequency conditions. In line with our prior analyses, both metrics varied with age (though repetition suppression only marginally (linear age: p = .067; quadratic age: p = .042); Appendix 3 y Tables 22 and 25), suggesting that older participants demonstrated better learning of the structure of the environment. We ran linear regressions examining the relations between each metric, age, and their interaction on neural activation in both the caudate and PFC. We observed no significant effects or interactions of average repetition suppression indices on neural activation (ps > .15; Appendix 3 Tables 23 and 24). We did, however, observe a significant effect of frequency distance on PFC activation (β = .42, SE = .12, p = .0012), such that participants who believed that average frequencies of the high- and low-frequency items were further apart also demonstrated greater PFC activation during encoding of pairs with high- vs. low-frequency items. Here, we did not observe a significant effect of age on PFC activation (β = -.03, SE = .13, p = .82), suggesting that age-related variance in PFC activation may be related to age differences in explicit frequency beliefs. Importantly, however, even when we accounted for both PFC activation and frequency distances, we continued to observe an effect of age on memory difference scores (β = .56, SE = .20, p = .006), which, together with our prior analyses, suggest that developmental differences in value-guided memory are not driven solely by age differences in beliefs about the structure of the environment but also depend on the use of those beliefs to guide encoding.”
We have added the full model results to Appendix 3: Full Model Specification and Results.
Given these results, we have now revised our interpretation of our neural data. Our memory analyses demonstrate that across our age range, we observed age-related differences in both the acquisition of knowledge of the structure of the environment and in its use. Originally, we interpreted the PFC activation as reflecting the use of learned value to guide memory. However, the strong relation we found between frequency distance and PFC activation suggests that the age differences in PFC activation that we observed may also be related to age differences in knowledge of the structure of the environment that governs when control processes should be engaged most strongly. However, these results must be interpreted cautiously. Participants provided explicit frequency reports after they completed the encoding and retrieval tasks, and so explicit frequency reports may have been influenced not only by participants’ memories of online frequency learning, but also by the strength with which they encoded the item and its paired associate, and the experience of successfully retrieving it.
We have now revised our discussion to consider these results. On p. 23, we now write,
“Our neural results further suggest that developmental differences in memory were driven by both knowledge of the structure of the environment and use of that knowledge to guide encoding.”
On p. 24, we write,
“The development of adaptive memory requires not only the implementation of encoding and retrieval strategies, but also the flexibility to up- or down-regulate the engagement of control in response to momentary fluctuations in information value (Castel et al., 2007, 2013; Hennessee et al., 2017). Importantly, value-based modulation of lateral PFC engagement during encoding mediated the relation between age and memory selectivity, suggesting that developmental change in both the representation of learned value and value-guided cognitive control may underpin the emergence of adaptive memory prioritization. Prior work examining other neurocognitive processes, including response inhibition (Insel et al., 2017) and selective attention (Störmer et al., 2014), has similarly found that increases in the flexible upregulation of control in response to value cues enhance goal-directed behavior across development (Davidow et al., 2018), and may depend on the engagement of both striatal and prefrontal circuitry (Hallquist et al., 2018; Insel et al., 2017). Here, we extend these past findings to the domain of memory, demonstrating that value signals derived from the structure of the environment increasingly elicit prefrontal cortex engagement and strengthen goal-directed encoding across childhood and into adolescence.”
And on p. 25, we have added an additional paragraph:
“Further, we also demonstrate that in the absence of explicit value cues, the engagement of prefrontal control processes may reflect beliefs about information value that are learned through experience. Here, we found that differential PFC activation during encoding of high- vs. low-value information reflected individual and age-related differences in beliefs about the structure of the environment; participants who represented the average frequencies of the low- and high-frequency items as further apart also demonstrated greater value-based modulation of lateral PFC activation. It is important to note, however, that we collected explicit frequency reports after associative encoding and retrieval. Thus the relation between PFC activation and explicit frequency reports may be bidirectional — while participants may have increased the recruitment of cognitive control processes to better encode information they believed was more valuable, the engagement of more elaborative or deeper encoding strategies that led to stronger memory traces may have also increased participants’ subjective sense of an item’s frequency (Jonides & Naveh-Benjamin, 1987).”
Third, more discussion is warranted on the nature of age-related changes given that some findings followed quadratic functions and others showed linear. Further interpretation of the quadratic versus linear fits would provide greater insight into the relative rates of maturation across discrete neurobehavioral processes.
We agree with the reviewer that more discussion is warranted here. While many cognitive processes tend to improve with increasing age, the significant interaction between quadratic age and frequency condition on memory accuracy could reflect a number of different patterns of developmental variance. Because quadratic curves are U-shaped, the significant interaction between quadratic age and frequency condition could reflect a peak in value-guided memory in adolescence. However, the combination of linear and quadratic effects can also capture “plateauing” effects, where the influence of age on a particular cognitive process decreases at a particular developmental timepoint. To determine how to interpret the quadratic effect of age on value-guided memory — and specifically, to test for the presence of an adolescent peak — we ran an additional analysis.
To test for an adolescent peak in value-guided memory, we first fit our memory accuracy model without any age terms, and then extracted the random slope across frequency conditions for each subject. We then conducted a ‘two lines test’ (Simonsohn, 2018) to examine the relation between age and these random slopes. In brief, the two-lines test fits the data with two linear models — one with a positive slope and one with a negative slope, algorithmically determining the breakpoint in the estimates where the signs of the slopes change. When we analyzed our memory data in this way, we found a robust, positive relation between age and value-guided memory (see newly added Appendix 2 Figure 3, also below) from childhood to mid- adolescence, that peaked around age 16 (age 15.86). From age ~16 to early adulthood, however, we observed only a marginal negative relation between age and value-guided memory (p = .0567). Thus, our findings do not offer strong evidence in support of an adolescent peak in value-guided memory — instead, they suggest that improvements in value-guided memory are strongest from childhood to adolescence.
Appendix 2 - Figure 3. Results from the two-lines test (Simonsohn, 2018) revealed that the influence of frequency condition on memory accuracy increased throughout childhood and early adolescence, and did not significantly decrease from adolescence into early adulthood.
To more clearly demonstrate the relation between age and value-guided memory, we have now included the results of the two-lines test in the results section of our main text. On p. 12 - 13, we write:
“In line with our hypothesis, we observed a main effect of frequency condition on memory, χ2(1) = 21.51, p <.001, indicating that individuals used naturalistic value signals to prioritize memory for high-value information. Critically, this effect interacted with both linear age (χ2(1) = 11.03, p < .001) and quadratic age (χ2(1) = 9.51, p = .002), such that the influence of frequency condition on memory increased to the greatest extent throughout childhood and early adolescence. To determine whether the interaction between quadratic age and frequency condition on memory accuracy reflected an adolescent peak in value-guided memory prioritization, we re-ran our memory accuracy model without including any age terms, and extracted each participant’s random slope across frequency conditions. We then submitted these random slopes to the “two-lines” test (Simonsohn, 2018), which fits two regression lines with oppositely signed slopes to the data, algorithmically determining where the sign flip should occur. The results of this analysis revealed that the influence of frequency condition on memory significantly increased from age 8 to age 15.86 (b = .03, z = 2.71, p = .0068; Appendix 2 – Figure 3), but only marginally decreased from age 15.86 to age 25 (b = -.02, z = 1.91, p = .0576). Thus, the interaction between frequency condition and quadratic age on memory performance suggests that the biggest age differences in value-guided memory occurred through childhood and early adolescence, with older adolescents and adults performing similarly.”
That said, this developmental trajectory is likely specific to the particular demands of our task. In our previous behavioral study that used a very similar paradigm (Nussenbaum, Prentis, & Hartley, 2018), we observed only a linear relation between age and value-guided memory.
Although the task used in our behavioral study was largely similar to the task we employed here, there were subtle differences in the design that may have extended the age range through which we observed improvements in memory prioritization. In particular, in our previous behavioral study, the memory test required participants to select the correct associate from a grid of 20 options (i.e., 1 correct and 19 incorrect options), whereas here, participants had to select the correct associate from a grid of 4 options (1 correct and 3 incorrect options). In our prior work, the need to differentiate the ‘correct’ option from many more foils may have increased the demands on either (or both) memory encoding or memory retrieval, requiring participants to encode and retrieve more specific representations that would be less confusable with other memory representations. By decreasing the task demands in the present study, we may have shifted the developmental curve we observed toward earlier developmental timepoints.
We originally did not emphasize our quadratic findings in the discussion of our manuscript because, given the marginal decrease in memory selectivity we observed from age 16 to age 25 and the different age-related findings across our two studies, we did not want to make strong claims about the specific shape of developmental change. However, we agree with the reviewer that these points are worthy of discussion within the manuscript. We have now amended our discussion on p. 25 accordingly:
“We found that memory prioritization varied with quadratic age, and our follow-up tests probing the quadratic age effect did not reveal evidence for significant age-related change in memory prioritization between late adolescence and early adulthood. However, in our prior behavioral work using a very similar paradigm (Nussenbaum et al., 2020), we found that memory prioritization varied with linear age only. In line with theoretical proposals (Davidow et al., 2018), subtle differences in the control demands between the two tasks (e.g., reducing the number of ‘foils’ presented on each trial of the memory test here relative to our prior study), may have shifted the age range across which we observed differences in behavior, with the more demanding variant of our task showing more linear age-related improvements into early adulthood. In addition, the specific control demands of our task may have also influenced the age at which value- guided memory emerged. Future studies should test whether younger children can modulate encoding based on the value of information if the mnemonic demands of the task are simpler.”
We thank the reviewer for this helpful suggestion, and believe our additions that expand on the quadratic age effects help clarify our developmental findings.
Although hippocamapal and PHC results did not show a main effect of value, it seems by the introduction that this region would be critical for the processes under study. I would suggest including these regions as ROIs of interest guiding age-related differences during the memory encoding and retrieval phases. Even reporting negative findings for these regions would be helpful to readers, especially given the speculation of the negative findings in the discussion.
Thank you for this suggestion. We have now examined how differential neural activation within the hippocampus and parahippocampal cortex during encoding of high- vs. low-value information varies with age. To do so, we followed the same approach as with our PFC and caudate ROI analyses. Specifically, we first identified the voxel within both the hippocampus and parahippocampal cortex with the highest z-statistic from our group-level 5 > 1 encoding contrast. We then drew a 5-mm sphere around these voxels and examined how mean beta weights within these spheres varied with age.
We did not observe any relation between differential hippocampal or parahippocampal cortex activation during encoding of high- vs. low-value information and age (ps > .50). We agree with the reviewer that these results are informative, and have now added them to Appendix 2: Supplementary Analyses, which we refer to in the main text (p. 15). In Appendix 2, we write:
“Hippocampal and parahippocampal cortex activation during encoding A priori, we expected that regions in the medial temporal lobe that have been linked to successful memory formation, including the hippocampus and parahippocampal cortex (Davachi, 2006), may be differentially engaged during encoding of high- vs. low- value information. Further, we hypothesized that the differential engagement of these regions across age may contribute to age differences in value-guided memory. Though we did not see any significant clusters of activation in the hippocampus or parahippocampal cortex in our group level high value vs. low value encoding contrast, we conducted additional ROI analyses to test these hypotheses. As with our other ROI analyses, we first identified the peak voxel (based on its z-statistic; hippocampus: x = 24, y = 34, z = 23; parahippocampal cortex: x = 22, y = 41, z = 16) in each region from our group-level contrast, and then drew 5-mm spheres around them. We then examined how average parameter estimates within these spheres related to both age and memory difference scores.
First, we ran a linear regression modeling the effects of age, WASI scores, and their interaction on hippocampal activation. We did not observe a main effect of age on hippocampal activation, (β = .00, SE = .10, p > .99). We did, however, observe a significant age x WASI score interaction effect (β = .30, SE = .10, p = .003). Next, we conducted another linear regression to examine the effects of hippocampal activation, age, WASI scores, and their interaction on memory difference scores. In contrast to our prefrontal cortex activation results, activation in the hippocampus did not relate to memory difference scores, (β = -.02, SE = .03, p = .50).
We repeated these analyses with our parahippocampal cortex sphere. Here, we did not observe any significant effects of age on parahippocampal activation (β = -.07, SE = .11, p = .50), nor did we observe any effects of parahippocampal activation on memory difference scores (β = .01, SE = .03, p = .25).”
Reviewer #3:
This paper investigated age differences in the neurocognitive mechanisms of value-based memory encoding and retrieval across children, adolescents and young adults. It used a novel experimental paradigm in combination with fMRI to disentangle age differences in determining the value of information based on its frequency from the usage of these learned value signals to guide memory encoding. During value learning, younger participants demonstrated a stronger effect of item repetition on response accuracy, whereas repetition suppression effects in a parahippocampal ROI were strongest in adults. Item frequency modulated memory accuracy such that associative memory was better for previously high-frequency value items. Notably, this effect increased with age. Differences in memory accuracy between low- and high-frequency items were associated with left lateral PFC activation which also increased with age. Accordingly, a mediation analyses revealed that PFC activation mediated the relation between age and memory benefit for high- vs. low-frequency items. Finally, both participants' representations of item frequency (which were more likely to deviate in younger children) and repetition suppression in the parahippocampal ROI were associated with higher memory accuracy. Together, these results data add to the still scarce literature examining how information value influences memory processes across development.
Overall, the conclusions of the paper are well supported by the data, but some aspects of the data analysis need to be clarified and extended.
Empirical findings directly comparing cross-sectional and longitudinal effects have demonstrated that cross-sectional analyses of age differences do not readily generalize to longitudinal research (e.g., Raz et al., 2005; Raz & Lindenberger, 2012). Formal analyses have demonstrated that proportion of explained age-related variance in cross-sectional mediation models may stem from various factors, including similar mean age trends, within-time correlations between a mediator and an outcome, or both (Lindenberger et al., 2011; see also Hofer, Flaherty, & Hoffman, 2006; Maxwell & Cole, 2007). Thus, the results of the mediation analysis showing that PFC activation explains age-related variance in memory difference scores, cannot be taken to imply that changes in PFC activation are correlated with changes in value-guided memory. While the general limitations of a cross-sectional study are noted in the Discussion of the manuscript, it would be important to discuss the critical limitations of the mediation analysis. While the main conclusions of the paper do not critically depend on this analysis, it would be important to alert the reader to the limited information value in performing cross-sectional mediation analyses of age variance.
Thank you for raising this critical point. We have expanded our discussion to specifically note the limitations of our mediation analysis and to more strongly emphasize the need for future longitudinal studies to reveal how changes in neural circuitry may support the emergence of motivated memory across development. Specifically, on p. 26, we now write:
“One important caveat is that our study was cross-sectional — it will be important to replicate our findings in a longitudinal sample to more directly measure how developmental changes in cognitive control within an individual contribute to changes in their ability to selectively encode useful information. Our mediation results, in particular, must be interpreted with caution as simulations have demonstrated that in cross-sectional samples, variables can emerge as significant mediators of age-related change due largely to statistical artifact (Hofer, Flaherty, & Hoffman, 2006; Lindenberger et al., 2011). Indeed, our finding that PFC activation mediates the relation between age and value-guided memory does not necessarily imply that within an individual, PFC development leads to improvements in memory selectivity. Longitudinal work in which individuals’ neural activity and memory performance is sampled densely within developmental windows of interest is needed to elucidate the complex relations between age, brain development, and behavior (Hofer, Flaherty, & Hoffman, 2006; Lindenberger et al., 2011).”
It would be helpful to provide more information on how chance memory performance was handled during data analysis, especially as it is more likely to occur in younger participants. Related to this, please connect the points that belong to the same individual in Figure 3 to facilitate evaluation of individual differences in the memory difference scores.
Thank you for raising this important point. On each memory test trial, participants viewed the item (either a postcard or picture) above images of four possible paired associates (see Figure 1 on p. 6). On each memory test trial, participants had 6 seconds to select one of these items. If participants did not make a response within 6 seconds, that trial was considered ‘missed.’ Missed trials were excluded from behavioral analyses and regressed out in neural analyses. If participants selected the correct associate, memory accuracy was coded as ‘1;’ if they selected an incorrect associate, accuracy was coded as ‘0.’ On each trial, there was 1 correct option and 3 incorrect options. As such, chance-level memory performance was 25%. We have now clarified this on p. 34 and included a dashed line indicating chance-level performance within Fig. 4 (formerly Figure 3) on p. 12. In addition, we have also updated Figure 4 (see below) to connect the points belonging to the same participants, as suggested by the reviewer.
Figure 4. Participants demonstrated prioritization of memory for high-value information, as indicated by higher memory accuracy for associations involving items in the five- relative to the one-frequency condition (χ2(1) = 19.73, p <.001). The effects of item frequency on associative memory increased throughout childhood and into adolescence (linear age x frequency condition: χ2(1) = 10.74, p = .001; quadratic age x frequency condition: χ2(1) = 9.27, p = .002).
Out of 90 participants, 2 children performed at or below chance (<= 25% memory accuracy). Interpreting the behavior of the participants who responded to fewer than 12 out of 48 trials correctly is challenging. On the one hand, they might not have remembered anything and responded correctly on these trials due to randomly guessing. On the other hand, they may have implemented an encoding strategy of focusing only on a small number of pairs. Thus, a priori, based on the analysis approach we implemented in our prior, behavioral study (Nussenbaum et al., 2019), we decided to include all participants in our memory analyses, regardless of their overall accuracy. However, when we exclude these two participants from our memory analyses, our main findings still hold. Specifically, we continue to observe main effects of frequency condition and age, and interactions between frequency condition and both linear and quadratic age on associative memory accuracy (ps < .012).
We have now clarified these details about chance-level performance in the methods section of our manuscript on p. 34.
“For our memory analyses, trials were scored as ‘correct’ if the participant selected the correct association from the set of four possible options presented during the memory test, ‘incorrect’ if the participant selected an incorrect association, and ‘missed’ if the participant failed to respond within the 6-second response window. Missed trials were excluded from all analyses. Because participants had to select the correct association from four possible options, chance-level performance was 25%. Two child participants performed at or below chance-level on the memory test. They were included in all analyses reported in the manuscript; however, we report full details of the results of our memory analyses when we exclude these two participants in Appendix 3 (Table 15). Importantly, our main findings remain unchanged.”
In Appendix 3, we include a table with the full results from our memory model without these two participants:
Appendix Table 15: Associative memory accuracy by frequency condition (below chance subjects excluded)
I would like to see some consideration of how the different signatures of value learning, repetition suppression and reported item frequency, are related to the observed PFC and caudate effects during memory encoding. Such a discussion would help the reader connect the findings on learning and using information value across development.
Thank you for this valuable suggestion. We agree that it would be interesting to link frequency- learning behavior to neural activity at encoding. As such, we have now conducted additional analyses to explore these relations.
In the original version of our manuscript, we examined behavior at the item level through mixed- effects models, and neural activation during encoding at the participant level. Thus, to examine the relation between frequency-learning metrics and neural activation at encoding, we created two additional participant-level metrics. For each participant we computed their average repetition suppression index, and a measure of frequency distance. The average repetition suppression index reflects the overall extent to which the participant demonstrated repetition suppression in response to the fifth presentation of the high-frequency items, and is computed by averaging each participant’s repetition suppression indices across items. We hypothesized that participants who demonstrated the greatest degree of repetition suppression might be the most sensitive to the difference between the 1- and 5-frequency items, and therefore, show the greatest differences in striatal and PFC activation during encoding of high- vs. low-value information. The frequency distance metric reflects the average distance between participants’ explicit frequency reports for items that appeared once and items that appeared five times, and is computed by averaging their explicit frequency reports for items in each frequency condition, and then subtracting the average reports in the low-frequency condition from those in the high- frequency condition. We hypothesized that participants with the largest frequency distances might similarly be the most sensitive to the difference between the 1- and 5-frequency items, and therefore, show the greatest differences in striatal and PFC activation during encoding of high- vs. low-value information.
We first wanted to confirm that the relations we observed between repetition suppression, frequency reports, and age, could also be observed at the participant level. In line with our prior, behavioral analyses, we found that age related to both mean repetition suppression indices (marginally; linear age: p = .067; quadratic age: p = .042); and frequency distances (linear and quadratic age: ps < .001).
In addition, we further tested whether these two metrics related to memory performance. In contrast to our item-level findings, we did not observe a significant relation between repetition suppression indices and memory (p = .83). We did observe an effect of frequency distance on memory performance. Specifically, we observed significant interactions between frequency distance and age (p = .014) and frequency distance and quadratic age (p = .021) on memory difference scores, such that the influence of frequency distance on memory difference scores increased with increasing age from childhood to adolescence.
We next examined how mean repetition suppression indices and frequency distances related to differential neural activation during encoding of high- and low-value pairs. In line with our memory findings, we did not observe any significant relations between mean repetition suppression indices and neural activation in the caudate or prefrontal cortex during encoding (ps > .15).
Frequency distance did not relate to caudate activation during encoding nor did we observe a frequency distance x age interaction effect (ps > .16). Frequency distance did, however, relate to differential PFC activation during encoding of high- vs. low-value pairs. Specifically, we observed a main effect of frequency distance on PFC activation (p = .0012), such that participants whose explicit reports of item frequency, were on average, more distinct across frequency conditions, demonstrated increased PFC activation during encoding of pairs involving high- vs. low-frequency items. Interestingly, when we included frequency distance in our model, we no longer observed a significant effect of age on differential PFC activation, nor did we observe a significant frequency distance x age interaction (ps > .13). These findings suggest that PFC activation during encoding may have, in part, reflected participants’ beliefs about the structure of the environment, with participants demonstrating stronger differential engagement of control processes across conditions when their representations of the conditions themselves were more distinct.
Finally, we examined how age, frequency distance, and PFC activation related to memory difference scores. Here, even when controlling for both frequency distance and PFC activation, we continued to observe main effects of age and quadratic age on memory difference scores (linear age: p = .006; quadratic age: p = .001). In line with our analysis of the relation between frequency reports and memory, these results suggest that age-related variance in value-guided memory may depend on both knowledge of the structure of the environment and use of that knowledge to effectively control encoding.
We have now added these results to our manuscript on p. 13 - 14. We write:
“Given the relations we observed between memory and both repetition suppression and frequency reports, we examined whether they related to neural activation in both our caudate and PFC ROI during encoding. To do so, we computed each participant’s average repetition suppression index, and their “frequency distance” — or the average difference in their explicit reports for items in the high- and low-frequency conditions. We expected that participants with greater average repetition suppression indices and greater frequency distances represented the high- and low-frequency items as more distinct from one another and therefore would show greater differences in neural activation at encoding across frequency conditions. In line with our prior analyses, both metrics varied with age (though repetition suppression only marginally (linear age: p = .067; quadratic age: p = .042); Appendix 3 Tables 22 and 25), suggesting that older participants demonstrated better learning of the structure of the environment. We ran linear regressions examining the relations between each metric, age, and their interaction on neural activation in both the caudate and PFC. We observed no significant effects or interactions of average repetition suppression indices on neural activation (ps > .15; Appendix 3 Tables 23 and 24). We did, however, observe a significant effect of frequency distance on PFC activation (β = .42, SE = .12, p = .0012), such that participants who believed that average frequencies of the high- and low-frequency items were further apart also demonstrated greater PFC activation during encoding of pairs with high- vs. low-frequency items. Here, we did not observe a significant effect of age on PFC activation (β = -.03, SE = .13, p = .82), suggesting that age-related variance in PFC activation may be related to age differences in explicit frequency beliefs. Importantly, however, even when we accounted for both PFC activation and frequency distances, we continued to observe an effect of age on memory difference scores (β = .56, SE = .20, p = .006), which, together with our prior analyses, suggest that developmental differences in value-guided memory are not driven solely by age differences in beliefs about the structure of the environment but also depend on the use of those beliefs to guide encoding.”
We have added the full model results to Appendix 3.
Given these results, we have now revised our interpretation of our neural data. Our memory analyses demonstrate that across our age range, we observed age-related differences in both the acquisition of knowledge of the structure of the environment and in its use. Originally, we interpreted the PFC activation as reflecting the use of learned value to guide memory. However, the strong relation we found between frequency distance and PFC activation suggests that the age differences in PFC activation that we observed may also be related to age differences in knowledge of the structure of the environment that governs when control processes should be engaged most strongly. However, these results must be interpreted cautiously. Participants provided explicit frequency reports after they completed the encoding and retrieval tasks, and so explicit frequency reports may have been influenced not only by participants’ memories of online frequency learning, but also by the strength with which they encoded the item and its paired associate, and the experience of successfully retrieving it.
We have now revised our discussion to consider these results. On p. 23, we now write,
“Our neural results further suggest that developmental differences in memory were driven by both knowledge of the structure of the environment and use of that knowledge to guide encoding.”
n p. 24, we write,
“The development of adaptive memory requires not only the implementation of encoding and retrieval strategies, but also the flexibility to up- or down-regulate the engagement of control in response to momentary fluctuations in information value (Castel et al., 2007, 2013; Hennessee et al., 2017). Importantly, value-based modulation of lateral PFC engagement during encoding mediated the relation between age and memory selectivity, suggesting that developmental change in both the representation of learned value and value-guided cognitive control may underpin the emergence of adaptive memory prioritization. Prior work examining other neurocognitive processes, including response inhibition (Insel et al., 2017) and selective attention (Störmer et al., 2014), has similarly found that increases in the flexible upregulation of control in response to value cues enhance goal-directed behavior across development (Davidow et al., 2018), and may depend on the engagement of both striatal and prefrontal circuitry (Hallquist et al., 2018; Insel et al., 2017). Here, we extend these past findings to the domain of memory, demonstrating that value signals derived from the structure of the environment increasingly elicit prefrontal cortex engagement and strengthen goal-directed encoding across childhood and into adolescence.”
And on p. 25, we have added an additional paragraph:
“Further, we also demonstrate that in the absence of explicit value cues, the engagement of prefrontal control processes may reflect beliefs about information value that are learned through experience. Here, we found that differential PFC activation during encoding of high- vs. low-value information reflected individual and age-related differences in beliefs about the structure of the environment; participants who represented the average frequencies of the low- and high-frequency items as further apart also demonstrated greater value-based modulation of lateral PFC activation. It is important to note, however, that we collected explicit frequency reports after associative encoding and retrieval. Thus the relation between PFC activation and explicit frequency reports may be bidirectional — while participants may have increased the recruitment of cognitive control processes to better encode information they believed was more valuable, the engagement of more elaborative or deeper encoding strategies that led to stronger memory traces may have also increased participants’ subjective sense of an item’s frequency (Jonides & Naveh-Benjamin, 1987).”
A point worthy of discussion are the implications of the finding that younger participants demonstrated greater deviations in their frequency reports for the development of value learning, given that frequency reports were found to predict associative memory accuracy.
Thank you for raising this important point. Indeed, one of our main findings is that older participants are better both at learning the structure of their environments and also at using structured knowledge to strategically prioritize memory. In our original manuscript, we described results of a model that included participants’ explicit frequency reports as a predictor of memory. Model comparison revealed that participants’ frequency reports — which we interpret as reflecting their beliefs about the structure of the environment — predicted memory more strongly than the item’s true frequency. In other words, participants’ beliefs about the structure of the environment (even if incorrect) more strongly influenced their memory encoding than the true structure of the environment. Critically, however, frequency reports interacted with age to predict memory (Fig 8). Even when we accounted for age-related differences in knowledge of the structure of the environment, older participants demonstrated a stronger influence of frequency on memory, suggesting they were better able to use their beliefs to control subsequent associative encoding. We have now clarified our interpretation of this model in our discussion on p. 23:
“Importantly, though we observed age-related differences in participants’ learning of the structure of their environment, the strengthening of the relation between frequency reports and associative memory with increasing age suggests that age differences in learning cannot fully account for age differences in value-guided memory. Even when accounting for individual differences in participants’ explicit knowledge of the structure of the environment, older participants demonstrated a stronger relation between their beliefs about item frequency and associative memory, suggesting that they used their beliefs to guide memory to a greater degree than younger participants.”
As noted by the reviewer, however, our initial memory analysis did not account for age-related differences in participants’ initial, online learning of item frequency, and our neural analyses further did not account for age differences in explicit frequency reports. We have now run additional control analyses to account for the potential influence of individual differences in frequency learning on associative memory. Specifically, for each participant, we computed three metrics: 1.) their overall accuracy during frequency-learning, 2.) their overall accuracy for the last presentation of each item during frequency-learning (as suggested by Reviewer 2), and 3.) the mean magnitude of the error in their frequency reports. We then included these metrics as covariates in our memory analyses.
When we include these control variables in our model, we continue to observe a robust effect of frequency condition (p < .001) as well as robust interactions between frequency condition and linear and quadratic age (ps < .003) on associative memory accuracy. We also observed a main effect of frequency error magnitude on memory accuracy (p < .001). Here, however, we no longer observe main effects of age or quadratic age on overall memory accuracy. Given the relation we observed between frequency error magnitudes and age, the results from this model suggests that there may be age-related improvements in overall memory that influence both memory for associations as well as learning of and memory for item frequencies. The fact that age no longer relates to overall memory when controlling for frequency error magnitudes suggest that age-related variance in memory for item frequencies and memory for associations are strongly related within individuals. Importantly, however, age-related variance in memory for item frequencies did not explain age-related variance in the influence of frequency condition on associative memory, suggesting that there are developmental differences in the use of knowledge of environmental structure to prioritize valuable information in memory that persist even when controlling for age-related differences in initial learning of environmental regularities. Given the importance of this analysis in elucidating the relation between the learning of environmental structure and value-guided memory, we have now updated the results in the main text of our manuscript to include them. Specifically, on p. 13, we now write:
“Because we observed age-related differences in participants’ online learning of item frequencies and in their explicit frequency reports, we further examined whether these age differences in initial learning could account for the age differences we observed in associative memory. To do so, we ran an additional model in which we included each participant’s mean frequency learning accuracy, mean frequency learning accuracy on the last repetition of each item, and explicit report error magnitude as covariates. Here, explicit report error magnitude predicted overall memory performance, χ2(1) =13.05, p < .001, and we did not observe main effects of age or quadratic age on memory performance (ps > .20). However, we continued to observe a main effect of frequency condition, χ2(1) = 19.65 p < .001, as well as significant interactions between frequency condition and both linear age χ2(1) = 10.59, p = .001, and quadratic age χ2(1) = 9.15, p = .002. Thus, while age differences in initial learning related to overall memory performance, they did not account for age differences in the use of environmental regularities to strategically prioritize memory for valuable information.”
In addition, as suggested by the reviewer, we also included the three covariates as control variables in our mediation analysis. When controlling for online frequency learning and explicit frequency report errors, PFC activity continued to mediate the relation between age and memory difference scores. We have now included these results on p. 16 - 17 of the main text:
“Further, when we included quadratic age, WASI scores, online frequency learning accuracy, online frequency learning accuracy on the final repetition of each item, and mean explicit frequency report error magnitudes as control variables in the mediation analysis, PFC activation continued to mediate the relation between linear age and memory difference scores (standardized indirect effect: .56, 95% confidence interval: [.06, 1.35], p = .023; standardized direct effect; 1.75, 95% confidence interval: [.12, .3.38], p = .034).”
We also refer to these analyses when we interpret our findings in our discussion. On p. 23, we write:
“In addition, we continued to observe a robust interaction between age and frequency condition on associative memory, even when controlling for age-related change in the accuracy of both online frequency learning and explicit frequency reports. Thus, though we observed age differences in the learning of environmental regularities and in their influence on subsequent associative memory encoding, our developmental memory effects cannot be fully explained by differences in initial learning.”
We thank the reviewer for this constructive suggestion, as we believe these control analyses strengthen our interpretation of age differences in both the learning and use of environmental regularities to prioritize memory.
Author response:
The following is the authors’ response to the previous reviews.
Reviewer #1 (Public review):
Summary:
This work shows that a specific adenosine deaminase protein in Dictyostelium generates the ammonia that is required for tip formation during Dictyostelium development. Cells with an insertion in the ADGF gene aggregate but do not form tips. A remarkable result, shown in several different ways, is that the ADGF mutant can be rescued by exposing the mutant to ammonia gas. The authors also describe other phenotypes of the ADGF mutant such as increased mound size, altered cAMP signalling, and abnormal cell type differentiation. It appears that the ADGF mutant has defects in the expression of a large number of genes, resulting in not only the tip defect but also the mound size, cAMP signalling, and differentiation phenotypes.
Strengths:
The data and statistics are excellent.
(1) Weaknesses: The key weakness is understanding why the cells bother to use a diffusible gas like ammonia as a signal to form a tip and continue development.
Ammonia can come from a variety of sources both within and outside the cells and this can be from dead cells also. Ammonia by increasing cAMP levels, trigger collective cell movement thereby establishing a tip in Dictyostelium. A gaseous signal can act over long distances in a short time and for instance ammonia promotes synchronous development in a colony of yeast cells (Palkova et al., 1997; Palkova and Forstova, 2000). The slug tip is known to release ammonia probably favouring synchronized development of the entire colony of Dictyostelium. However, after the tips are established ammonia exerts negative chemotaxis probably helping the slugs to move away from each other ensuring equal spacing of the fruiting bodies (Feit and Sollitto, 1987).
It is well known that ammonia serves as a signalling molecule influencing both multicellular organization and differentiation in Dictyostelium (Francis, 1964; Bonner et al., 1989; Bradbury and Gross, 1989). Ammonia by raising the pH of the intracellular acidic vesicles of prestalk cells (Poole and Ohkuma, 1981; Gross et al, 1983), and the cytoplasm, is known to increase the speed of chemotaxing amoebae (Siegert and Weijer, 1989; Van Duijn and Inouye, 1991), inducing collective cell movement (Bonner et al., 1988, 1989), favoring tipped mound development.
Ammonia produced in millimolar concentrations during tip formation (Schindler and Sussman, 1977) could ward off other predators in soil. For instance, ammonia released by Streptomyces symbionts of leaf-cutting ants is known to inhibit fungal pathogens (Dhodary and Spiteller, 2021). Additionally, ammonia may be recycled back into amino acids, as observed during breast cancer proliferation (Spinelli et al., 2017). Such a process may also occur in starving Dictyostelium cells, supporting survival and differentiation. These findings suggest that ammonia acts as both a local and long-range regulatory signal, integrating environmental and cellular cues to coordinate multicellular development.
(2) The rescue of the mutant by adding ammonia gas to the entire culture indicates that ammonia conveys no positional information within the mound.
Ammonia reinforces or maintains the positional information by elevating cAMP levels, favoring prespore differentiation (Bradbury and Gross, 1989; Riley and Barclay, 1990; Hopper et al., 1993). Ammonia is known to influence rapid patterning of Dictyostelium cells confined in a restricted environment (Sawai et al., 2002). In adgf mutants that have low ammonia levels, both neutral red staining (a marker for prestalk and ALCs) (Figure. S3) and the prestalk marker ecmA/ ecmB expression (Figure. 7D) are higher than the WT and the mound arrest phenotype can be reversed by exposing the adgf mutant mounds to ammonia.
Prestalk cells are enriched in acidic vesicles, and ammonia, by raising the pH of these vesicles and the cytoplasm (Davies et al 1993; Van Duijn and Inouye 1991), plays an active role in collective cell movement during tip formation (Bonner et al., 1989).
(3) By the time the cells have formed a mound, the cells have been starving for several hours, and desperately need to form a fruiting body to disperse some of themselves as spores, and thus need to form a tip no matter what.
Exposure of adgf mounds to ammonia, led to tip development within 4 h (Figure. 5). In contrast, adgf controls remained at the mound stage for at least 30 h. This demonstrates that starvation alone is not the trigger for tip development and ammonia promotes the transition from mound to tipped mound formation.
Many mound arrest mutants are blocked in development and do not proceed to form fruiting bodies (Carrin et al., 1994). Further, not all the mound arrest mutants tested in this study were rescued by ADA enzyme (Figure. S4A), and they continue to stay as mounds.
(4) One can envision that the local ammonia concentration is possibly informing the mound that some minimal number of cells are present (assuming that the ammonia concentration is proportional to the number of cells), but probably even a minuscule fruiting body would be preferable to the cells compared to a mound. This latter idea could be easily explored by examining the fate of the ADGF cells in the mound - do they all form spores? Do some form spores?
Or perhaps the ADGF is secreted by only one cell type, and the resulting ammonia tells the mound that for some reason that cell type is not present in the mound, allowing some of the cells to transdifferentiate into the needed cell type. Thus, elucidating if all or some cells produce ADGF would greatly strengthen this puzzling story.
A fraction of adgf mounds form bulkier spore heads by the end of 36 h as shown in Figure. 2H. This late recovery may be due to the expression of other ADA isoforms. Mixing WT and adgf mutant cell lines results in a chimeric slug with mutants occupying the prestalk region (Figure. 8) and suggests that WT ADGF favours prespore differentiation. However, it is not clear if ADGF is secreted by a particular cell type, as adenosine can be produced by both cell types, and the activity of three other intracellular ADAs may vary between the cell types. To address whether adgf expression is cell type-specific, prestalk and prespore cells will be separated by fluorescence activated cell sorter (FACS), and thereafter, adgf expression will be examined in each population.
Reviewer #2 (Public review):
Summary:
The paper describes new insights into the role of adenosine deaminase-related growth factor (ADGF), an enzyme that catalyses the breakdown of adenosine into ammonia and inosine, in tip formation during Dictyostelium development. The ADGF null mutant has a pre-tip mound arrest phenotype, which can be rescued by the external addition of ammonia. Analysis suggests that the phenotype involves changes in cAMP signalling possibly involving a histidine kinase dhkD, but details remain to be resolved.
Strengths:
The generation of an ADGF mutant showed a strong mound arrest phenotype and successful rescue by external ammonia. Characterization of significant changes in cAMP signalling components, suggesting low cAMP signalling in the mutant and identification of the histidine kinase dhkD as a possible component of the transduction pathway. Identification of a change in cell type differentiation towards prestalk fate
(1) Weaknesses: Lack of details on the developmental time course of ADGF activity and cell type type-specific differences in ADGF expression.
adgf expression was examined at 0, 8, 12, and 16 h (Figure. 1), and the total ADA activity was assayed at 12 and 16 h (Figure. 3). Previously, the 12 h data was not included, and it’s been added now (Figure. 3A). The adgf expression was found to be highest at 16 h and hence, the ADA assay was carried out at that time point. Since the ADA assay will also report the activity of other three isoforms, it will not exclusively reflect ADGF activity.
Mixing WT and adgf mutant cell lines results in a chimeric slug with mutants occupying the prestalk region (Figure. 8) suggesting that WT adgf favours prespore differentiation. To address whether adgf expression is cell type-specific, prestalk and prespore cells will be separated by fluorescence activated cell sorter (FACS), and thereafter, adgf expression will be examined in each population.
(2) The absence of measurements to show that ammonia addition to the null mutant can rescue the proposed defects in cAMP signalling.
The adgf mutant in comparison to WT has diminished acaA expression (Fig. 6B) and reduced cAMP levels (Fig. 6A) both at 12 and 16 h of development. The cAMP levels were measured at 8 h and 12 h in the mutant.
We would like to add that ammonia is known to increase cAMP levels (Riley and Barclay, 1990; Feit et al., 2001) in Dictyostelium. Exposure to ammonia increases acaA expression in WT (Figure. 7B) and is likely to increase acaA expression/ cAMP levels in the mutant also (Riley and Barclay, 1990; Feit et al., 2001) thereby rescuing the defects in cAMP signalling. Based on the comments, cAMP levels will also be measured in the mutant after the rescue with ammonia.
(3) No direct measurements in the dhkD mutant to show that it acts upstream of adgf in the control of changes in cAMP signalling and tip formation.
cAMP levels will be quantified in the dhkD mutant after treatment with ammonia. The histidine kinases dhkD and dhkC are reported to modulate phosphodiesterase RegA activity, thereby maintaining cAMP levels (Singleton et al., 1998; Singleton and Xiong, 2013). By activating RegA, dhkD ensures proper cAMP distribution within the mound, which is essential for the patterning of prestalk and prespore cells, as well as for tip formation (Singleton and Xiong, 2013). Therefore, ammonia exposure to dhkD mutants is likely to regulate cAMP signalling and thereby tip formation.
Reviewer #1 (Recommendations for the authors):
(1) Lines: 47,48 - "The gradient of these morphogens along the slug axis determines the cell fate, either as prestalk (pst) or as prespore (psp) cells." - many workers have shown that this is not true - intrinsic factors such as cell cycle phase drive cell fate.
Thank you for pointing this out. We have removed the line and rephrased as “Based on cell cycle phases, there exists a dichotomy of cell types, that biases cell fate as prestalk or prespore (Weeks and Weijer, 1994; Jang and Gomer, 2011).
(2) Line 48 - PKA - please explain acronyms at first use.
Corrected
(3) Line 56 - The relationship between adenosine deaminase and ADGF is a bit unclear, please clarify this more.
Adenosine deaminase (ADA) is intracellular, whereas adenosine deaminase related growth factor (ADGF) is an extracellular ADA and has a growth factor activity (Li and Aksoy, 2000; Iijima et al., 2008).
(4) Figure 1 - where are these primers, and the bsr cassette, located with respect to the coding region start and stop sites?
The primer sequences are mentioned in the supplementary table S2. The figure legend is updated to provide a detailed description.
(5) Line 104 - 37.47% may be too many significant figures.
Corrected
(6) Line 123 - 1.003 Å may be too many significant figures.
Corrected
(7) Line 128 - Since the data are in the figure, you don't need to give the numbers, also too many significant figures.
Corrected
(8) Figure 3G - did the DCF also increase mound size? It sort of looks like it did.
Yes, the addition of DCF increases the mound size (now Figure. 2G).
(9) Figure 3I - the spore mass shown here for ADGF - looks like there are 3 stalks protruding from it; this can happen if a plate is handled roughly and the spore masses bang into each other and then merge
Thank you for pointing this out. The figure 3I (now Figure. 2I) is replaced.
(10) Lines 160-162 - since the data are in the figure, you don't need to give the numbers, also too many significant figures.
Corrected.
(11) Line 165 - ' ... that are involved in adenosine formation' needs a reference.
Reference is included.
(12) Line 205 - 'Addition of ADA to the CM of the mutant in one compartment.' - might clarify that the mutant is the ADGF mutant
Yes, revised to 'Addition of ADA to the CM of the adgf mutant in one compartment.'
(13) Lines 222-223 need a reference for caffeine acting as an adenosine antagonist.
Reference is included.
(14) Figure 8B - left - use a 0-4 or so scale so the bars are more visible.
Thank you for the suggestion. The scale of the y-axis is adjusted to 0-4 in Figure. 7B to enhance the visibility of the bars.
Reviewer #2 (Recommendations for the authors):
The paper describes new insights into the role of ADGF, an enzyme that catalyses the breakdown of adenosine in ammonia and inosine, in tip formation in Dictyostelium development.
A knockout of the gene results in a tipless mound stage arrest and the mounds formed are somewhat larger in size. Synergy experiments show that the effect of the mutation is non-cell autonomous and further experiments show that the mound arrest phenotype can be rescued by the provision of ammonia vapour. These observations are well documented. Furthermore, the paper contains a wide variety of experiments attempting to place the observed effects in known signalling pathways. It is suggested that ADGF may function downstream of DhkD, a histidine kinase previously implicated in ammonia signalling. Ammonia has long been described to affect different aspects, including differentiation of slug and culmination stages of Dictyostelium development, possibly through modulating cAMP signalling, but the exact mechanisms of action have not yet been resolved. The experiments reported here to resolve the mechanistic basis of the mutant phenotype need focusing and further work.
(1) The paper needs streamlining and editing to concentrate on the main findings and implications.
The manuscript will be revised extensively.
Below is a list of some more specific comments and suggestions.
(2) Introduction: Focus on what is relevant to understanding tip formation and the role of nucleotide metabolism and ammonia (see https://doi.org/10.1016/j.gde.2016.05.014).leading). This could lead to the rationale for investigating ADGF.
The manuscript will be revised extensively
(3) Lines 36-38 are not relevant. Lines 55-63 need shortening and to focus on ADGF, cellular localization, and substrate specificity.
The manuscript will be revised accordingly. Lines 36-38 will be removed, and the lines 55-63 will be shortened.
In humans, two isoforms of ADA are known including ADA1 and ADA2, and the Dictyostelium homolog of ADA2 is adenosine deaminase-related growth factor (ADGF). Unlike ADA that is intracellular, ADGF is extracellular and also has a growth factor activity (Li and Aksoy, 2000; Iijima et al., 2008). Loss-of-function mutations in ada2 are linked to lymphopenia, severe combined immunodeficiency (SCID) (Gaspar, 2010), and vascular inflammation due to accumulation of toxic metabolites like dATP (Notarangelo, 2016; Zhou et al., 2014).
(4) Results: This section would benefit from better streamlining by a separation of results that provide more mechanistic insight from more peripheral observations.
The manuscript will be revised and the peripheral observations (Figure. 2) will be shifted to the supplementary information.
(5) Line 84 needs to start with a description of the goal, to produce a knockout.
Details on the knockout will be elaborated in the revised manuscript. Line number 84 (now 75). Dictyostelium cell lines carrying mutations in the gene adgf were obtained from the genome wide Dictyostelium insertion (GWDI) bank and were subjected to further analysis to know the role of adgf during Dictyostelium development.
(6) Knockout data (Figure 1) can be simplified and combined with a description of the expression profile and phenotype Figure 3 F, G, and Figure 5. Higher magnification and better resolution photographs of the mutants would be desirable.
Thank you, as suggested the data will be simplified (section E will be removed) and combined with a description of the expression profile and, the phenotype images of Figure 3 F, G, and Figure 5 ( now Figure. 2 F, G, and Figure. 4) will be replaced with better images/ resolution.
(7) It would also be relevant to know which cells actually express ADGF during development, using in-situ hybridisation or promoter-reporter constructs.
To address whether adgf expression is cell type-specific, prestalk and prespore cells will be separated by fluorescence activated cell sorter (FACS), and thereafter, adgf expression will be examined in each population.
(8) Figure 2 - Information is less directly relevant to the topic of the paper and can be omitted (or possibly in Supplementary Materials).
Figure. 2 will be moved to supplementary materials.
(9) Figures 4A, B - It is shown that as could be expected ada activity is somewhat reduced and adenosine levels are slightly elevated. However, the fact that ada levels are low at 16hrs could just imply that differentiation of the ADGF- cells is blocked/delayed at an earlier time point. To interpret these data, it would be necessary to see an ada activity and adenosine time course comparison of wt and mutant, or to see that expression is regulated in a celltype specific manner that could explain this (see above). It would be good to combine this with the observation that ammonia levels are lower in the ADGF- mutant than wildtype and that the mutant phenotype, mound arrest can be rescued by an external supply of ammonia (Figure 6).
In Dictyostelium four isoforms of ADA including ADGF are present, and thus the time course of total ADA activity will also report the function of other isoforms. Further, a number of pathways, generate adenosine (Dunwiddie et al., 1997; Boison and Yegutkin, 2019). ADGF expression was examined at 0, 8, 12 and 16 h (Fig 1) and the ADA activity was assayed at 12 h, the time point where the expression gradually increases and reaches a peak at 16 h. Earlier, we have not shown the 12 h activity data which will be included in the revised version. ADGF expression was found to be highly elevated at 16 h and adenosine/ammonia levels were measured at the two points indicated in the mutant.
(10) Panel 4C could be combined with other measurements trying to arrive at more insight in the mechanisms by which ammonia controls tip formation.
Panel 4C (now 3C) illustrates the genes involved in the conversion of cAMP to adenosine. Since Figure. 3 focuses on adenosine levels and ADA activity in both WT and adgf mutants, we have retained Panel 3C in Figure. 3, for its relevance to the experiment.
(11) There is a large variety of experiments attempting to link the mutant phenotype and its rescue by ammonia to cAMP signalling, however, the data do not yet provide a clear answer.
It is well known that ammonia increases cAMP levels (Riley and Barclay, 1990; Feit et al., 2001) and adenylate cyclase activity (Cotter et al., 1999) in D. discoideum, and exposure to ammonia increases acaA expression (Fig 7B) suggesting that ammonia regulates cAMP signaling. To address the concerns, cAMP levels will be quantified in the mutant after ammonia treatment.
(12) The mutant is shown to have lower cAMP levels at the mound stage which ties in with low levels of acaA expression (Figures 7A and B), also various phosphodiesterases, the extracellular phosphodiesterase pdsa and the intracellular phosphodiesterase regA show increased expression. Suggesting a functional role for cAMP signalling is that the addition of di cGMP, a known activator of acaA, can also rescue the mound phenotype (Figure 7E). There appears to be a partial rescue of the mound arrest phenotype level by the addition of 8Br-cAMP (fig 7C), suggesting that intracellular cAMP levels rather than extracellular cAMP signalling can rescue some of the defects in the ADGF- mutant. Better images and a time course would be helpful.
The relevant images will be replaced and a developmental time course after 8-Br-cAMP treatment will be included in the revised manuscript (Figure. 6D).
(13) There is also the somewhat surprising observation that low levels of caffeine, an inhibitor of acaA activation also rescues the phenotype (Figure 7F).
With respect to caffeine action on cAMP levels, the reports are contradictory. Caffeine has been reported to increase adenylate cyclase expression thereby increasing cAMP levels (Hagmann, 1986) whereas Alvarez-Curto et al., (2007) found that caffeine reduced intracellular cAMP levels in Dictyostelium. Caffeine, although is a known inhibitor of ACA, is also known to inhibit PDEs (Nehlig et al., 1992; Rosenfeld et al., 2014). Therefore, if caffeine differentially affects ADA and PDE activity, it may potentially counterbalance the effects and rescue the phenotype.
(14) The data attempting to asses cAMP wave propagation in mounds (Fig 7H) are of low quality and inconclusive in the absence of further analysis. It remains unresolved how this links to the rescue of the ADGF- phenotype by ammonia. There are no experiments that measure any of the effects in the mutant stimulated with ammonia or di-cGMP.
The relevant images will be replaced (now Figure. 6H). Ammonia by increasing acaA expression (Figure. 7B), and cAMP levels (Figure. 7C) may restore spiral wave propagation, thereby rescuing the mutant.
(15) A possible way forward could also come from the observation that ammonia can rescue the wobbling mound arrest phenotype from the histidine kinase mutant dhkD null mutant, which has regA as its direct target, linking ammonia and cAMP signalling. This is in line with other work that had suggested that another histidine kinase, dhkC transduces an ammonia signal sensor to regA activation. A dhkC null mutant was reported to have a rapid development phenotype and skip slug migration (Dev. Biol. (1998) 203, 345). There is no direct evidence to show that dhkD acts upstream of ADGF and changes in cAMP signalling, for instance, measurements of changes in ADA activity in the mutant.
cAMP levels will be quantified in the dhkD mutant after ammonia treatment and accordingly, the results will be revised.
(16) The paper makes several further observations on the mutant. After 16 hrs of development the adgf- mutant shows increased expression of the prestalk cell markers ecmA and ecmB and reduced expression of the prespore marker pspA. In synergy experiments with a majority of wildtype, these cells will sort to the tip of the forming slug, showing that the differentiation defect is cell autonomous (Fig 9). This is interesting but needs further work to obtain more mechanistic insight into why a mutant with a strong tip/stalk differentiation tendency fails to make a tip. Here again, knowing which cells express ADGF would be helpful.
The adgf mutant shows increased prestalk marker expression in the mound but do not form a tip. It is well known that several mound arrest mutants form differentiated cells but are blocked in development with no tips (Carrin et al., 1994). This is addressed in the discussions (539). To address whether adgf expression is cell type-specific, prestalk and prespore cells will be separated by fluorescence activated cell sorter (FACS), and thereafter, adgf expression will be examined in each population.
(17) The observed large mound phenotype could as suggested possibly be explained by the low ctn, smlA, and high cadA and csA expression observed in the mutant (Figure 3). The expression of some of these genes (csA) is known to require extracellular cAMP signalling. The reported low level of acaA expression and high level of pdsA expression could suggest low levels of cAMP signalling, but there are no actual measurements of the dynamics of cAMP signalling in this mutant to confirm this.
The acaA expression was examined at 8 and 12 h (Figure. 6B) and cAMP levels were measured at 12 and 16 h in the adgf mutants (Figure. 6A). Both acaA expression and cAMP levels were reduced, suggesting that cells expressing adgf regulate acaA expression and cAMP levels. This regulation, in turn, is likely to influence cAMP signaling, collective cell movement within mounds, ultimately driving tip development. Exposure to ammonia led to increased acaA expression (Figure. 7B) in in WT. Based on the comments above, cAMP levels will be measured in the mutant before and after rescue with ammonia.
(18) Furthermore, it would be useful to quantify whether ammonia addition to the mutant reverses mound size and restores any of the gene expression defects observed.
Ammonia treatment soon after plating or six hours after plating, had no effect on the mound size (Figure. 5G).
(19) There are many experimental data in the supplementary data that appear less relevant and could be omitted Figure S1, S3, S4, S7, S8, S9, S10.
Figure S8, S9, S10 are omitted. We would like to retain the other figures
Figure S1 (now Figure. S2): It is widely believed that ammonia comes from protein (White and Sussman, 1961; Hames and Ashworth, 1974; Schindler and Sussman, 1977) and RNA (Walsh and Wright, 1978) catabolism. Figure. S2 shows no significant difference in protein and RNA levels between WT and adgf mutant strains, suggesting that adenosine deaminaserelated growth factor (ADGF) activity serves as a major source of ammonia and plays a crucial role in tip organizer development in Dictyostelium. Thus, it is important to retain this figure.
Figure S3 (now Figure. S4): The figure shows the treatment of various mound arrest mutants and multiple tip mutants with ADA enzyme and DCF, respectively, to investigate the pathway through which adgf functions. Additionally, it includes the rescue of the histidine kinase mutant dhkD with ammonia, indicating that dhkD acts upstream of adgf via ammonia signalling. Therefore, it is important to retain this figure.
Figure S4 (now Figure. S5): This figure represents the developmental phenotype of other deaminase mutants. Unlike adgf mutants, mutations in other deaminases do not result in complete mound arrest, despite some of these genes exhibiting strong expression during development. This underscores the critical role of adenosine deamination in tip formation. Therefore, let this figure be retained.
Figure S7 (now Figure. S8): Figure S8 presents the transcriptomic profile of ADGF during gastrulation and pre-gastrulation stages across different organisms, indicating that ADA/ADGF is consistently expressed during gastrulation in several vertebrates (Pijuan-Sala et al., 2019; Tyser et al., 2021). Notably, the process of gastrulation in higher organisms shares remarkable similarities with collective cell movement within the Dictyostelium mound (Weijer, 2009), suggesting a previously overlooked role of ammonia in organizer development. This implies that ADA may play a fundamental role in regulating morphogenesis across species, including Dictyostelium and vertebrates. Therefore, we would like to retain this figure.
(20). Given the current state of knowledge, speculation about the possible role of ADGF in organiser function in amniotes seems far-fetched. It is worth noting that the streak is not equivalent to the organiser. The discussion would benefit from limiting itself to the key results and implications.
The discussion is revised accordingly by removing the speculative role of ADGF in organizer function in amniotes. The lines “It is likely that ADA plays a conserved, fundamental role in regulating morphogenesis in Dictyostelium and other organisms including vertebrates” have been removed.
Reviewer #2 (Public review):
Summary:
The ADP-ribosyltransferase tankyrase controls many biological processes, many of which are relevant to human disease. This includes Wnt/beta-catenin signalling, which is dysregulated in many cancers, most notably colorectal cancer. Tankyrase is a positive regulator of Wnt/beta-catenin signalling in that it counters the activity of the beta-catenin destruction complex (DC). Catalytic inhibition of tankyrase not only blocks PAR-dependent ubiquitylation and degradation of AXIN1/2, the central scaffolding protein in the DC, but also tankyrase itself. As a result, blocking tankyrase gives rise to tankyrase accumulation, which may accentuate its non-catalytic functions, which have been proposed to drive Wnt/beta-catenin signalling. Most tankyrase catalytic inhibitors have shown limited efficacy and substantial toxicity in vivo. By developing tankyrase-directed PROTACs, the authors aim to block both catalytic and non-catalytic functions of tankyrase, aspiring to achieve a more complete inhibition of Wnt/beta-catenin signalling. The successfully developed PROTAC, based on the existing catalytic inhibitor IWR1, IWR1-POMA, induces the degradation of both TNKS and TNKS2, blocks beta-catenin-dependent transcription without stabilising the DC in puncta/degradasomes, and inhibits cancer cell growth in vitro. Mechanistically, this points to a scaffolding role of tankyrase in the DC, at least under conditions of tankyrase catalytic inhibition, in line with previous proposals.
Strengths:
The study clearly illustrates the incentive for developing a tankyrase degrader, namely, to abolish both catalytic and non-catalytic functions of tankyrase. By and large, the study achieves these ambitions, and the findings support the main conclusions, although the statement that a more complete inhibition of the pathway is achieved requires corroboration. The proteomics studies are powerful. IWR1-POMA constitutes a very useful tool to re-evaluate targeting of tankyrase in oncogenic Wnt/beta-catenin signalling. The paired compounds will benefit investigations of tankyrase scaffolding functions across many different biological systems controlled by tankyrase. The findings are exciting.
Weaknesses:
Although the results are promising and mostly compelling, the claim that the PROTACs provide "a deeper suppression of the WNT/β-catenin pathway activity" requires further corroboration, particularly at endogenous tankyrase levels.
There are also some other points that, if considered, would further improve the manuscript, as detailed below.
(1) Abstract and line 62: Many catalytic tankyrase inhibitors tend to display toxicity, which is likely on-target (e.g., 10.1177/0192623315621192; 10.1158/0008-5472). This constitutes the main limiting factor for these compounds. An incomplete inhibition of Wnt/beta-catenin signalling may contribute to the challenges, but this does not appear to be the dominant problem. A more prominent introduction to this important challenge is probably expected by the field.
(2) The authors do a good job in setting the scene for the need for tankyrase degraders. Their observations relating to the formation of puncta (degradasomes) being tankyrase-dependent are compatible with a previous study by Martino-Echarri et al. 2016 (10.1371/journal.pone.0150484): simultaneous silencing of TNKS and TNKS2 by RNAi abolishes degradasome formation. The paper is cited as reference 17, but only in passing, and deserves more prominence. (It includes an entire paragraph titled "Expression of tankyrases 1 and 2 is required for TNKSi-induced formation of axin puncta").
(3) Moreover, the scaffolding concept has been discussed comprehensively in other studies: 10.1111/bph.14038 and more recently 10.1042/BCJ20230230. There are also a few studies that focus on targeting the ankyrin repeat clusters of tankyrase to disengage substrates (10.1038/s41598-020-69229-y; 10.1038/s41598-019-55240-5) that illustrate the concept of blocking the scaffolding function. In that sense, the hypotheses are mature, and it is interesting to see some of them supported in this study. The authors could improve how they set their work into the context of these other efforts and proposals.
(4) In several places in the manuscript, the DC is referred to as "biomolecular condensate", at times even as a "classic example", implying that it operates through phase separation. This has not been demonstrated. In fact, super-resolution microscopy indicates that the puncta are not droplet-like (10.7554/eLife.08022), which would argue against the condensate hypothesis.
(5) It is beautiful to be able to use IWR1 and IWR1-POMA at identical concentrations for direct comparisons. However, this requires the two compounds to bind to tankyrase similarly well and reach the target to a comparable extent. How sure are authors that target engagement is comparable? Has this been evaluated?
(6) Figure 1F: It is not immediately apparent how IWR1-POMA shows more complete containment of Wnt/beta-catenin signalling. Most Wnt/beta-catenin targets lie close to the perfect diagonal, so I do not see how the statement "that IWR1-POMA controlled WNT/β-catenin signaling more effectively than IWR1" (in the legend of Figure 1F) is supported. Minimally, an expanded explanation would benefit the reader. Providing the colour-coding legend directly in the figure would help improve clarity. Also, the panel is very small and may benefit from a different presentation in the figure.
(7) Figure 2: The conclusion of a "deeper suppression" of signalling relies on overexpression of tankyrase in an otherwise tankyrase-null background. Have the authors attempted to measure reporter activity or endogenous gene expression without tankyrase overexpression, in Wnt3a-stimulated cells (in the context of a normal Wnt/beta-catenin pathway) or CRC cells at the basal level? Non-catalytic activity in a similar assay has previously been observed upon tankyrase overexpression (10.1016/j.molcel.2016.06.019). Whether or not there is a substantial scaffolding effect at endogenous tankyrase levels after tankyrase inhibition remains unconfirmed, and the PROTAC is a valuable tool to address this important question. The findings presented in Figure S7C and D go some way towards answering this question - these data could be presented more prominently, and similar assays could be performed in other cell systems.
(8) Line 237/238: "TNKS accumulation negatively impacts the catalytic activity of the DC (Figure 5D)" - the data do not show this. Beta-catenin levels are a surrogate readout for DC function (phosphorylation and ubiquitylation). Minimally, this requires rewording, with reference to beta-catenin levels.
(9) Line 303-304: Beta-catenin is thought to exchange at beta-catenin degradasomes; this is clear from previous FRAP assays and the observation that phospho-beta-catenin accumulates in degradasomes upon proteasome inhibition (10.1158/1541-7786.MCR-15-0125). However, degradasome size hasn't, to my knowledge, been related to activity. Can this be clarified, please?
(10) There are previous hypotheses/proposals that the sensitivity of CRC cells to tankyrase inhibition correlates with APC truncation or PIK3CA status (10.1158/1535-7163.MCT-16-0578; 10.1038/s41416-023-02484-8). Have the authors considered expanding their cell line panel (Figure S7) to sample a wider range of cell lines, including some that are wild-type with regard to APC or Wnt/beta-catenin signalling in general? This would be a valuable addition to the work. Quantitated colony formation data could be moved to the main body of the manuscript.
(11) The manuscript only mentions toxicity (i.e., therapeutic window) in the last sentence of the Discussion section. As this is THE main challenge with tankyrase inhibitors (as mentioned above), can the authors expand their discussion of this aspect? Is there an expectation that PROTACs may be less toxic?
(12) Figures 3, 4, 5A: For fluorescence microscopy experiments, can these be quantified, and can repeat data be included?
(13) Figure 4, S6: An additional channel illustrating the distribution of cells (e.g., nuclei, cytoskeleton, or membrane) would be helpful for orientation and context for the AXIN1 signal.
(14) How were cytosolic fractions of cells prepared to assess cytosolic beta-catenin levels? This detail is missing from the methods.
RRID:SCR_021758
DOI: 10.1021/acsabm.5c01736
Resource: Colorado State University Analytical Resources Core Facility (RRID:SCR_021758)
Curator: @scibot
SciCrunch record: RRID:SCR_021758
moved to https://bafybeihtxbr3mkagapvdagtjrllwmoptxu6tuiufgrxmlrdqxyqwueirwq.ipfs.dweb.link/?filename=index.html&urn=/hyperpost/🌐/♖/indy0/🌐/1/gyuri/⭕/0/
move to
https://bafybeihtxbr3mkagapvdagtjrllwmoptxu6tuiufgrxmlrdqxyqwueirwq.ipfs.dweb.link/?filename=index.html&urn=/hyperpost/%F0%9F%8C%90/%E2%99%96/indy0/%F0%9F%8C%90/1/gyuri/%E2%AD%95/0/
original draft
https://bafybeif3cet7g43jvug3hvaikdjzecuqdv2f2ldyno5ubjcaykfnlnzuve.ipfs.dweb.link?filename=index.html
dynamic listing in reverse chronological ordera list of all the recently annotated pages
Skip Stream
Use faceted search of annotations to get a slip stream of active Pages
Reviewer #3 (Public review):
Summary:
The manuscript by Shukla and colleagues presents a comprehensive study that addresses a central question in kinesin-1 regulation - how cargo binding to the kinesin light chain (KLC) tetratricopeptide repeat (TPR) domains triggers activation of full-length kinesin-1 (KHC). The authors combine AlphaFold3 modeling, biophysical analysis (fluorescence polarization, hydrogen-deuterium exchange), and electron microscopy to derive a mechanistic model in which the KLC-TPR domains dock onto coiled-coil 1 (CC1) of the KHC to form the "TPR shoulder," stabilizing the autoinhibited (λ-particle) conformation. Binding of a W/Y-acidic cargo motif (KinTag) or deletion of the CC1 docking site (TDS) dislocates this shoulder, liberating the motor domains and enhancing accessibility to cofactors such as MAP7. The results link cargo recognition to allosteric structural transitions and present a unified model of kinesin-1 activation.
Strengths:
(1) The study addresses a fundamental and long-standing question in kinesin-1 regulation using a multidisciplinary approach that combines structural modeling, quantitative biophysics, and electron microscopy.
(2) The mechanistic model linking cargo-induced dislocation of the TPR shoulder to activation of the motor complex is well supported by both structural and biochemical evidence.
(3) The authors employ elegant protein-engineering strategies (e.g., ElbowLock and ΔTDS constructs) that enable direct testing of model predictions, providing clear mechanistic insight rather than purely correlative data.
(4) The data are internally consistent and align well with previous studies on kinesin-1 regulation and MAP7-mediated activation, strengthening the overall conclusion.
Weaknesses:
(1) While the EM and HDX-MS analyses are informative, the conformational heterogeneity of the complex limits structural resolution, making some aspects of the model (e.g., stoichiometry or symmetry of TPR docking) indirect rather than directly visualized.
(2) The dynamics of KLC-TPR docking and undocking remain incompletely defined; it is unclear whether both TPR domains engage CC1 simultaneously or in an alternating fashion.
(3) The interplay between cargo adaptors and MAP7 is discussed but not experimentally explored, leaving open questions about the sequence and exclusivity of their interactions with CC1.
https://bafybeihtxbr3mkagapvdagtjrllwmoptxu6tuiufgrxmlrdqxyqwueirwq.ipfs.dweb.link/?filename=index.html&urn=/hyperpost/🌐/♖/indy0/🌐/1/gyuri/⭕/0/
able to share indy0pad created pages for annotations without needing to rely on via.hypothesis
Los israelitas se olvidaron otra vez de Jehová
(Jueces 10:14-16) Vayan a pedirles ayuda a los dioses que han elegido;+ que ellos los salven en tiempos de angustia”.+ 15 Sin embargo, los israelitas le dijeron a Jehová: “Hemos pecado. Haz con nosotros lo que sea bueno a tus ojos. Pero sálvanos esta vez, por favor”. 16 Y se deshicieron de los dioses extranjeros y volvieron a servir a Jehová.+ Entonces él ya no pudo soportar más* ver sufrir a Israel.+
theme: Knowledge Serving Commons
full set of wiki-articles, you can find them
https://bafybeicuznswtf2d3sevop4ripd5hy77nauwbicurnruv4ocf3w32ke7ha.ipfs.dweb.link/?filename=index.html&&urn=🧊/♖/hyperpost/~/indyweb/📓/20/25/11/4/do/🏛%EF%B8%8F/growingcommons.substack.com/p/structuring-knowledge-commons
SlipStream
or slip stream
wake of your trailblae
indyWiki0pad

practical.steps - indy0pad & indy0wiki.pad interplay
// experiment - indy0pad & indy0wiki.pad interplay
top-slice
autopoiesis of the IndyWeb
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public Reviews):
Weaknesses:
A limitation of the study is the reliance on standard techniques; however, this is a minor concern that does not diminish the overall impact or significance of the work.
We agree that standard techniques were utilized. We believe this approach enhances the reliability and reproducibility of our findings. These methods are well-validated in the field and allow for robust interpretation of the results presented.
Reviewer #2 (Public Reviews):
Weaknesses:
(1) Clarify the strain background of the DBA/2J GPNMB+ mice: While DBA/2J GPNMB+ is described as a control, it would help to explicitly state whether these are transgenically rescued mice or another background strain. Are they littermates, congenic, or a separate colony?
The following language was added to the manuscript, “The DBA/2J GPNMB+ mice are a coisogenic strain purchased from Jackson Laboratories. Jackon Laboratories generated these mice by knocking in the wild-type allele of Gpnmb into the DBA/2J background. By doing so, they rescued the phenotype of the DBA/2J mice. This description has been highlighted in our previous publications (Abdelmagid et al., 2014; Abdelmagid et al., 2015).”
(2) Provide exact sample sizes and variance in all figure legends: Some figures (e.g., Figure 2 panels) do not consistently mention how many replicates were used (biological vs. technical) for each experimental group. Standardizing this across all panels would improve reproducibility.
The manuscript has been updated to include replicates in each figure legend.
(3) Expand on potential sex differences: The DMM model is applied only in male mice, which is noted in the methods. It would be helpful if the authors added 1-2 lines in the discussion acknowledging potential sex-based differences in OA progression and GPNMB function.
To our knowledge there are no sexbased differences in OA progression and GPNMB function in the literature. It was initially reported that only male C57BL/6J mice (Jackson Laboratories) develop OA following DMM however, recent literature has shown that both male and female mice develop the disease (Hwang et al., 2021; Ma et al., 2007). For the purpose of this manuscript, only male mice were used to provide preliminary results, however, we plan to repeat the included studies in female mice in the near future.
(4) Visual clarity in schematic (Figure 7): The proposed mechanism is helpful, but the text within the schematic is somewhat dense and could be made more readable with spacing or enlarged font. Also, label the MAPK/ERK pathway explicitly in panel B.
We updated the schematic diagram in figure 7 and the figure legend.
Reviewer #1 (Recommendations for the Authors):
Several concerns must be addressed to improve the clarity and scientific rigor of the manuscript:
(1) Abstract: Specify which MMPs and MAPKs are modulated by osteoactivin.
We specified the MMPs and clarified that GPNMB plays a role in pERK inhibition following inflammation induced by IL-1β stimulation.
(2) Human explant validation: The regulation of MMP-9, MMP-13, and IL-6 should be validated in the human cartilage explant model to support the claim that "GPNMB has an anti-inflammatory role in human primary chondrocytes" (line 123). Additionally, the anatomical origin of the explants must be stated.
Thank you very much for the recommendation. We agree that validating the explant culture for MMP-9, MMP-13, and IL-6 would strengthen our data. Unfortunately, this experiment has been terminated and we no longer have access to the tissue. Human explants were obtained from discarded knee articular cartilage following arthroplasty. The manuscript has been updated to include this information.
(3) DBA/2J GPNMB expression: GPNMB is known to be produced as a truncated protein in DBA/2J cells. The manuscript should address why its expression is reduced. Does this involve mRNA instability? Also, the nomenclature "DBA/2J GPNMB+" versus "DBA/2J" is confusing, especially since both mRNA and protein are still detectable, albeit at reduced levels. Figure 2C is not convincing; therefore, Figures 2C and 2D can be omitted.
The following language was added to the manuscript, “Our results are consistent with the literature which shows that that the GPNMB gene in DBA/2J mice carries a nonsense mutation that leads to reduced RNA stability (Anderson et al., 2008).” We can appreciate that the nomenclature "DBA/2J GPNMB+" versus "DBA/2J" could be confusing. However, this is the standard language used in multiple publications, and we want to remain consistent with the literature. Based on your recommendation we have removed Figure 2 C and D and updated the methods and results sections accordingly.
(4) Figures 2J-L: The claim that gene expression changes are "significantly higher in DBA/2J animals compared to fold changes seen in chondrocytes from DBA/2J GPNMB+ controls" is not supported by the current presentation. The data should be plotted on the same graphs, and appropriate statistical analysis (e.g., two-way ANOVA) must be performed.
Graphs for figure 2 have been updated and the appropriate analyses have been performed.
(5) Figure 6: The GPNMB expression data in the presence and absence of IL-1β at 0 and 10 minutes are missing.
We apologize for the confusion. We corrected the mistake and removed the mention of the timepoints 0 and 10 minutes.
Reviewer #2 (Recommendations for the Authors):
Consider unifying terminology around "GPNMB" and "osteoactivin": The term "osteoactivin" is used in some contexts and "GPNMB" in others. Since the focus is GPNMB's role in cartilage, suggest using a single term throughout to prevent confusion.
Thank you for your comment. We include osteoactivin for clarification purposes once in the abstract, introduction and discussion.
In summary, we believe we have addressed all comments/concerns raised by the reviewers. We appreciate the opportunity to improve the quality of our manuscript.
References
Abdelmagid, S. M., Belcher, J. Y., Moussa, F. M., Lababidi, S. L., Sondag, G. R., Novak, K. M., Sanyurah, A. S., Frara, N. A., Razmpour, R., & Del Carpio-Cano, F. E. (2014). Mutation in osteoactivin decreases bone formation in vivo and osteoblast differentiation in vitro. The American journal of pathology, 184(3), 697-713.
Abdelmagid, S. M., Sondag, G. R., Moussa, F. M., Belcher, J. Y., Yu, B., Stinnett, H., Novak, K., Mbimba, T., Khol, M., Hankenson, K. D., Malcuit, C., & Safadi, F. F. (2015). Mutation in Osteoactivin Promotes Receptor Activator of NFκB Ligand (RANKL)-mediated Osteoclast Differentiation and Survival but Inhibits Osteoclast Function. J Biol Chem, 290(33), 2012820146. https://doi.org/10.1074/jbc.M114.624270
Anderson, M. G., Nair, K. S., Amonoo, L. A., Mehalow, A., Trantow, C. M., Masli, S., & John, S. W. (2008). GpnmbR 150Xallele must be present in bone marrow derived cells to mediate DBA/2J glaucoma. BMC genetics, 9(1), 1-14.
Hwang, H., Park, I., Hong, J., Kim, J., & Kim, H. (2021). Comparison of joint degeneration and pain in male and female mice in DMM model of osteoarthritis. Osteoarthritis and Cartilage, 29(5), 728738.
Ma, H.-L., Blanchet, T., Peluso, D., Hopkins, B., Morris, E., & Glasson, S. (2007). Osteoarthritis severity is sex dependent in a surgical mouse model. Osteoarthritis and Cartilage, 15(6), 695-700.
In this three-part diagram, tension is graphed on the Y axis, and time is graphed on the X axis. Tension is introduced at the end of the first act or the beginning of second, it rises and rises throughout the second act, and then it is released in a climactic moment. The third act addresses the aftermath and the results that spring from this release of tension.
I really like how this three-part diagram connects the structure of tension to the flow of time because it makes the shape of a story feel almost mathematical. The idea that tension is introduced right at the transition into the second act feels accurate to most stories I enjoy—there’s always that moment where things shift from setup to real conflict. I agree that the gradual rise through Act Two is what keeps audiences engaged, because it mirrors how stakes build in real life. What stood out to me most is the idea that the third act isn’t just a resolution, but a space to examine the consequences of the climax, which makes the story feel more meaningful instead of just abruptly ending. Overall, this model helped me visualize why the “And, But, Therefore” structure works so well in narrative writing.
Author Response
Reviewer #1 (Public Review):
In this manuscript, the authors find CpGs within 500Kb of a gene that associate with transcript abundance (cis-eQTMs) in children from the HELIX study. There is much to admire about this work. With two notable exceptions, their work is solid and builds/improves on the work that came before it. Their catalogue of eQTMs could be useful to many other researchers that utilize methylation data from whole blood samples in children. Their annotation of eQTMs is well thought out and exhaustive. As this portion of the work is descriptive, most of their methods are appropriate.
Unfortunately, their use of results from a model that does not account for cell-type proportions across samples diminishes the utility and impact of their findings. I believe that their catalog of eQTMs contains a great deal of spurious results that primarily represent the differences in cell-type proportions across samples.
Lastly, the authors postulate that the eQTM gene associations found uniquely in their unadjusted model (in comparison to results from a model that does account for cell type proportion) represent cell-specific associations that are lost when a fully-adjusted model is assumed. To test this hypothesis, the authors appear to repurpose methods that were not intended for the purposes used in this manuscript. The manuscript lacks adequate statistical validation to support their repurposing of the method, as well as the methodological detail needed to peer review it. This section is a distraction from an otherwise worthy manuscript. But provide evidences that enriched for cell sp CpGs.
Major points
- Line 414-475: In this section, the authors are suggesting that CpGs that are significant without adjusting for cell type are due to methylation-expression associations that are found only in one cell type, while association found in the fully adjusted model are associations that are shared across the cell types. I do not agree with this hypothesis, as I do not agree that the confounding that occurs when cell-type proportions are not accounted for would behave in this way. Although restricting their search for eQTMs to only those CpGs proximal to a gene will reduce the number of spurious associations, a great deal of the findings in the authors' unadjusted model likely reflect differences in cell-type proportions across samples alone. The Reinius manuscript, cited in this paper, indicates that geneproximal CpGs can have methylation patterns that vary across cell types.
Following reviewers’ recommendations, we have reconsidered our initial hypothesis about the role of cellular composition in the association between methylation and gene expression. Although we still think that some of the eQTMs only found in the model unadjusted for cellular composition could represent cell specific effects, we acknowledge that the majority might be confounded by the extensive gene expression and DNA methylation differences between cell types. Also, we recognize that more sophisticated statistical tests should be applied to prove our hypothesis. Because of this, we have decided to report the eQTMs of the model adjusted for cellular composition in the main manuscript and keep the results of the model unadjusted for cellular composition only in the online catalogue.
- Line 476-488: Their evidence due to F-statistics is tenuous. The authors do not give enough methodological detail to explain how they're assessing their hypothesis in the results or methods (lines 932-946) sections. The methods they give are difficult to follow. The results in figure S19A are not compelling. The citation in the methods (by Reinius) do not make sense, because Reinius et al did not use F-statistics as a proxy for cell type specificity. The citation that the authors give for this method in the results does not appear to be appropriate for this analysis, either. Jaffe and Irizarry state that a CpG with a high Fstatistic indicates that the methylation at that CpG varies across cell type. They suggest removing these CpGs from significant results, or estimating and correcting for cell type proportions, as their presence would be evidence of statistical confounding. The authors of this manuscript indicate that they find higher F-statistics among the eQTMs uniquely found in the unadjusted model, which seems to only strengthen the idea that the unadjusted model is suffering from statistical confounding.
We recognize the miss-interpretation of the F-statistic in relation to cellular composition. We have deleted all this part from the updated version of the manuscript.
- The methods used to generate adjusted p-values in this manuscript are not appropriate as they are written. Further, they are nothing like the methods used in the paper cited by the authors. The Bonder paper used permutations to estimate an empirical FDR and cites a publication by Westra et al for their method (below). The Westra paper is a better one to cite, because the methods are more clear. Neither the Bonder nor the Westra paper uses the BH procedure for FDR.
Westra, H.-J. et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 45, 1238-1243 (2013).
We apologize for this misleading citation. Although Bonder et al applied a permutation approach to adjust for multiple testing, our approach was inspired by the method applied in the GTEx project (GTEx consortium, 2020), using CpGs instead of SNPs. The citation has been corrected in the manuscript. Moreover, we have explained in more detail the whole multiple-testing processes in the Material and Methods section (page 14, line 316):
“To ensure that CpGs paired to a higher number of Genes do not have higher chances of being part of an eQTM, multiple-testing was controlled at the CpG level, following a procedure previously applied in the Genotype-Tissue Expression (GTEx) project (Gamazon et al., 2018). Briefly, our statistic used to test the hypothesis that a pair CpGGene is significantly associated is based on considering the lowest p-value observed for a given CpG and all its pairs Gene (e.g. those in the 1 Mb window centered at the TSS). As we do not know the distribution of this statistic under the null, we used a permutation test. We generated 100 permuted gene expression datasets and ran our previous linear regression models obtaining 100 permuted p-values for each CpG-Gene pair. Then, for each CpG, we selected among all CpG-Gene pairs the minimum p-value in each permutation and fitted a beta distribution that is the distribution we obtain when dealing with extreme values (e.g. minimum) (Dudbridge and Gusnanto, 2008). Next, for each CpG, we took the minimum p-value observed in the real data and used the beta distribution to compute the probability of observing a lower p-value. We defined this probability as the empirical p-value of the CpG. Then, we considered as significant those CpGs with empirical p-values to be significant at 5% false discovery rate using BenjaminiHochberg method. Finally, we applied a last step to identify all significant CpG-Gene pairs for all eCpGs. To do so, we defined a genome-wide empirical p-value threshold as the empirical p-value of the eCpG closest to the 5% false discovery rate threshold. We used this empirical p-value to calculate a nominal p-value threshold for each eCpG, based on the beta distribution obtained from the minimum permuted p-values. This nominal p-value threshold was defined as the value for which the inverse cumulative distribution of the beta distribution was equal to the empirical p-value. Then, for each eCpG, we considered as significant all eCpG-Gene variants with a p-value smaller than nominal p-value.”
References:<br /> GTEx consortium, The GTEx Consortium atlas of genetic regulatory effects across human tissues, Science (2020) Sep 11;369(6509):1318-1330. doi: 10.1126/science.aaz1776.
Reviewer #2 (Public Review):
Strength:
Comprehensive analysis Considering genetic factors such as meQTL and comparing results with adult data are interesting.
We thank the reviewer for his/her positive feedback on the manuscript. We agree that the analysis of genetic data and the comparison with eQTMs described in adults are two important points of the study.
Weakness:
- Manuscript is not summarized well. Please send less important findings to supplementary materials. The manuscript is not well written, which includes every little detail in the text, resulting in 86 pages of the manuscript.
Following reviewers’ comments, we have simplified the manuscript. Now only the eQTMs identified in the model adjusted for cellular composition are reported. In addition, functional enrichment analyses have been simplified without reporting all odds ratios (OR) and p-values, which can be seen in the Figures.
- Any possible reason that the eQTM methylation probes are enriched in weak transcription regions? This is surprising.
Bonder et al also found that blood eQTMs were slightly enriched for weak transcription regions (TxWk). Weak transcription regions are highly constitutive and found across many different cell types (Roadmap Epigenetics Consortium, 2015). However, hematopoietic stem cells and immune cells have lower representation of TxWk and other active states, which may be related to their capacity to generate sub-lineages and enter quiescence.
Given that we analyzed whole blood and that ROADMAP chromatin states are only available for blood specific cell types, each CpG in the array was annotated to one or several chromatin states by taking a state as present in that locus if it was described in at least 1 of the 27 bloodrelated cell types. By applying this strategy we may be “over-representing” TxWk chromatin states, in the case TxWk are cell-type specific. As a result, even if each blood cell type might have few TxWk, many positions can be TxWk in at least one cell type, inflating the CpGs considered as TxWk. This might have affected some of the enrichments.
On the other hand, CpG probe reliability depends on methylation levels and variance. TxWk regions show high methylation levels, which tend to be measured with more error. This also might have impacted the results, however the analysis considering only reliable probes (ICC >0.4) showed similar enrichment for TxWk.
Besides these, we do not have a clear answer for the question raised by the reviewer.
References:
Bonder MJ, Luijk R, Zhernakova D V, Moed M, Deelen P, Vermaat M, et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet [Internet]. 2017 [cited 2017 Nov 2];49:131–8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27918535
Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh KH, Feizi S, Karlic R, Kim AR, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJ, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai LH, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M. Integrative analysis of 111 reference human epigenomes. Nature. 2015 Feb 19;518(7539):317-30. doi: 10.1038/nature14248. PMID: 25693563; PMCID: PMC4530010.
- The result that the magnitude of the effect was independent of the distance between the CpG and the TC TSS is surprising. Could you draw a figure where x-axis is the distance between the CpG site and TC TSS and y-axis is p-value?
As suggested by the reviewer, we have taken a more detailed look at the relationship between the effect size and the distance between the CpG and the TC’s TSS. First, we confirmed that the relative orientation (upstream or downstream) did not affect the strength of the association (p-value=0.68). Second, we applied a linear regression between the absolute log2 fold change and the log10 of the distance (in absolute value), finding that they were inversely related. We have updated the manuscript with this information (page 22, line 504):
“We observed an inverse linear association between the eCpG-eGene’s TSS distance and the effect size (p-value = 7.75e-9, Figure 2B); while we did not observe significant differences in effect size due to the relative orientation of the eCpG (upstream or downstream) with respect to the eGene’s TSS (p-value = 0.68).”
Results are shown in Figure 2B. Of note, we winsorized effect size values in order to improve the visualization. The winsorizing process is also explained in Figure 2 legend. Moreover, we have done the plot suggested by the reviewer (see below). It shows that associations with smallest p-values are found close to the TC’s TSS. Nonetheless, as this pattern is also observed for the effect sizes, we have decided to not include it in the manuscript.
- Concerned about too many significant eQTMs. Almost half of genes are associated with methylation. I wonder if false positives are well controlled using the empirical p-values. Using empirical p-value with permutation may mislead since especially you only use 100 permutations. I wonder the result would be similar if they compare their result with the traditional way, either adjusting p-values using p-values from entire TCs or adjusting pvalues using a gene-based method as commonly used in GWAS. Compare your previous result with my suggestion for the first analysis.
Despite the number of genes (TCs) whose expression is associated with DNA methylation is quite high, we do not think this is due to not correctly controlling false positives. Our approach is based on the method used by GTEx (GTEx consortium) and implemented in the FastQTL package (Ongen et al. 2016), to control for positives in the eQTLs discovery. As in GTEx, we run 100 permutations to estimate the parameters of a beta distribution, which we used to model the distribution of p-values for each CpG. Then, to correct for the number of TCs among significant CpGs, we applied False Discovery Rate (FDR) at a threshold < 0.05. Finally, we defined the final set of significant eQTMs using the beta distribution defined in a previous step.
For illustration, we compared the number of eQTMs with our approach to what we would obtain by uniquely applying the FDR method (adjusted p-value <0.05), getting fewer associations with our approach: eQTMs (45,203 with FDR vs 39,749 with our approach), eCpGs (24,611 vs 21,966) and eGenes (9,937 vs 8,886). Among the 8,886 significant eGenes, 6,288 of them are annotated to coding genes, thus representing 27% of the 23,054 eGenes coding for a gene included in the array.
References:
GTEx consortium, The GTEx Consortium atlas of genetic regulatory effects across human tissues, Science (2020) Sep 11;369(6509):1318-1330. doi: 10.1126/science.aaz1776.
Ongen et al. Fast and efficient QTL mapper for thousands of molecular phenotypes, Bioinformatics (2016) May 15;32(10):1479-85. doi: 10.1093/bioinformatics/btv722. Epub 2015 Dec 26.
- I recommend starting with cell type specific results. Without adjusting cell type, the result doesn't make sense.
As suggested by other reviewers, we have withdrawn the model unadjusted for cellular composition.
Reviewer #3 (Public Review):
Although several DNA methylation-gene expression studies have been carried out in adults, this is the first in children. The importance of this is underlined by the finding that surprisingly few associations are observed in both adults and children. This is a timely study and certain to be important for the interpretation of future omic studies in blood samples obtained from children.
We agree with the reviewer that eQTMs in children are important for interpreting EWAS findings conducted in child cohorts such as those of the Pregnancy And Childhood Epigenetics (PACE) consortium.
It is unfortunate that the authors chose to base their reporting on associations unadjusted for cell count heterogeneity. They incorrectly claim that associations linked to cell count variation are likely to be cell-type-specific. While possible, it is probably more likely that the association exists entirely due to cell type differences (which tend to be large) with little or no association within any of the cell types (which tend to be much smaller). In the interests of interpretability, it would be better to report only associations obtained after adjusting for cell count variation.
Following reviewers’ recommendations, we have reconsidered our initial hypothesis about the role of cellular composition in the association between methylation and gene expression. Although we still think that some of the eQTMs only found in the model unadjusted for cellular composition could represent cell specific effects, we acknowledge that the majority might be confounded by the extensive gene expression and DNA methylation differences between cell types. Also, we recognize that more sophisticated statistical tests should be applied to prove our hypothesis. Because of this we have decided to report the eQTMs of the model adjusted for cellular composition in the main manuscript and keep the results of the model unadjusted for cellular composition only in the online catalogue.
Several enrichments could be related to variation in probe quality across the DNA methylation arrays.
For example, enrichment for eQTM CpG sites among those that change with age could simply be due to the fact age and eQTM effects are more likely to be observed for CpG sites with high quality probes than low quality probes. It is more informative to instead ask if eQTM CpG sites are more likely to have increasing rather than decreasing methylation with age. This avoids the probe quality bias since probes with positive associations with age would be expected to have roughly the same quality as those with negative associations with age. There are several other analyses prone to the probe quality bias.
See answer to question 2, below.
Author Response:
Reviewer #1 (Public Review):
In this report, Shekhar et al, have profiled developing retinal ganglion cells from embryonic and postnatal mouse retina to explore the diversification of this class of neurons into specific subtypes. In mature retina, scRNAseq and other methods have defined approximately 45 different subtypes of RGCs, and the authors ask whether these arise from a common postmitotic precursor, or many ditinct subtypes of precursors. The overall message, is that subtype diversification arises as a "gradual, asynchronus fate restriction of postmitotic multipotential precursors. The authors find that over time, clusters of cells become "decoupled" as they split into subclusters. This process of fate decoupling is associated with changes in the expression of specific transcription factors. This allows them to both predict lineage relationships among RGC subtypes and the time during development when these specification events occur. Although this conclusion based almost entirely on a computational analysis of the relationships among cells sampled at discrete times, the evidence presented supports the overall conclusion. Future experimental validation of the proposed lineage relationships of RGC subtypes will be needed, but this report clearly outlines the overall pattern of diversification in this cell class.
We thank the reviewer for their thoughtful assessment of our study.
Reviewer #2 (Public Review):
The manuscript "Diversification of multipotential postmitotic mouse retinal ganglion cell precursors into discrete types" by Shekhar and colleagues represents an in-depth analysis of an additional transcriptomic datasets of retinal single-cells. It explores the progression of retinal ganglion cells diversity during development and describes some of aspects of fate acquisition in these postmitotic neurons. Altogether the findings provide another resource on which the neural development community will be able to generate new hypotheses in the field of retinal ganglion cell differentiation. A key point that is made by the authors regards the progression of the number of ganglion cell types in the mouse retina, i.e., how, and when neuronal "classes diversify into subclasses and types" (also p. 125). In particular, the authors would like to address whether postmitotic neurons follow either a predetermination or a stepwise progression (Fig. 2a). This is indeed a fascinating question, and the analysis, including the one based on the Waddington-OT method is conceptually interesting.
Comments and questions:
Is the transcriptomic diversity, based on highly variable genes (the number of which is not detailed in the study) a robust proxy to assess cell types? One could argue that early on predetermined cell types are specified by a small set of determinants, both at the proteomic and transcriptomic level, and that it takes several days or week to generate the cascade that allows the detection of transcriptional diversity at the level of >100 gene expression levels.
We had tested the dependence of our results on the number of highly variable genes (HVGs) used. This analysis, shown in Figure 2h, demonstrates that results are robust over the range tested – 1244-3003 total HVGs. Since the analysis in the paper employs 2800 HVGs (~800- 1500 at each stage), we are confident that we are in comfortable excess of the number at which we would need to worry. We have expanded the discussion to avoid confusion on this point. We also address the possibility that a small set of determinants are sufficient to define cell state in a transcriptomic study. This is a common argument, but we believe it is a tenuous one. We believe that the only way a small number of genes can truly define cell state is if they are expressed at very high levels. If these are expressed at high levels, they should be detected in our data and should drive the clustering. If they are expressed at extremely low levels, then given the nature of molecular fluctuations in cells, they cannot be expected to serve as a stable scaffold for differentiation. Indeed, a small set of determinants (usually transcription factors) may be necessary to specify a cell type. However, sufficiency of specification requires the expression of a usually much larger of number downstream regulators.
Since there are many RGC subsets (45) that share a great number of their gene expression, is it possible that a given RGC could transition from one subset to another between P5 and P56? Or even responding to a state linked to sustained activity? Was this possibility tested in the model?
We cannot address the possibility that cells swap types postnatally so that the cells comprising type X at P5 are not the same ones that comprise type X at P56. It does seem pretty unlikely, as the cell types are well-separated in transcriptional space (~250 DE genes on average). Regarding activity, we have made some initial tests by preventing visually evoked activity from birth to P56 in three different ways (dark-rearing and two mutant lines). We find no statistically significant effect on diversification. These results are currently being prepared for publication.
The authors state that early during development there is less diversity than later. This statement seems obvious but how much. Can this be due to differential differentiation stage? At E16 RGC are a mix of cells born from E11 to E16, with the latter barely located in the GCL. Does this tend to show a continuum that is may be probably lost when the analysis is performed on cells isolated a long time after they were born (postnatal stages)? Alternatively, would it be possible to compare RGC that have been label with birth dating methods?
Regarding the amount of diversification, we quantified this using the Rao diversity index (Figure 2h), which suggests an overall increase in 2-fold transcriptional diversity at P56 compared to the early stages. The continuum is likely because cells at early stage are close to the precursor stage and not very differentiated. Regarding combining RNA-seq with birthdating, although elegant methods now make this combination possible, it falls beyond the scope of this study.
Comparing data produced by different methods can be challenging. Here the authors compared transcriptomic diversity between embryonic dataset produced with 10X genomics (E13 to P0) and, on the other hand, postnatal P5 that were produced using a different drop-seq procedure). Is it possible to control that the differences observed are not due to the different methods?
It is correct that most of the P5 data was produced using Drop-seq, but that dataset also includes transcriptomes obtained by the 10X method. The relative frequency of RGC clusters and the average gene expression values obtained using either method was highly correlated (Reviewer Fig. 1). This is now pointed out in the “Methods.”
Reviewer Fig. 1. Comparison between the relative frequency of types (left) and the average gene expression levels (right) at P5 between 10X data (y-axis) and Drop-seq data (x-axis). R corresponds to the Pearson correlation coefficient. The axes are plotted in the logarithmic scale.
It might be important to control the conclusion that diversity is lower at E13 vs P5 when we see that thrice less cells (5900 vs 180000) were analyzed at early stage (BrdU, EdU, CFSE...)? A simple downsampling prior to the analysis may help.
Although we collected different numbers of cells at different ages, we noted in the text that they do not influence the number of clusters. Regarding P5 specifically, Rheaume et al. (who we now discuss) obtained very similar results to ours with only 6000 cells (3x lower).
Ipsilateral RGC: It is striking that the DEG between C-RGC and I-RGC reflect a strong bias with cells scored as" ipsi" are immature RGC while the other ("contra") are much more mature. This bias comes from the way ipsilateral RGC were "inferred" using non-specific markers. Can the author try again the analysis by identifying RGC using more robust markers? (eg. EphB1). Would it be possible to select I-RGC and C-RGC that share same level of differentiation? Previous studies already identified I-RGC signature using more specific set-up (Wang et al., 2016 from retrogradely labelled RGC; Lo Giudice et al., 2019 with I-RGC specific transgenic mouse).
We are not sure how the reviewer concludes that the putative I-RGCs are more immature than the putative C-RGCs. As discussed earlier, insofar as expression levels of pan-RGC markers are indicative of maturational stage, we found no evidence that clustering is driven by maturation gradients. Thus, we expect our putative I-RGCs and C-RGCs to not differ in differentiation state. Following the reviewer’s suggestion, we now include EphB1(Ephb1) in our I-RGC signature. The impact of replacing Igfbp5 with Ephb1 on the inferred proportion of I-RGCs within each terminal type was minimal (Reviewer Fig. 2). We would like to note that to assemble our IRGC/C-RGC signatures we relied on data presented Wang et al. (2016). Outside of wellestablished markers (e.g. Zic2, and Isl2), we chose the RNA-seq hits in Wang et al. that had been validated histologically in the same paper or that are correlated with Zic2 expression in our data. This nominated Igfbp5, Zic1, Fgf12, and Igf1.
Reviewer Fig. 2. Comparison of inferred I-RGC frequency within each terminal type (points) using two I-RGC signature reported in the paper. For the y-axis we used Zic2 and EphB1.
It would be important to discuss how their findings differs from the others (including Rheaume et al., 2018). To make a strong point, I-RGC shall be isolated at a stage of final maturation (P5?) and using retrograde labelling, which is a robust method to ensure the ipsilateral identity of postnatal RGCs.
We cite Rheaume et al. in several places. In fact, there is good transcriptional correspondence between our dataset and theirs (Figure S1i), despite the differences in the number of cells profiled (~6000 vs ~18000) and technologies (10X vs. Drop-seq/10X). We now mention this is the text. Note also that we had compared our P56 data with Rheaume et al.’s, P5 data in an earlier publication (Tran et al., 2019) and observed a similar tight correspondence between clusters. Zic1 is expressed in I-RGCs (Wang et al., 2016) at early stages, and in our dataset its expression at E13 and E14 is similar to that of Zic2 (Supplementary Fig. 8); Postnatally, however, it marks W3B RGCs (Tran et al., 2019), many of which project contralaterally (Kim et al., J. Neurosci. 2010). Regarding retrograde labeling, as noted above, additional experiments would take a prohibitively long time (up to a year) to complete.
It is unclear how good Zic1 and Igf1 can be used as I-RGC marker. Can the author specify how specific to I-RGC they are? Have they been confirmed as marker using retrograde labelling experiments?
We have relied on previous work, primarily from the Mason lab, to choose I-RGC and C-RGC markers. Igf1 is a C-RGC marker that is expressed in a complementary fashion with Igfbp5, an I-RGC marker as noted in Wang et al, 2016. They also perform ISH to show that Igf1 is not expressed in the VT crescent, while Igfbp5 is (see Fig. 5 in Wang et al., 2016). Similarly, Zic1 is also cited in Wang et al. as an RNA-seq hit for I-RGCs. Although Zic1 was not validated using ISH, we found its expression pattern to be highly correlated with Zic2 at E13 (Supplementary Fig. 8c).
The enrichment procedure may deplete the RGC subpopulation that express low levels of Thy1 or L1CAM. A comparison on that point could be done with the other datasets analysed in the study.
We presume the reviewer is referring to the data of Lo Guidice and Clark/Blackshaw, which we show in comparison to ours in Figure S1. In both of those studies, all retinal cells were analyzed, whereas we enriched RGCs. As noted in the text, RGCs comprise a very small fraction of all retinal cells, so Lo Giudice and Clark/Blackshaw lacked the resolution to resolve RGC diversity at later time points. Indeed, there is no whole retina dataset available in which RGCs are numerous enough for comprehensive subtyping. Our approach to this issue was to collect RGCs with both Thy1 and L1 at E13, E14, E16 and P0, with the idea that the markers might have complementary strengths and weaknesses. In fact, at each age, all clusters are present in both collection types, although frequencies vary. This concordance supports the idea that neither marker excludes particular types. We now stress this point in results and in the Supplementary Fig. 2 legend.
In supplemental Fig. S1e: why are cells embedded from "Clark" datasets only clusters on the right side of the UMAP while the others are more evenly distributed?
Actually, both the Clark et al. and Lo Giudice et al. datasets are predominantly clustered on the right side of the UMAP. This reflects the methodological difference noted above: they profiled the whole retina, whereas we isolated RGCs. Thus, their datasets contain a much higher abundance of RPCs and non-neurogenic precursors compared to ours. The right clusters represent RPCs due to their expression of Fgf15 and other markers, while the left clusters represent RGCs based on their expression of Nefl. Indeed, a main reason for including these plots was to illustrate the relative abundance of RGCs in our data (also see Supplementary Fig. S1h).
What could explain that CD90 and L1CAM population are intermingled at E14, distinct at E16, and then more mixed at P0?
We believe the reviewer is referring to Supplementary Figs. S2a-c. Given the temporal expression level changes in Thy1 and L1cam (Supplementary Fig. S1c) in RGCs, a likely possibility is that they enrich RGC precursor subsets at different relative frequencies. We now note this in the Supplementary Fig. 2 legend.
On Fig. 6: the E13 RGC seems to be segregated in early born RGC expressing Eomes and later born expressing neurod2. Thus, fare coupling with P5 seems to suggest that Eomes population at P5 may have been generated first, and Neurod2 generated later. Is that possible?
That the Eomes RGCs are specified before Neurod2 RGCs is one of our conclusions from the fate decoupling analysis (Figures 6f-h). Whether this is because the former arise from early born cells and the latter arise from later born cells is not clear. There is disagreement in the literature on whether ipRGCs are born at a different time than other RGCs, so we prefer not to make a comment.
Methods: The Methods section is extensive, and yet it is presented in a rather complex manner so that it is difficult to understand for a broad audience. It would be valuable if the authors could simplify or better explain some parts (the WOT section in particular).
We believe that the sections on animals, molecular biology and histology are quite straightforward, but agree that the sections describing the computational analysis are hard going. We have modified them in several places as requested. As regards better explanation of the WOT, we now precede that section with an “overview” as a way of making it easier to follow. (We had already included an overview of the clustering procedures.) We have also provided further detail on some of the reviewer’s subsequent questions on this section, including the use of HVGs, the Classifier, and the strategy for inferring I-RGCs (see below). Perhaps most important, we have worked to make the “Results” and “Discussion” sections accessible to a broad audience.
*Highly variable genes (HVG) used for clustering and dimensionality reduction: how many of them and what are they? Are they the same used for each stage?
Since clustering was performed at each stage independently, we determined HVGs at each stage separately using a statistical method introduced in one of our previous studies (Pandey et al., Current Biology, 2018). The total number of HVGs at each stage were as follows: E13: N=1094 E14: N=834 E16: N=822 P0: N=881 P5: N=1105 P56: N=1510
We note that these are not necessarily the same at each stage due to the temporal variation in gene expression. Together these correspond to 2854 unique genes (union of all HVGs). The WOT analysis was done using this full set.
*In the methods p9: "The common features G = GR ∩ GT are used to train a third classifier ClassR on the reference atlas AR. This ensures that inferred transcriptomic correspondences are based on "core" gene expression programs that underlie cell type identity rather than maturation-associated genes." Could the authors explain the relevance of using a third model and, more importantly, is there any genes that eliminated through the procedure that could be important to drive the diversification process? If so, would it be possible to estimate their number and the relative impact?
The rationale for this was as follows. Our goal is to map cells from one time point to a type at another time point. The naïve way to do this would be to use a classifier trained entirely at either of the time point. However, the features of such a classifier is likely to contain genes that are not expressed at the earlier time point, and likely to generate spurious mappings (since the set of cluster specific genes are not identical). Therefore, we sought to train a classifier that is trained using genes that are part of conserved transcriptional signatures at both time points, which corresponds to the third model.
When this filtering was not performed, the temporal correspondences in the supervised classification model were less specific than those reported. In particular, ARI values dropped by about 15% on average. The simple reason for this is that a cluster specific gene at E13 (for e.g.) may no longer be expressed at E14, and vice-versa. Thus, by restricting the features to a common set of cluster specific genes, we obtained the “best possible” transcriptomic correspondences between clusters at consecutive time points. We note that the correspondences obtained in this way (Figure 3) were recovered through WOT when the results of the latter were collapsed at the cluster level (Supplementary Fig. 5).
*Methods page 15: Inference of ipsilaterally-projecting RGC types. Wouldn't it be more valuable to consider more markers to distinguish RGC precursors?
As indicated before, we used I-RGC genes and C-RGC genes reported in Wang et al., 2016 (Table 2), in addition to the well-known markers Zic2 and Isl2. Here, we prioritized genes that had been histologically validated (Figs. 4 and 5), which were expressed in our data (Sema3e and Tbx20 were not considered as these undetectable at E13 in our data). Following the reviewer’s earlier suggestion, we also noted that including Ephb1 in our signature minimally impacts the results.
Discussion: *Is there somewhat a plasticity that allow the RGC subgroups to switch over time? (IF we were to record the transcriptome of the same cell over time, will one observe that the cell belong to another cluster / subgroup?
One can only speculate. Other than long-term in vivo imaging combined with vital type-specific markers we know of no way to experimentally address the possibility that cells swap types postnatally so that the cells comprising type x at P5 are not the same ones that comprise type x at P56. It does seem pretty unlikely though.
*While the data appears technically rigorous, and the number of cells sequenced very high, the results seem redundant with several prior studies and the discrepancies are not sufficiently discussed.
We are confused by this point, since the reviewer does not cite the papers to which s/he refers. To our knowledge there is no study at present that has described RGC diversification, so it is not clear what would be discrepant.
Author Response
Reviewer #1 (Public Review):
This manuscript provides a comprehensive investigation of the effects of the genetic ablation of three different transcription factors (Srf, Mrtfa, and Mrtfb) in the inner ear hair cells. Based on the published data, the authors hypothesized that these transcription factors may be involved in the regulation of the genes essential for building the actin-rich structures at the apex of hair cells, the mechanosensory stereocilia and their mechanical support - the cuticular plate. Indeed, the authors found that two of these transcription factors (Srf and Mrtfb) are essential for the proper formation and/or maintenance of these structures in the auditory hair cells. Surprisingly, Srf- and Mrtfb- deficient hair cells exhibited somewhat similar abnormalities in the stereocilia and in the cuticular plates even though these transcription factors have very different effects on the hair cell transcriptome. Another interesting finding of this study is that the hair cell abnormalities in Srfdeficient mice could be rescued by AAV-mediated delivery of Cnn2, one of the downstream targets of Srf. However, despite a rather comprehensive assessment of the novel mouse models, the authors do not have yet any experimentally testable mechanistic model of how exactly Srf and Mrtfb contribute to the formation of actin cytoskeleton in the hair cells. The lack of any specific working model linking Srf and/or Mrtfb with stereocilia formation decreases the potential impact of this study.
Major comments:
Figures 1 & 3: The conclusion on abnormalities in the actin meshwork of the cuticular plate was based largely on the comparison of the intensities of phalloidin staining in separate samples from different groups. In general, any comparison of the intensity of fluorescence between different samples is unreliable, no matter how carefully one could try matching sample preparation and imaging conditions. In this case, two other techniques would be more convincing: 1) quantification of the volume of the cuticular plates from fluorescent images; and 2) direct examination of the cuticular plates by transmission electron microscopy (TEM).
In fact, the manuscript provides no single TEM image of the F-actin abnormalities either in the cuticular plate or in the stereocilia, even though these abnormalities seem to be the major focus of the study. Overall, it is still unclear what exactly Srf or Mrtfb deficiencies do with F-actin in the hair cells.
Yes, we agree. As suggested by the reviewer, to directly examine the defects in F-actin organization within the cuticular plate of mutant mice, we conducted Transmission Electron Microscopy (TEM) analyses. The results, as presented in the revised Figures 1 and 4 (panels F, G, and E, F, respectively), provide crucial insights into the structural changes in the cuticular plate. Meanwhile, the comparison of the volume of the phalloidin labeled cuticular plate after 3-D reconstruction using Imaris software was conducted and shown in Author response image 1. The results of the cuticular plate (CP) volume were consistent with the relative F-actin intensity change of the cuticular plate in the revised Figures 1B and 4B. For the TEM analysis of the stereocilia, we regret that due to time constraints, we were unable to collect TEM images of stereocilia with sufficient quality for a meaningful comparison. However, we believe that the data we have presented sufficiently addresses the primary concerns, and we appreciate the reviewers’ understanding of these limitations.
Author response image 1.
Figures 2 & 4 represent another example of how deceiving could be a simple comparison of the intensity of fluorescence between the genotypes. It is not clear whether the reduced immunofluorescence of the investigated molecules (ESPN1, EPS8, GNAI3, or FSCN2) results from their mis-localization or represents a simple consequence of the fact that a thinner stereocilium would always have a smaller signal of the protein of interest, even though the ratio of this protein to the number of actin filaments remains unchanged. According to my examination of the representative images of these figures, loss of Srf produces mis-localization of the investigated proteins and irregular labeling in different stereocilia of the same bundle, while loss of Mrtfb does not. Obviously, a simple quantification of the intensity of fluorescence conceals these important differences.
Yes, we agree. In addition to the quantification of tip protein intensity, we have added a few more analyses in the revised Figure 3 and Figure 6, such as the percentage of row 1 tip stereocilia with tip protein staining and the percentage of IHCs with tip protein staining on row 2 tip. Using the results mentioned above, the differences in the expression level, the row-specific distribution and the irregular labeling of tip proteins between the control and the mutants can be analyzed more thoroughly.
Reviewer #2 (Public Review):
The analysis of bundle morphology using both confocal and SEM imaging is a strength of the paper and the authors have some nice images, especially with SEM. Still, the main weakness is that it is unclear how significant their findings are in terms of understanding bundle development; the mouse phenotypes are not distinct enough to make it clear that they serve different functions so the reader is left wondering what the main takeaway is.
Based on the reviewer’s comments, in this revised manuscript, we put more emphasis on describing the effects of SRF and MRTFB on key tip proteins’ localization pattern during stereocilia development, represented by ESPN1, EPS8 and GNAI3, as well as the effects of SRF and MRTFB on the F-actin organization of cuticular plate using TEM. We have made substantial efforts to interpret the mechanistic underpinnings of the roles of SRF and MRTFB in hair cells. This is reflected in the revised Figures 1, 3, 4, 6, and 10, where we provide more comprehensive insights into the mechanisms at play.
We interpret our data in a way that both SRF and MRTF regulate the development and maintenance of the hair cell’s actin cytoskeleton in a complementary manner. Deletion of either gene thus results in somewhat similar phenotypes in hair cell morphology, despite the surprising lack of overlap of SRF and MRTFB downstream targets in the hair cell.
In Figure 1 and 3, changes in bundle morphology clearly don't occur until after P5. Widening still occurs to some extent but lengthening does not and instead the stereocilia appear to shrink in length. EPS8 levels appear to be the most reduced of all the tip proteins (Srf mutants) so I wonder if these mutants are just similar to an EPS8 KO if the loss of EPS8 occurred postnatally (P0-P5).
To address this question, we performed EPS8 staining on the control and Srf cKO hair cells at P4 and P10. We found that the dramatic decrease of the row 1 tip signal for EPS8 started since P4 in Srf cKO IHCs. Although the major hair bundle phenotype of Eps8 KO, including the defects of row 1 stereocilia lengthening and additional rows of short stereocilia also appeared in Srf cKO IHCs, there are still some bundle morphology differences between Eps8 KO and Srf cKO. For example, firstly, both Eps8 KO OHCs and IHCs showed additional rows of short stereocilia, but we only observed additional rows of short stereocilia in Srf cKO IHCs. Secondly, in Valeria Zampini’s study, SEM and TEM images did not show an obvious reduction of row 2 stereocilia widening (P18-P35), while our analysis of SEM images confirmed that the width of row 2 IHC stereocilia was drastically reduced by 40% in Srf cKO (P15). Generally, we think although Srf cKO hair bundles are somewhat similar to Eps8 KO, the Srf cKO hair bundle phenotype might be governed by multiple candidate genes cooperatively.
Reference:
Valeria Zampini, et al. Eps8 regulates hair bundle length and functional maturation of mammalian auditory hair cells. PLoS Biol. 2011 Apr;9(4): e1001048.
A major shortcoming is that there are few details on how the image analyses were done. Were SEM images corrected for shrinkage? How was each of the immunocytochemistry quantitation (e.g., cuticular plates for phalloidin and tip staining for antibodies) done? There are multiple ways of doing this but there are few indications in the manuscript.
We apologize for not making the description of the procedure of images analyses clear enough. As described in Nicolas Grillet group’s study, live and mildly-fixed IHC stereocilia have similar dimensions, while SEM preparation results in a hair bundle at a 2:3 scale compared to the live preparation. In our study, the hair cells selected for SEM imaging and measurements were located in the basal turn (30-32kHz), while the hair cells selected for fluorescence-based imaging and measurements were located in the middle turn (20-24kHz) or the basal turn (32-36kHz). Although our SEM imaging and fluorescence-based imaging of basal turn’s hair bundles were not from the same area exactly, the control hair bundles with SEM imaging have reduced row 1 stereocilia length by 10%-20%, compared to the control hair bundles with fluorescence-based imaging (revised Figure 2 and Figure 5). Generally, our stereocilia dimensions data showed appropriate shrinkage caused by the SEM preparation.
Recognizing the need for clarity, we have provided a detailed description of our image quantification and analysis procedures in the “Materials and Methods” section, specifically under “Immunocytochemistry.” This will aid readers in understanding our methodologies and ensure transparency in our approach.
Reference:
Katharine K Miller, et al. Dimensions of a Living Cochlear Hair Bundle. Front Cell Dev Biol. 2021 Nov 25:9:742529.
The tip protein analysis in Figs 2 and 4 is nice but it would be nice for the authors to show the protein staining separately from the phalloidin so you could see how restricted to the tips it is (each in grayscale). This is especially true for the CNN2 labeling in Fig 7 as it does not look particularly tip specific in the x-y panels. It would be especially important to see the antibody staining in the reslices separate from phalloidin.
Thank you for the suggestions. We have shown tip proteins staining in grayscale separately from the phalloidin in the revised Figure 3 and Figure 6. To clearly show the tip-specific localization of CNN2, we conducted CNN2 staining at different ages during hair bundle development and showed CNN2 labeling in grayscale and in reslices in revised Figure 9-figure supplement 1B.
In Fig 6, why was the transcriptome analysis at P2 given that the phenotype in these mice occurs much later? While redoing the transcriptome analysis is probably not an option, an alternative would be to show more examples of EPS8/GNAI/CNN2 staining in the KO, but at younger ages closer to the time of PCR analysis, such as at P5. Pinpointing when the tip protein intensities start to decrease in the KOs would be useful rather than just showing one age (P10).
We agree with the reviewer. To address this question, we have performed ESPN1, EPS8 and GNAI3 staining on the control and the mutant’s hair cells at P4, P10 and P15 (the revised Figures 3 and 6). According to the new results, we found that the dramatic decreases of the row 1 tip signal for ESPN1 and EPS8 started since P4 in Srf cKO IHCs, is consistent with the appearance of the mild reduction of row 1 stereocilia length in P5 Srf cKO IHCs. For Mrtfb cKO hair cells, the obvious reduction of the row 1 tip signal for ESPN1 was observed until P10. However, a few genes related to cell adhesion and regulation of actin cytoskeleton were significantly down-regulated in P2 Mrtfb deficient hair cell transcriptome. We think that in hair cells the MRTFB may not play a major role in the regulation of stereocilia development, so the morphological defects of stereocilia happened much later in the Mrtfb mutant than in the Srf mutant.
While it is certainly interesting if it turns out CNN2 is indeed at tips in this phase, the experiments do not tell us that much about what role CNN2 may be playing. It is notable that in Fig 7E in the control+GFP panel, CNN2 does not appear to be at the tips. Those images are at P11 whereas the images in panel A are at P6 so perhaps CNN2 decreases after the widening phase. An important missing control is the Anc80L65-Cnn2 AAV in a wild-type cochlea.
We agree with the reviewer. We have conducted more immunostaining experiments to confirm the expression pattern of CNN2 during the stereocilia development, from P0 to P11. The results were included in the revised Figure 9-figure supplement 1B. As the reviewer suggested, CNN2 expression pattern in control cochlea injected with Anc80L65-Cnn2 AAV has also been provided in revised Figure 9E.
Author Response
Reviewer #1 (Public Review):
This is an awesome comprehensive manuscript. Authors start by sorting putative stromal cellcontaining BM non-hematopoietic (CD235a-/CD45-) plus additional CD271+/CD235a/CD45- populations to identify nine individual stromal identities by scRNA-seq. The dual sorting strategy is a clever trick as it enriches for rare stromal (progenitor) cell signals but may suffer a certain bias towards CD271+ stromal progenitors. The lack of readable signatures already among CD45-/CD45- sorts might argue against this fear. This reviewer would appreciate a brief discussion on number & phenotype of putative additional MSSC phenotypes in light of the fact that the majority of 'blood lineage(s)'-negative scRNA-seq signatures identified blood cell progenitor identities (glycophorin A-negative & leukocyte common antigen-negative). The nine stromal cell entities share the CXCL12, VCAN, LEPR main signature. Perhaps the authors could speculate if future studies using VCAN or LEPRbased sort strategies could identify additional stromal progenitor identities?
We would like to thank the reviewer for critically evaluating our work and for the generally positive evaluation of the paper. We apologize for delayed resubmission as it took a long time for a specific antibody to arrive to complete the confocal microscopy analyses.
The reviewer asks for a brief discussion on the cell numbers and phenotypes of MSSC phenotypes. The cell numbers and percentages of MSSC in sorted CD45low/-CD235a- and CD45low/-CD235a-CD271+ cells can be found in Supplementary File 3 and we have added a summary of the phenotypes of MSSC in the new Supplementary File 7.
Due to the extremely low frequency of stromal cells in human bone marrow, we chose a sorting strategy that also included CD45low cells (Fig 1A) to ensure that no stromal cells were excluded from the analysis. Although stromal elements are certainly enriched using this approach, the CD45low population contains several different hematopoietic cell types. These include CD34+ HSPCs which are characterized by low CD45 expression2, as well as the CD45low-expressing fractions of other hematopoietic cell populations such as B cells, T cells, NK cells, megakaryocytes, monocytes, dendritic cells, and granulocytes. Furthermore, CD235a- late-stage erythroid progenitors, which are negative for CD45, are represented as well. Of note, our data are consistent with previously reported murine studies showing the presence of a number of hematopoietic populations in CD45- cells, which accounted for the majority of CD45-Ter119-CD31- murine BM cells3,4. However, despite a certain enrichment of stromal elements in the CD45low cell fraction, frequencies were still too low to allow for a detailed analysis of this important bone marrow compartment. This prompted us to adopt the stromal cell-enrichment strategy as described in the manuscript to achieve a better resolution of the stromal compartment. In fact, sorting based on CD45low/-CD235a-CD271+ allowed us to sufficiently enrich bone marrow stromal cells to be clearly detectable in scRNAseq analysis. According to the reviewer’s suggestion, a brief discussion on this issue is now included in the Discussion (page 28, lines 10-15).
The reviewer also suggested using VCAN or LEPR-based sorting strategy to identify additional stromal identities in future studies.
However, as an extracellular matrix protein, FACS analysis of cellular VCAN expression can only be achieved based on its intracellular expression after fixation and permeabilization5,6. Additionally, while VCAN is highly and ubiquitously expressed by stromal clusters, VCAN is also expressed by monocytes (cluster 36). Therefore, VCAN is not an optimal marker to isolate viable stromal cells.
LEPR is the marker that was reported to identify the majority of colony-forming cells in adult murine bone marrow7. We have previously reported that the majority of human adult bone marrow CFU-Fs is contained in the LEPR+ fraction 8. In our current scRNAseq surface marker profiling analysis, group A cells showed high expression of several canonical stromal markers including VCAM1, PDGFRB, ENG (CD73), as well as LEPR (Fig. 4A). However, the four stromal clusters in Group A could not be separated based on the expression of LEPR. Therefore, we chose not to use LEPR as a marker to prospectively isolate the different stromal cell types.
The authors furthermore localized CD271+, CD81+ and NCAM/CD56+ cells in BM sections in situ. Finally, referring to the strong background of the group in HSC research, in silico prediction by CellPhoneDB identified a wide range of interactions between stromal cells and hematopoietic cells. Evidence for functional interdependence of FCU-F forming cells is completing the novel and more clear bone marrow stromal cell picture.
We thank the reviewer for the positive comments.
An illustrative abstract naming the top9 stromal identities in their top4 clusters by their "top10 markers" + functions would be highly appreciated.
We thank the reviewer for the suggestion. A summary of the characteristics of stromal clusters is now shown in the new Supplementary File 7, which we hope matches the reviewer’s expectations.
Reviewer #2 (Public Review):
Knowledge about composition and function of the different subpopulations of the hematopoietic niche of the BM is limited. Although such knowledge about the mouse BM has been accumulating in recent years, a thorough study of the human BM still needs to be performed. The present manuscript of Li and coworkers fills this gap by performing single cell RNA sequencing (scRNAseq) on control BM as well as CD271+ BM cells enriched for non-hematopoietic niche cells.
We apologize for delayed resubmission as it took a long time for a specific antibody to arrive to complete the confocal microscopy analyses. We thank the reviewer for the critical expert review and overall positive comments.
Based on their scRNAseq, the authors propose 41 different BM cell populations, ten of which represented non-hematopoietic cells, including one endothelial cell cluster. The nine remaining skeletal subpopulations were subdivided into multipotent stromal stem cells (MSSC), four distinct populations of osteoprogenitors, one cluster of osteoblasts and three clusters of pre-fibroblasts. Using bioinformatic tools, the authors then compare their results and divisions of subpopulations to some previously published work from others and attempt to delineate lineage relationships using RNA velocity analyses. From these, they propose different paths from which MSSC enter the progenitor stages, and might differentiate into pre-osteoblasts and -fibroblasts.
It is of interest to note, that apparently adipo-primed cells may also differentiate into osteolineage cells, something that should be further explored or validated. Furthermore, although this analysis yields a large adipo-primed populations, pre-adipocytes and mature adipocytes appear not to be included in the data set the authors used, which should also be explained.
We thank the reviewer for this comment. We chose to annotate Cluster 5 as adipoprimed cluster based on the higher expression of adipogenic differentiation markers as well as a group of stress-related transcription factors (FOS, FOSB, JUNB, EGR1) (Fig. 2B-C, Figure 2-figure supplement 1C) some of which had been shown to mark bone marrow adipogenic progenitors1. Although at considerably lower levels compared to adipogenic genes, osteogenic genes were also expressed in cluster 5 cells (Fig. 2B and D), indicating the multi-potent potential of this cluster. Therefore, our initial annotation of these cells as adipoprimed progenitors was too narrow as it did not include the possible osteogenic differentiation potential. We apologize for the confusion caused by the inappropriate annotation and, in order to avoid any further confusion, cluster 5 has now been re-annotated as ‘highly adipocytic gene-expressing progenitors (HAGEPs), which we believe is a better representation of the cells. We furthermore agree with the reviewer that in-vivo differentiation needs to be performed to address potential differentiation capacities in future studies.
With regard to the lack of adipocytes in our data set, we described in the Materials and Methods section that human bone marrow cells were isolated based on density gradient centrifugation. After centrifugation, the mononuclear cell-containing monolayers were harvested for further analysis. However, the resulting supernatant containing mature adipocytic cells was discarded14. Therefore, adipocyte clusters were not identified in our dataset. We have amended the manuscript accordingly (page 5, line 7).
Regarding the pre-adipocytes, we are not aware of any specific markers for pre-adipocytes in the bone marrow. We examined the only known markers (ICAM1, PPARG, FABP4) that have been shown to mark committed pre-adipocytes in human adipose tissue15. As illustrated in Fig. R1 (below), low expression of all three markers was not restricted to a single distinct cluster but could be found in almost all stromal clusters. These data thus allow us to neither confirm nor exclude the presence of pre-adipocytes in the dataset. Due to the lack of specific markers for pre-adipocytes and the absence of mature adipocytes in the current dataset, it is therefore difficult to identify a well-defined pre-adipocytes cluster.
Figure R1. UMAP illustration of the normalized expression of the markers for pre-adipocytes in stromal clusters.
In addition, based on a separate analysis of surface molecules, the authors propose new markers that could be used to prospectively isolate different human subpopulations of BM niche cells by using CD52, CD81 and NCAM1 (=CD56). Indeed, these analyses yield six different populations with differential abilities to form fibroblast-like colonies and differentiate into adipo-, osteo-, and chondrogenic lineages. To explore how the scRNAseq data may help to understand regulatory processes within the BM, the authors predict possible interactions between hematopoietic and non-hematopoietic subpopulations in the BM. These should be further validated, to support statements as the suggestion in the abstract that separate CXCL12- and SPP1-regulated BM niches might exist.
We agree with the reviewer that functional validation of the CellPhoneDB results using for example in vivo humanized mouse models would be needed to demonstrate the presence of different niches in the bone marrow. At this point of time we only put forward the hypothesis that different niche types exist while we will work on providing experimental proof in our future studies.
The scRNAseq analysis is indeed a strong and important resource, also for later studies meant to increase knowledge about the hematopoietic niche of the BM. Although the analyses using different bioinformatic tools is very helpful, they remain mostly speculative, since validatory experiments, as already mentioned, are missing. As such, I feel the authors did not succeed in achieving their goals of understanding how non-hematopoietic cells of the BM regulate the different hematopoietic processes within the BM. Nevertheless, they have created valuable resources, both in the scRNAseq data they generated, as well as the different predictions about different cell populations, their lineage relationships, and how they might interact with hematopoietic cells.
We thank the reviewer for the appreciation of the value of this dataset. We agree with the reviewer that it is of great importance to validate the contribution of potential driver genes for stromal cell differentiation and verify the in vitro data and in-silico prediction using in-vivo models. As the main goal of the current study was to formulate hypotheses based on the scRNAseq data for future studies, we believe that in vivo validation experiments using engineered human bone marrow models or humanized bone marrow ossicles are out of the scope of the current study, but certainly need to be performed in the future.
The impact of this work is difficult to envision, since validations still need to be performed. Also, it has the born in mind that humans are not mice, which can be studied in neat homogeneous inbred populations. Human populations on the other hand, are quite diverse, so that the data generated in this manuscript and others will probably have to be combined to extrapolate data relevant to the whole of the human population. However, as it is equally difficult to generate reliable scRNAseq data from human BM, it seems likely that the data will indeed an important resource, when more data from different donors become available.
We thank the reviewer for the generally positive evaluation of this study.
Taken at point value, the authors provide evidence that human counterparts exist to several BM populations described in mice. In my opinion, the lineage relationships predicted using the RNA velocity analyses need more substance, as it seems the differentiation-paths may diverge from what is known from mice. If so, this issue should be studied more stringently. Similarly, the paper would have been strengthened considerably if a relevant experimental validation would have been attempted, perhaps by using genetically modified (knockdown) MSSC, similar to Battula et al. (doi: 10.1182/blood-2012-06-437988).
In the study from Welner’s group, stromal differentiation trajectory was inferred based on scRNAseq analysis of murine bone marrow cells using Velocyto16. Velocyto identified MSCs as the ‘source’ cell state with pre-adipocytes, pro-osteoblasts, and prochondrocytes being end states. In our study, the MSSC population was predicted to be at the apex of the trajectory and the pre-osteoblast cluster was placed close to the terminal state of differentiation, which is consistent with the murine study. However, different stromal cell types were identified in mice compared with humans. For example, we have identified prefibroblasts in our dataset which are absent in the murine study, while a well-defined murine pre-adipocyte population was not identified in our human dataset. Therefore, it is not surprising to find some discrepancies between human and murine stromal differentiation trajectories. Of course and as mentioned before, critical in-vivo functional validations need to be carried out to address these important issues in the future.
In summary, this is a very interesting but also descriptive paper with highly important resources. However, to prospectively identify or isolate human non-hematopoietic/nonendothelial niche populations, more stringent validations should have been performed to strengthen the validity of the different analyses that have been performed. As such, it remains an open question which niche subpopulations has the most impact on the different hematopoietic processes important for normal and stress hematopoiesis, as well as malignancies.
Thank you for this comment. We completely agree that more stringent validations are necessary but are outside of the aim of our current hypothesis-generating study. Accordingly, we are planning functional verification studies using genetically manipulated stromal cells in combination with in-vivo humanized ossicles. Furthermore, other groups will hopefully use our database and contribute with functional studies in model systems that are currently not available to us, e.g. iPS-derived bone marrow in-vitro proxies.
Specific remarks
• Since CD45, CD235a, and CD271 are used as distinguishing markers in the sample preparation of the scRNAseq, it would be helpful to highlight these markers in the different analyses (Figures 1D, 2B, 2C-F, and 4A), and restrict the analyses to those cells that also not express CD45, CD235a (why use CD71?) and highly express CD271.
Thank you for this comment. As shown in Fig. R2, we have modified figures Fig. 1D, 2B, and 4A showing now also the expression of PTPRC (CD45), GYPA (CD235a), and NGFR (CD271) on the top (Fig. 1D and 2B) or right (Fig. 4A) panel of the figures. To complement Fig. 2C-F, we have generated new stacked violin plots showing the expression level of three markers by all 9 stromal clusters (Fig. R2B). As we believe that including these three markers in the figures does not provide a better strategy to improve the analyses, we decided to leave the original figures unchanged in this respect.
Figure R2. (A) Modified Fig. 1D, 2B and 4A with PTPRC (CD45), GYPA (CD235a) and NGFR (CD271) expression. (B) Stacked violin plots of PTPRC, GYPA and NGFR expressed by stromal clusters to complement Fig. 2C-F.
With regard to cell exclusion based on CD45, as shown in the modified Figure corresponding to Fig 1A in the manuscript (Fig R2A), CD45 gene expression is observed also in the endothelial cluster, basal cluster, and neuronal cluster (Fig. R2A). These clusters represent non-hematopoietic clusters that we would like to keep in our dataset for further analysis, such as cell-cell interaction. Therefore, we choose to not restrict the analysis to solely CD45 nonexpressing cells.
With regard to CD235a (GYPA), expression of CD235a is not detected in any of the nonhematopoietic clusters. Thus, CD235a-expressing cell exclusion is not necessary.
For CD271, according to our previous results (own unpublished data, belonging to a dataset of which only significantly expressed genes were reported in Li et al.8), protein expression of CD271 is not necessarily reflected by gene expression. In the other words, stromal cells with CD271 protein expression do not always have high mRNA expression. A significant fraction of stromal cells would be excluded if we restrict the analyses only to those cells that show high CD271 gene expression, which would not reflect the real cellular composition of human bone marrow stroma. In order to not risk losing stromal cells, we therefore kept our previous analyses which included stromal cells with various CD271 expression levels.
With regard to using CD71 as an exclusion marker, please see also the comments to reviewer 1. Briefly, according to our data, CD71 (TFRC)-expressing erythroid precursors could still be found after excluding CD45 and CD235a positive cells (Figure 1-figure supplement 1B and R3). As furthermore shown in Figure 1-figure supplement 1G and R2, CD71 expression in the stromal clusters is negligible. Therefore, we believe that this justifies the use of CD71 as an additional marker to exclude erythroid cells. We have amended the discussion to address this issue (page 19, lines 7-8).
Figure R3. FACS plots illustrating the expression of (A) CD71 (TFRC) vs CD271 in CD45- CD235a- cells and (B) FSC-A vs CD81 in CD45-CD235a-CD271+CD71+ cells following exclusion of doublets and dead cells.
• Despite a distinct neuronal cluster (39), there does not seem to be a distinctive marker for these cells. Is this true?
Yes, the reviewer is correct that there is no significantly-expressed distinctive marker for neuronal cells. Multiple markers indicating the presence of different cell types were identified in cluster 39 (Supplementary File 4). Among them, several neuronal markers (NEUROD1, CHGB, ELAVL2, ELAVL3, ELAVL4, STMN2, INSM1, ZIC2, NNAT) were found to be enriched in this cluster (Supplementary File 4 and Fig. 1D) with higher fold changes compared to other identified genes. However, the expression of these genes was not statistically significant, which is mainly due to the heterogeneity of the cluster and thus does not allow us to draw any firm conclusions.
Several genes including MALAT1, HNRNPH1, AC010970.1, and AD000090.1 were identified to be statistically highly expressed by cluster 39 (Supplementary File 4). The expression of these genes is not restricted to any specific cell type. It is therefore impossible to annotate the cluster based on this and our data thus indicated that cluster 39 is a heterogeneous population containing multiple cell types. Based on the expression of neuronal markers, we nevertheless chose to annotate Cluster 39 as “neuronal” as the prominent expression of neuronal markers indicated the presence of neurons in this cluster. To be more accurate, the annotation of cluster 39 has been changed to ‘neuronal cell-containing cluster’ to correctly reflect the presence of non-neuronal gene expressing cells as well (page 29, lines 3-8).
• Since based on 2C and 2D, the authors are unable to distinguish adipo- from osteogenic cells, would the authors use the same molecules to distinguish different populations of 2C-D, or would they use other markers, if so which and why.
We agree with the reviewer that at the first glance adipo-primed (cluster 5, now annotated as “highly adipocytic gene-expressing progenitors”, HAGEPs), balanced progenitors (cluster 16), and pre-osteoblasts (cluster 38) shared a similar expression pattern according to the violin plots in Fig. 2C and 2D. However, as illustrated in the heatmap (Fig. 2B), the expression patterns of adipo-primed (HAGEP) and balanced progenitors were quite different in terms of their expression of adipogenic and osteogenic markers. Both adipogenic and osteogenic marker expression was detected in HAGEPs, balanced progenitors, and preosteoblasts. Thus, as violin plots are summarizing the overall expression levels of a certain marker in a certain cluster, these plots tend to make it more difficult to detect differential expression patterns between different clusters. In this case, the heatmap shown in Fig. 2B is a good complement to the violin plots as it is demonstrating the different expression patterns of every cell in the different stromal clusters.
Additionally, cluster 5 showed the expression of a group of stress-related transcription factors (FOS, FOSB, JUNB, EGR1) (Fig. 2B and Figure 2-figure supplement 1C), some of which had been shown to mark bone marrow adipogenic progenitors1. The expression of the abovementioned stress-related transcription factors (putative adipogenic progenitor markers) was generally lower in cluster 38 compared to cluster 5, further demonstrating that clusters were different.
Furthermore, there was a gradual upregulation of more mature osteogenic markers such as RUNX1, CDH11, EBF1, and EBF3 from cluster 5 to cluster 16 and finally cluster 38. As shown in Fig. 2D, the expression of these markers was higher in cluster 38 compared to cluster 5. Therefore, cluster 38 was annotated as pre-osteoblasts.
Most of the stromal clusters form a continuum (Fig. 2A), which correlates very well with the gradual transition of different cellular states during stromal cell development. It is highly unlikely that abrupt and dramatic gene expression changes would occur during the cellular state transition of cells of the same lineage. Therefore, it is not surprising to find the differences in gene expression profiles between stromal clusters share a certain level of similarities.
In summary, we rely on several factors to distinguish different stromal clusters, which include canonical adipo-, osteo- and chondrogenic markers, stress markers, heatmap, violin plots, and the gradual up-regulation of certain lineage-specific markers.
To directly answer the reviewer’s question, we believe that we are able to distinguish different stromal clusters based on our data.
• In de Jong et al., an inflammatory MSC population (iMSC) is defined. Since the Schneider group showed that inflammatory S100A8 and A9 are expressed by inflamed MSC, is it possible that the some of the designated pre-fibroblasts actually correspond to these S100A8/A9-expressing iMSC?
We thank the reviewer for raising this interesting question.
First of all, we would like to point out that scRNAseq was performed using viably frozen bone marrow aspirates in de Jong’s study while freshly isolated bone marrows were used in our study. There might be discrepancies between frozen and fresh bone marrow samples in terms of cellular composition including stromal composition and, importantly, processinginduced stress-related gene expression profiles.
To investigate if designated pre-fibroblasts actually correspond to iMSCs as suggested by the reviewer, we have re-examined the expression of some of the key iMSC genes as reported by de Jong et al 17. As shown in Fig. R6, the markers that can distinguish iMSC from other MSC clusters in de Jong et al. study were not exclusively expressed by pre-fibroblasts, but also by other stromal cell types including HAGEPs, balanced progenitors, and pre-osteoblasts.
In the study by R. Schneider’s group18, significant upregulation of S100A8/S100A9 was observed in stromal cells from patients with myelofibrosis. Furthermore, base-line expression of S100A8/A9 was also observed in the fibroblast clusters in the control group, which correlates very well with our data of S100A8/9 expression in pre-fibroblasts in normal donors (Fig. 2F). Our data thus indicate – in line with Schneider’s findings - that there is a baseline level expression of S100A8/9 in fibroblasts in hematologically normal samples and that the expression of S100A8/9 is not restricted to inflamed MSC.
In summary, the gene expression profiles observed in our study do not indicate the presence of iMSC in the healthy bone marrow.
• Figure 3A: Do human adipo-primed cells (cluster 5) indeed differentiate into osteogenic cells (clusters 6, 38, and 39). This would be highly unexpected. Can the authors substantiate this "reliable outcome of the RNA velocity analysis"?
Please refer to our previous responses regarding this topic. Briefly, as shown in Fig. 2B and D, both osteogenic and adipogenic genes are expressed in cluster 5, indicating the multi-potent potentials of this cluster. Although the cluster was initially annotated as adipo-primed progenitors, this was not intended to exclude the osteogenic differentiation potential of these progenitors. Nevertheless, this annotation did not correctly reflect the differentiation potential and might thus have caused confusion, for which we apologize. In order to more correctly describe the characteristics of these cells, cluster 5 has now been reannotated as ‘highly adipocytic gene-expressing progenitors (HAGEPs)’.
In general, the outcome of the RNA velocity analysis needs to be corroborated by in-vivo differentiation experiments. But we believe that functional verification, which would be extensive, is out of the scope of the current study and we will address these questions in future studies.
• How statistically certain are the authors, that the populations in Figure 4B as defined by flow cytometry, correspond to MSSC, adipo-primed cells, osteoprogenitors, etc., as defined by scRNAseq?
To address this question, we sorted the A1-A4 populations and performed RT- PCR to examine the CD81 expression level in each cluster. As shown in Figure 4-figure supplement 1B, CD81 expression levels were higher in A1 and A2 compared with A3 and A4, which is consistent with the scRNAseq data that showed the highest CD81 expression in MSSCs compared to other clusters (Supplementary File 4).
The phenotypes defined in this study allowed us to isolate different stromal cell types which demonstrated significant functional differences as described in the manuscript (page 19, lines 17-25; page 20, lines 1-11). These results, in combination with the quantitative real-time PCR results (Figure 4-figure supplement 1B), demonstrated that the A1-A4 subsets in FACS are functionally distinct populations and are likely to be – at least in large parts – identical or equivalent to the transcriptionally identified clusters in group A stromal cells. However, at this point, we do not have performed the required experiments (scRNAseq of sorted cells) that would provide sufficient proof to confirm this statement statistically.
• The immunohistochemistry results shown do not allow distinct conclusions as the colors give unequivocal mix-colors, and surface expression cannot be distinguished from intracellular expression. Please use a 3D (confocal) method for such statements.
We thank the reviewer for the suggestion and we have performed additional confocal microscopy analysis of human bone marrow biopsies as suggested by the reviewer. Representative confocal images are now presented in the middle and right panel of Fig. 6E. We also include a separate file (Supplemental confocal image file). Here, confocal scans of all maker combinations are shown as ortho views in addition to detailed intensity profile analyses of the cells of interest clearly distinguishing surface staining from intracellular staining.
Confocal analysis of bone marrow biopsies confirmed our findings presented in the manuscript. As observed in the scanning images, CD271-expressing cells were negative for CD45 and were located in perivascular, endosteal, and peri-adipocytic regions. CD271/CD81double positive cells could be found either in the peri-adipocytic regions or perivascular regions while CD271/NCAM1 double-positive cells were exclusively situated at the bone-lining endosteal regions. The results of the confocal analysis have been added to the revised manuscript (page 21, lines 15-17).
• Figure 5A: as all cells seem to interact with all other cells, this figure does not convey relevant information about BM regions using for instance CXCL12 or SPP1. Please reanalyze to show specificity of the interactions of the single clusters. Also, since it is unlikely the CellPhoneDB2-predicted interactions are restricted to hematopoietic responders, please also describe the possible interactions between non-hematopoietic cells.
Fig. 5A was used to demonstrate the complexity of the interactions between hematopoietic cells and stromal cells.
To gain a more detailed understanding of the interactions, we also performed an analysis with the top-listed ligand-receptor pairs as shown in Fig. 5B-C and Figure 5-figure supplement 1B. Here, each dot represents the interaction of a specific ligand-receptor pair listed on the x-axis between the two individual clusters indicated in the y-axis, which we believe shows what the reviewer is asking for.
The specificity of the interactions between single clusters were shown in Fig. 5B-C and Figure 5-figure supplement 1B. The CXCL12- and SPP1-mediated interactions between MSSC/OC and hematopoietic clusters clearly suggested stromal cell type-specific interactions.
Regarding non-hematopoietic cells, both inter- and intra-stromal interactions were identified to be operative between different stromal subsets as well as within the same stromal cell population as shown in Figure 5-figure supplement 3B. In addition, we have also analyzed the interaction pattern between endothelial cells and hematopoietic cells as shown in Fig. 7A, and thus we believe that we have sufficiently described these interactions as requested by the reviewer.
Author Response
Reviewer #1 (Public Review):
This study used a multi-day learning paradigm combined with fMRI to reveal neural changes reflecting the learning of new (arbitrary) shape-sound associations. In the scanner, the shapes and sounds are presented separately and together, both before and after learning. When they are presented together, they can be either consistent or inconsistent with the learned associations. The analyses focus on auditory and visual cortices, as well as the object-selective cortex (LOC) and anterior temporal lobe regions (temporal pole (TP) and perirhinal cortex (PRC)). Results revealed several learning-induced changes, particularly in the anterior temporal lobe regions. First, the LOC and PRC showed a reduced bias to shapes vs sounds (presented separately) after learning. Second, the TP responded more strongly to incongruent than congruent shape-sound pairs after learning. Third, the similarity of TP activity patterns to sounds and shapes (presented separately) was increased for non-matching shape-sound comparisons after learning. Fourth, when comparing the pattern similarity of individual features to combined shape-sound stimuli, the PRC showed a reduced bias towards visual features after learning. Finally, comparing patterns to combined shape-sound stimuli before and after learning revealed a reduced (and negative) similarity for incongruent combinations in PRC. These results are all interpreted as evidence for an explicit integrative code of newly learned multimodal objects, in which the whole is different from the sum of the parts.
The study has many strengths. It addresses a fundamental question that is of broad interest, the learning paradigm is well-designed and controlled, and the stimuli are real 3D stimuli that participants interact with. The manuscript is well written and the figures are very informative, clearly illustrating the analyses performed.
There are also some weaknesses. The sample size (N=17) is small for detecting the subtle effects of learning. Most of the statistical analyses are not corrected for multiple comparisons (ROIs), and the specificity of the key results to specific regions is also not tested. Furthermore, the evidence for an integrative representation is rather indirect, and alternative interpretations for these results are not considered.
We thank the reviewer for their careful reading and the positive comments on our manuscript. As suggested, we have conducted additional analyses of theoretically-motivated ROIs and have found that temporal pole and perirhinal cortex are the only regions to show the key experience-dependent transformations. We are much more cautious with respect to multiple comparisons, and have removed a series of post hoc across-ROI comparisons that were irrelevant to the key questions of the present manuscript. The revised manuscript now includes much more discussion about alternative interpretations as suggested by the reviewer (and also by the other reviewers).
Additionally, we looked into scanning more participants, but our scanner has since had a full upgrade and the sequence used in the current study is no longer supported by our scanner. However, we note that while most analyses contain 17 participants, we employed a within-subject learning design that is not typically used in fMRI experiments and increases our power to detect an effect. This is supported by the robust effect size of the behavioural data, whereby 17 out of 18 participants revealed a learning effect (Cohen’s D = 1.28) and which was replicated in a follow-up experiment with a larger sample size.
We address the other reviewer comments point-by-point in the below.
Reviewer #2 (Public Review):
Li et al. used a four-day fMRI design to investigate how unimodal feature information is combined, integrated, or abstracted to form a multimodal object representation. The experimental question is of great interest and understanding how the human brain combines featural information to form complex representations is relevant for a wide range of researchers in neuroscience, cognitive science, and AI. While most fMRI research on object representations is limited to visual information, the authors examined how visual and auditory information is integrated to form a multimodal object representation. The experimental design is elegant and clever. Three visual shapes and three auditory sounds were used as the unimodal features; the visual shapes were used to create 3D-printed objects. On Day 1, the participants interacted with the 3D objects to learn the visual features, but the objects were not paired with the auditory features, which were played separately. On Day 2, participants were scanned with fMRI while they were exposed to the unimodal visual and auditory features as well as pairs of visual-auditory cues. On Day 3, participants again interacted with the 3D objects but now each was paired with one of the three sounds that played from an internal speaker. On Day 4, participants completed the same fMRI scanning runs they completed on Day 2, except now some visual-auditory feature pairs corresponded with Congruent (learned) objects, and some with Incongruent (unlearned) objects. Using the same fMRI design on Days 2 and 4 enables a well-controlled comparison between feature- and object-evoked neural representations before and after learning. The notable results corresponded to findings in the perirhinal cortex and temporal pole. The authors report (1) that a visual bias on Day 2 for unimodal features in the perirhinal cortex was attenuated after learning on Day 4, (2) a decreased univariate response to congruent vs. incongruent visual-auditory objects in the temporal pole on Day 4, (3) decreased pattern similarity between congruent vs. incongruent pairs of visual and auditory unimodal features in the temporal pole on Day 4, (4) in the perirhinal cortex, visual unimodal features on Day 2 do not correlate with their respective visual-auditory objects on Day 4, and (5) in the perirhinal cortex, multimodal object representations across Days 2 and 4 are uncorrelated for congruent objects and anticorrelated for incongruent. The authors claim that each of these results supports the theory that multimodal objects are represented in an "explicit integrative" code separate from feature representations. While these data are valuable and the results are interesting, the authors' claims are not well supported by their findings.
We thank the reviewer for the careful reading of our manuscript and positive comments. Overall, we now stay closer to the data when describing the results and provide our interpretation of these results in the discussion section while remaining open to alternative interpretations (as also suggested by Reviewer 1).
(1) In the introduction, the authors contrast two theories: (a) multimodal objects are represented in the co-activation of unimodal features, and (b) multimodal objects are represented in an explicit integrative code such that the whole is different than the sum of its parts. However, the distinction between these two theories is not straightforward. An explanation of what is precisely meant by "explicit" and "integrative" would clarify the authors' theoretical stance. Perhaps we can assume that an "explicit" representation is a new representation that is created to represent a multimodal object. What is meant by "integrative" is more ambiguous-unimodal features could be integrated within a representation in a manner that preserves the decodability of the unimodal features, or alternatively the multimodal representation could be completely abstracted away from the constituent features such that the features are no longer decodable. Even if the object representation is "explicit" and distinct from the unimodal feature representations, it can in theory still contain featural information, though perhaps warped or transformed. The authors do not clearly commit to a degree of featural abstraction in their theory of "explicit integrative" multimodal object representations which makes it difficult to assess the validity of their claims.
Due to its ambiguity, we removed the term “explicit” and now make it clear that our central question was whether crossmodal object representations require only unimodal feature-level representations (e.g., frogs are created from only the combination of shape and sound) or whether crossmodal object representations also rely on an integrative code distinct from the unimodal features (e.g., there is something more to “frog” than its original shape and sound). We now clarify this in the revised manuscript.
“One theoretical view from the cognitive sciences suggests that crossmodal objects are built from component unimodal features represented across distributed sensory regions.8 Under this view, when a child thinks about “frog”, the visual cortex represents the appearance of the shape of the frog whereas the auditory cortex represents the croaking sound. Alternatively, other theoretical views predict that multisensory objects are not only built from their component unimodal sensory features, but that there is also a crossmodal integrative code that is different from the sum of these parts.9,10,11,12,13 These latter views propose that anterior temporal lobe structures can act as a polymodal “hub” that combines separate features into integrated wholes.9,11,14,15” – pg. 4
For this reason, we designed our paradigm to equate the unimodal representations, such that neural differences between the congruent and incongruent conditions provide evidence for a crossmodal integrative code different from the unimodal features (because the unimodal features are equated by default in the design).
“Critically, our four-day learning task allowed us to isolate any neural activity associated with integrative coding in anterior temporal lobe structures that emerges with experience and differs from the neural patterns recorded at baseline. The learned and non-learned crossmodal objects were constructed from the same set of three validated shape and sound features, ensuring that factors such as familiarity with the unimodal features, subjective similarity, and feature identity were tightly controlled (Figure 2). If the mind represented crossmodal objects entirely as the reactivation of unimodal shapes and sounds (i.e., objects are constructed from their parts), then there should be no difference between the learned and non-learned objects (because they were created from the same three shapes and sounds). By contrast, if the mind represented crossmodal objects as something over and above their component features (i.e., representations for crossmodal objects rely on integrative coding that is different from the sum of their parts), then there should be behavioral and neural differences between learned and non-learned crossmodal objects (because the only difference across the objects is the learned relationship between the parts). Furthermore, this design allowed us to determine the relationship between the object representation acquired after crossmodal learning and the unimodal feature representations acquired before crossmodal learning. That is, we could examine whether learning led to abstraction of the object representations such that it no longer resembled the unimodal feature representations.” – pg. 5
Furthermore, we agree with the reviewer that our definition and methodological design does not directly capture the structure of the integrative code. With experience, the unimodal feature representations may be completely abstracted away, warped, or changed in a nonlinear transformation. We suggest that crossmodal learning forms an integrative code that is different from the original unimodal representations in the anterior temporal lobes, however, we agree that future work is needed to more directly capture the structure of the integrative code that emerges with experience.
“In our task, participants had to differentiate congruent and incongruent objects constructed from the same three shape and sound features (Figure 2). An efficient way to solve this task would be to form distinct object-level outputs from the overlapping unimodal feature-level inputs such that congruent objects are made to be orthogonal from the representations before learning (i.e., measured as pattern similarity equal to 0 in the perirhinal cortex; Figure 5b, 6, Supplemental Figure S5), whereas non-learned incongruent objects could be made to be dissimilar from the representations before learning (i.e., anticorrelation, measured as patten similarity less than 0 in the perirhinal cortex; Figure 6). Because our paradigm could decouple neural responses to the learned object representations (on Day 4) from the original component unimodal features at baseline (on Day 2), these results could be taken as evidence of pattern separation in the human perirhinal cortex.11,12 However, our pattern of results could also be explained by other types of crossmodal integrative coding. For example, incongruent object representations may be less stable than congruent object representations, such that incongruent objects representation are warped to a greater extent than congruent objects (Figure 6).” – pg. 18
“As one solution to the crossmodal binding problem, we suggest that the temporal pole and perirhinal cortex form unique crossmodal object representations that are different from the distributed features in sensory cortex (Figure 4, 5, 6, Supplemental Figure S5). However, the nature by which the integrative code is structured and formed in the temporal pole and perirhinal cortex following crossmodal experience – such as through transformations, warping, or other factors – is an open question and an important area for future investigation.” – pg. 18
(2) After participants learned the multimodal objects, the authors report a decreased univariate response to congruent visual-auditory objects relative to incongruent objects in the temporal pole. This is claimed to support the existence of an explicit, integrative code for multimodal objects. Given the number of alternative explanations for this finding, this claim seems unwarranted. A simpler interpretation of these results is that the temporal pole is responding to the novelty of the incongruent visual-auditory objects. If there is in fact an explicit, integrative multimodal object representation in the temporal pole, it is unclear why this would manifest in a decreased univariate response.
We thank the reviewer for identifying this issue. Our behavioural design controls unimodal feature-level novelty but allows object-level novelty to differ. Thus, neural differences between the congruent and incongruent conditions reflects sensitivity to the object-level differences between the combination of shape and sound. However, we agree that there are multiple interpretations regarding the nature of how the integrative code is structured in the temporal pole and perirhinal cortex. We have removed the interpretation highlighted by the reviewer from the results. Instead, we now provide our preferred interpretation in the discussion, while acknowledging the other possibilities that the reviewer mentions.
As one possibility, these results in temporal pole may reflect “conceptual combination”. “hummingbird” – a congruent pairing – may require less neural resources than an incongruent pairing such as “bark-frog”.
“Furthermore, these distinct anterior temporal lobe structures may be involved with integrative coding in different ways. For example, the crossmodal object representations measured after learning were found to be related to the component unimodal feature representations measured before learning in the temporal pole but not the perirhinal cortex (Figure 5, 6, Supplemental Figure S5). Moreover, pattern similarity for congruent shape-sound pairs were lower than the pattern similarity for incongruent shape-sound pairs after crossmodal learning in the temporal pole but not the perirhinal cortex (Figure 4b, Supplemental Figure S3a). As one interpretation of this pattern of results, the temporal pole may represent new crossmodal objects by combining previously learned knowledge. 8,9,10,11,13,14,15,33 Specifically, research into conceptual combination has linked the anterior temporal lobes to compound object concepts such as “hummingbird”.34,35,36 For example, participants during our task may have represented the sound-based “humming” concept and visually-based “bird” concept on Day 1, forming the crossmodal “hummingbird” concept on Day 3; Figure 1, 2, which may recruit less activity in temporal pole than an incongruent pairing such as “barking-frog”. For these reasons, the temporal pole may form a crossmodal object code based on pre-existing knowledge, resulting in reduced neural activity (Figure 3d) and pattern similarity towards features associated with learned objects (Figure 4b).”– pg. 18
(3) The authors ran a neural pattern similarity analysis on the unimodal features before and after multimodal object learning. They found that the similarity between visual and auditory features that composed congruent objects decreased in the temporal pole after multimodal object learning. This was interpreted to reflect an explicit integrative code for multimodal objects, though it is not clear why. First, behavioral data show that participants reported increased similarity between the visual and auditory unimodal features within congruent objects after learning, the opposite of what was found in the temporal pole. Second, it is unclear why an analysis of the unimodal features would be interpreted to reflect the nature of the multimodal object representations. Since the same features corresponded with both congruent and incongruent objects, the nature of the feature representations cannot be interpreted to reflect the nature of the object representations per se. Third, using unimodal feature representations to make claims about object representations seems to contradict the theoretical claim that explicit, integrative object representations are distinct from unimodal features. If the learned multimodal object representation exists separately from the unimodal feature representations, there is no reason why the unimodal features themselves would be influenced by the formation of the object representation. Instead, these results seem to more strongly support the theory that multimodal object learning results in a transformation or warping of feature space.
We apologize for the lack of clarity. We have now overhauled this aspect of our manuscript in an attempt to better highlight key aspects of our experimental design. In particular, because the unimodal features composing the congruent and incongruent objects were equated, neural differences between these conditions would provide evidence for an experience-dependent crossmodal integrative code that is different from its component unimodal features.
Related to the second and third points, we were looking at the extent to which the original unimodal representations change with crossmodal learning. Before crossmodal learning, we found that the perirhinal cortex tracked the similarity between the individual visual shape features and the crossmodal objects that were composed of those visual shapes – however, there was no evidence that perirhinal cortex was tracking the unimodal sound features on those crossmodal objects. After crossmodal learning, we see that this visual shape bias in perirhinal cortex was no longer present – that is, the representation in perirhinal cortex started to look less like the visual features that comprise the objects. Thus, crossmodal learning transformed the perirhinal representations so that they were no longer predominantly grounded in a single visual modality, which may be a mechanism by which object concepts gain their abstraction. We have now tried to be clearer about this interpretation throughout the paper.
Notably, we suggest that experience may change both the crossmodal object representations, as well as the unimodal feature representations. For example, we have previously shown that unimodal visual features are influenced by experience in parallel with the representation of the conjunction (e.g., Liang et al., 2020; Cerebral Cortex). Nevertheless, we remain open to the myriad possible structures of the integrative code that might emerge with experience.
We now clarify these points throughout the manuscript. For example:
“We then examined whether the original representations would change after participants learned how the features were paired together to make specific crossmodal objects, conducting the same analysis described above after crossmodal learning had taken place (Figure 5b). With this analysis, we sought to measure the relationship between the representation for the learned crossmodal object and the original baseline representation for the unimodal features. More specifically, the voxel-wise activity for unimodal feature runs before crossmodal learning was correlated to the voxel-wise activity for crossmodal object runs after crossmodal learning (Figure 5b). Another linear mixed model which included modality as a fixed factor within each ROI revealed that the perirhinal cortex was no longer biased towards visual shape after crossmodal learning (F1,32 = 0.12, p = 0.73), whereas the temporal pole, LOC, V1, and A1 remained biased towards either visual shape or sound (F1,30-32 between 16.20 and 73.42, all p < 0.001, η2 between 0.35 and 0.70).” – pg. 14
“To investigate this effect in perirhinal cortex more specifically, we conducted a linear mixed model to directly compare the change in the visual bias of perirhinal representations from before crossmodal learning to after crossmodal learning (green regions in Figure 5a vs. 5b). Specifically, the linear mixed model included learning day (before vs. after crossmodal learning) and modality (visual feature match to crossmodal object vs. sound feature match to crossmodal object). Results revealed a significant interaction between learning day and modality in the perirhinal cortex (F1,775 = 5.56, p = 0.019, η2 = 0.071), meaning that the baseline visual shape bias observed in perirhinal cortex (green region of Figure 5a) was significantly attenuated with experience (green region of Figure 5b). After crossmodal learning, a given shape no longer invoked significant pattern similarity between objects that had the same shape but differed in terms of what they sounded like. Taken together, these results suggest that prior to learning the crossmodal objects, the perirhinal cortex had a default bias toward representing the visual shape information and was not representing sound information of the crossmodal objects. After crossmodal learning, however, the visual shape bias in perirhinal cortex was no longer present. That is, with crossmodal learning, the representations within perirhinal cortex started to look less like the visual features that comprised the crossmodal objects, providing evidence that the perirhinal representations were no longer predominantly grounded in the visual modality.” – pg. 13
“Importantly, the initial visual shape bias observed in the perirhinal cortex was attenuated by experience (Figure 5, Supplemental Figure S5), suggesting that the perirhinal representations had become abstracted and were no longer predominantly grounded in a single modality after crossmodal learning. One possibility may be that the perirhinal cortex is by default visually driven as an extension to the ventral visual stream,10,11,12 but can act as a polymodal “hub” region for additional crossmodal input following learning.” – pg. 19
(4) The most compelling evidence the authors provide for their theoretical claims is the finding that, in the perirhinal cortex, the unimodal feature representations on Day 2 do not correlate with the multimodal objects they comprise on Day 4. This suggests that the learned multimodal object representations are not combinations of their unimodal features. If unimodal features are not decodable within the congruent object representations, this would support the authors' explicit integrative hypothesis. However, the analyses provided do not go all the way in convincing the reader of this claim. First, the analyses reported do not differentiate between congruent and incongruent objects. If this result in the perirhinal cortex reflects the formation of new multimodal object representations, it should only be true for congruent objects but not incongruent objects. Since the analyses combine congruent and incongruent objects it is not possible to know whether this was the case. Second, just because feature representations on Day 2 do not correlate with multimodal object patterns on Day 4 does not mean that the object representations on Day 4 do not contain featural information. This could be directly tested by correlating feature representations on Day 4 with congruent vs. incongruent object representations on Day 4. It could be that representations in the perirhinal cortex are not stable over time and all representations-including unimodal feature representations-shift between sessions, which could explain these results yet not entail the existence of abstracted object representations.
We thank the reviewer for this suggestion and have conducted the two additional analyses. Specifically, we split the congruent and incongruent conditions and also investigated correlations between unimodal representations on Day 4 with crossmodal object representations on Day 4. There was no significant interaction between modality and congruency in any ROI across or within learning days. One possible explanation for these findings is that both congruent and incongruent crossmodal objects are represented differently from their underlying unimodal features, and all of these representations can transform with experience.
However, the new analyses also revealed that perirhinal cortex was the only region without a modality-specific bias after crossmodal learning (e.g., Day 4 Unimodal Feature runs x Day 4 Crossmodal Object runs; now shown in Supplemental Figure S5). Overall, these results are consistent with the notion of a crossmodal integrative code in perirhinal cortex that has changed with experience and is different from the component unimodal features. Nevertheless, we explore alternative interpretations for how the crossmodal code emerges with experience in the discussion.
“To examine whether these results differed by congruency (i.e., whether any modality-specific biases differed as a function of whether the object was congruent or incongruent), we conducted exploratory linear mixed models for each of the five a priori ROIs across learning days. More specifically, we correlated: 1) the voxel-wise activity for Unimodal Feature Runs before crossmodal learning to the voxel-wise activity for Crossmodal Object Runs before crossmodal learning (Day 2 vs. Day 2), 2) the voxel-wise activity for Unimodal Feature Runs before crossmodal learning to the voxel-wise activity for Crossmodal Object Runs after crossmodal learning (Day 2 vs Day 4), and 3) the voxel-wise activity for Unimodal Feature Runs after crossmodal learning to the voxel-wise activity for Crossmodal Object Runs after crossmodal learning (Day 4 vs Day 4). For each of the three analyses described, we then conducted separate linear mixed models which included modality (visual feature match to crossmodal object vs. sound feature match to crossmodal object) and congruency (congruent vs. incongruent)….There was no significant relationship between modality and congruency in any ROI between Day 2 and Day 2 (F1,346-368 between 0.00 and 1.06, p between 0.30 and 0.99), between Day 2 and Day 4 (F1,346-368 between 0.021 and 0.91, p between 0.34 and 0.89), or between Day 4 and Day 4 (F1,346-368 between 0.01 and 3.05, p between 0.082 and 0.93). However, exploratory analyses revealed that perirhinal cortex was the only region without a modality-specific bias and where the unimodal feature runs were not significantly correlated to the crossmodal object runs after crossmodal learning (Supplemental Figure S5).” – pg. 14
“Taken together, the overall pattern of results suggests that representations of the crossmodal objects in perirhinal cortex were heavily influenced by their consistent visual features before crossmodal learning. However, the crossmodal object representations were no longer influenced by the component visual features after crossmodal learning (Figure 5, Supplemental Figure S5). Additional exploratory analyses did not find evidence of experience-dependent changes in the hippocampus or inferior parietal lobes (Supplemental Figure S4c-e).” – pg. 14
“The voxel-wise matrix for Unimodal Feature runs on Day 4 were correlated to the voxel-wise matrix for Crossmodal Object runs on Day 4 (see Figure 5 in the main text for an example). We compared the average pattern similarity (z-transformed Pearson correlation) between shape (blue) and sound (orange) features specifically after crossmodal learning. Consistent with Figure 5b, perirhinal cortex was the only region without a modality-specific bias. Furthermore, perirhinal cortex was the only region where the representations of both the visual and sound features were not significantly correlated to the crossmodal objects. By contrast, every other region maintained a modality-specific bias for either the visual or sound features. These results suggest that perirhinal cortex representations were transformed with experience, such that the initial visual shape representations (Figure 5a) were no longer grounded in a single modality after crossmodal learning. Furthermore, these results suggest that crossmodal learning formed an integrative code different from the unimodal features in perirhinal cortex, as the visual and sound features were not significantly correlated with the crossmodal objects. * p < 0.05, ** p < 0.01, *** p < 0.001. Horizontal lines within brain regions indicate a significant main effect of modality. Vertical asterisks denote pattern similarity comparisons relative to 0.” – Supplemental Figure S5
“We found that the temporal pole and perirhinal cortex – two anterior temporal lobe structures – came to represent new crossmodal object concepts with learning, such that the acquired crossmodal object representations were different from the representation of the constituent unimodal features (Figure 5, 6). Intriguingly, the perirhinal cortex was by default biased towards visual shape, but that this initial visual bias was attenuated with experience (Figure 3c, 5, Supplemental Figure S5). Within the perirhinal cortex, the acquired crossmodal object concepts (measured after crossmodal learning) became less similar to their original component unimodal features (measured at baseline before crossmodal learning); Figure 5, 6, Supplemental Figure S5. This is consistent with the idea that object representations in perirhinal cortex integrate the component sensory features into a whole that is different from the sum of the component parts, which might be a mechanism by which object concepts obtain their abstraction…. As one solution to the crossmodal binding problem, we suggest that the temporal pole and perirhinal cortex form unique crossmodal object representations that are different from the distributed features in sensory cortex (Figure 4, 5, 6, Supplemental Figure S5). However, the nature by which the integrative code is structured and formed in the temporal pole and perirhinal cortex following crossmodal experience – such as through transformations, warping, or other factors – is an open question and an important area for future investigation.” – pg. 18
In sum, the authors have collected a fantastic dataset that has the potential to answer questions about the formation of multimodal object representations in the brain. A more precise delineation of different theoretical accounts and additional analyses are needed to provide convincing support for the theory that “explicit integrative” multimodal object representations are formed during learning.
We thank the reviewer for the positive comments and helpful feedback. We hope that our changes to our wording and clarifications to our methodology now more clearly supports the central goal of our study: to find evidence of crossmodal integrative coding different from the original unimodal feature parts in anterior temporal lobe structures. We furthermore agree that future research is needed to delineate the structure of the integrative code that emerges with experience in the anterior temporal lobes.
Reviewer #3 (Public Review):
This paper uses behavior and functional brain imaging to understand how neural and cognitive representations of visual and auditory stimuli change as participants learn associations among them. Prior work suggests that areas in the anterior temporal (ATL) and perirhinal cortex play an important role in learning/representing cross-modal associations, but the hypothesis has not been directly tested by evaluating behavior and functional imaging before and after learning cross- modal associations. The results show that such learning changes both the perceived similarities amongst stimuli and the neural responses generated within ATL and perirhinal regions, providing novel support for the view that cross-modal learning leads to a representational change in these regions.
This work has several strengths. It tackles an important question for current theories of object representation in the mind and brain in a novel and quite direct fashion, by studying how these representations change with cross-modal learning. As the authors note, little work has directly assessed representational change in ATL following such learning, despite the widespread view that ATL is critical for such representation. Indeed, such direct assessment poses several methodological challenges, which the authors have met with an ingenious experimental design. The experiment allows the authors to maintain tight control over both the familiarity and the perceived similarities amongst the shapes and sounds that comprise their stimuli so that the observed changes across sessions must reflect learned cross-modal associations among these. I especially appreciated the creation of physical objects that participants can explore and the approach to learning in which shapes and sounds are initially experienced independently and later in an associated fashion. In using multi-echo MRI to resolve signals in ventral ATL, the authors have minimized a key challenge facing much work in this area (namely the poor SNR yielded by standard acquisition sequences in ventral ATL). The use of both univariate and multivariate techniques was well-motivated and helpful in testing the central questions. The manuscript is, for the most part, clearly written, and nicely connects the current work to important questions in two literatures, specifically (1) the hypothesized role of the perirhinal cortex in representing/learning complex conjunctions of features and (2) the tension between purely embodied approaches to semantic representation vs the view that ATL regions encode important amodal/crossmodal structure.
There are some places in the manuscript that would benefit from further explanation and methodological detail. I also had some questions about the results themselves and what they signify about the roles of ATL and the perirhinal cortex in object representation.
We thank the reviewer for their positive feedback and address the comments in the below point-by-point responses.
(A) I found the terms "features" and "objects" to be confusing as used throughout the manuscript, and sometimes inconsistent. I think by "features" the authors mean the shape and sound stimuli in their experiment. I think by "object" the authors usually mean the conjunction of a shape with a sound---for instance, when a shape and sound are simultaneously experienced in the scanner, or when the participant presses a button on the shape and hears the sound. The confusion comes partly because shapes are often described as being composed of features, not features in and of themselves. (The same is sometimes true of sounds). So when reading "features" I kept thinking the paper referred to the elements that went together to comprise a shape. It also comes from ambiguous use of the word object, which might refer to (a) the 3D- printed item that people play with, which is an object, or (b) a visually-presented shape (for instance, the localizer involved comparing an "object" to a "phase-scrambled" stimulus---here I assume "object" refers to an intact visual stimulus and not the joint presentation of visual and auditory items). I think the design, stimuli, and results would be easier for a naive reader to follow if the authors used the terms "unimodal representation" to refer to cases where only visual or auditory input is presented, and "cross-modal" or "conjoint" representation when both are present.
We thank the reviewer for this suggestion and agree. We have replaced the terms “features” and “objects” with “unimodal” and “crossmodal” in the title, text, and figures throughout the manuscript for consistency (i.e., “crossmodal binding problem”). To simplify the terminology, we have also removed the localizer results.
(B) There are a few places where I wasn't sure what exactly was done, and where the methods lacked sufficient detail for another scientist to replicate what was done. Specifically:
(1) The behavioral study assessing perceptual similarity between visual and auditory stimuli was unclear. The procedure, stimuli, number of trials, etc, should be explained in sufficient detail in methods to allow replication. The results of the study should also minimally be reported in the supplementary information. Without an understanding of how these studies were carried out, it was very difficult to understand the observed pattern of behavioral change. For instance, I initially thought separate behavioral blocks were carried out for visual versus auditory stimuli, each presented in isolation; however, the effects contrast congruent and incongruent stimuli, which suggests these decisions must have been made for the conjoint presentation of both modalities. I'm still not sure how this worked. Additionally, the manuscript makes a brief mention that similarity judgments were made in the context of "all stimuli," but I didn't understand what that meant. Similarity ratings are hugely sensitive to the contrast set with which items appear, so clarity on these points is pretty important. A strength of the design is the contention that shape and sound stimuli were psychophysically matched, so it is important to show the reader how this was done and what the results were.
We agree and apologize for the lack of sufficient detail in the original manuscript. We now include much more detail about the similarity rating task. The methodology and results of the behavioral rating experiments are now shown in Supplemental Figure S1. In Figure S1a, the similarity ratings are visualized on a multidimensional scaling plot. The triangular geometry for shape (blue) and sound (red) indicate that the subjective similarity was equated within each unimodal feature across individual participants. Quantitatively, there was no difference in similarity between the congruent and incongruent pairings in Figure S1b and Figure S1c prior to crossmodal learning. In addition to providing more information on these methods in the Supplemental Information, we also now provide a more detailed description of the task in the manuscript itself. For convenience, we reproduce these sections below.
“Pairwise Similarity Task. Using the same task as the stimulus validation procedure (Supplemental Figure S1a), participants provided similarity ratings for all combinations of the 3 validated shapes and 3 validated sounds (each of the six features were rated in the context of every other feature in the set, with 4 repeats of the same feature, for a total of 72 trials). More specifically, three stimuli were displayed on each trial, with one at the top and two at the bottom of the screen in the same procedure as we have used previously27. The 3D shapes were visually displayed as a photo, whereas sounds were displayed on screen in a box that could be played over headphones when clicked with the mouse. The participant made an initial judgment by selecting the more similar stimulus on the bottom relative to the stimulus on the top. Afterwards, the participant made a similarity rating between each bottom stimulus with the top stimulus from 0 being no similarity to 5 being identical. This procedure ensured that ratings were made relative to all other stimuli in the set.”– pg. 28
“Pairwise similarity task and results. In the initial stimulus validation experiment, participants provided pairwise ratings for 5 sounds and 3 shapes. The shapes were equated in their subjective similarity that had been selected from a well-characterized perceptually uniform stimulus space27 and the pairwise ratings followed the same procedure as described in ref 27. Based on this initial experiment, we then selected the 3 sounds from the that were most closely equated in their subjective similarity. (a) 3D-printed shapes were displayed as images, whereas sounds were displayed in a box that could be played when clicked by the participant. Ratings were averaged to produce a similarity matrix for each participant, and then averaged to produce a group-level similarity matrix. Shown as triangular representational geometries recovered from multidimensional scaling in the above, shapes (blue) and sounds (orange) were approximately equated in their subjective similarity. These features were then used in the four-day crossmodal learning task. (b) Behavioral results from the four-day crossmodal learning task paired with multi-echo fMRI described in the main text. Before crossmodal learning, there was no difference in similarity between shape and sound features associated with congruent objects compared to incongruent objects – indicating that similarity was controlled at the unimodal feature-level. After crossmodal learning, we observed a robust shift in the magnitude of similarity. The shape and sound features associated with congruent objects were now significantly more similar than the same shape and sound features associated with incongruent objects (p < 0.001), evidence that crossmodal learning changed how participants experienced the unimodal features (observed in 17/18 participants). (c) We replicated this learning-related shift in pattern similarity with a larger sample size (n = 44; observed in 38/44 participants). *** denotes p < 0.001. Horizontal lines denote the comparison of congruent vs. incongruent conditions. – Supplemental Figure S1
(2) The experiences through which participants learned/experienced the shapes and sounds were unclear. The methods mention that they had one minute to explore/palpate each shape and that these experiences were interleaved with other tasks, but it is not clear what the other tasks were, how many such exploration experiences occurred, or how long the total learning time was. The manuscript also mentions that participants learn the shape-sound associations with 100% accuracy but it isn't clear how that was assessed. These details are important partly b/c it seems like very minimal experience to change neural representations in the cortex.
We apologize for the lack of detail and agree with the reviewer’s suggestions – we now include much more information in the methods section. Each behavioral day required about 1 hour of total time to complete, and indeed, participants rapidly learned their associations with minimal experience. For example:
“Behavioral Tasks. On each behavioral day (Day 1 and Day 3; Figure 2), participants completed the following tasks, in this order: Exploration Phase, one Unimodal Feature 1-back run (26 trials), Exploration Phase, one Crossmodal 1-back run (26 trials), Exploration Phase, Pairwise Similarity Task (24 trials), Exploration Phase, Pairwise Similarity Task (24 trials), Exploration Phase, Pairwise Similarity Task (24 trials), and finally, Exploration Phase. To verify learning on Day 3, participants also additionally completed a Learning Verification Task at the end of the session. – pg. 27
“The overall procedure ensured that participants extensively explored the unimodal features on Day 1 and the crossmodal objects on Day 3. The Unimodal Feature and the Crossmodal Object 1-back runs administered on Day 1 and Day 3 served as practice for the neuroimaging sessions on Day 2 and Day 4, during which these 1-back tasks were completed. Each behavioral session required less than 1 hour of total time to complete.” – pg. 27
“Learning Verification Task (Day 3 only). As the final task on Day 3, participants completed a task to ensure that participants successfully formed their crossmodal pairing. All three shapes and sounds were randomly displayed in 6 boxes on a display. Photos of the 3D shapes were shown, and sounds were played by clicking the box with the mouse cursor. The participant was cued with either a shape or sound, and then selected the corresponding paired feature. At the end of Day 3, we found that all participants reached 100% accuracy on this task (10 trials).” – pg. 29
(3) I didn't understand the similarity metric used in the multivariate imaging analyses. The manuscript mentions Z-scored Pearson's r, but I didn't know if this meant (a) many Pearson coefficients were computed and these were then Z-scored, so that 0 indicates a value equal to the mean Pearson correlation and 1 is equal to the standard deviation of the correlations, or (b) whether a Fisher Z transform was applied to each r (so that 0 means r was also around 0). From the interpretation of some results, I think the latter is the approach taken, but in general, it would be helpful to see, in Methods or Supplementary information, exactly how similarity scores were computed, and why that approach was adopted. This is particularly important since it is hard to understand the direction of some key effects.
The reviewer is correct that the Fisher Z transform was applied to each individual r before averaging the correlations. This approach is generally recommended when averaging correlations (see Corey, Dunlap, & Burke, 1998). We are now clearer on this point in the manuscript:
“The z-transformed Pearson’s correlation coefficient was used as the distance metric for all pattern similarity analyses. More specifically, each individual Pearson correlation was Fisher z-transformed and then averaged (see 61).” – pg. 32
(C) From Figure 3D, the temporal pole mask appears to exclude the anterior fusiform cortex (or the ventral surface of the ATL generally). If so, this is a shame, since that appears to be the locus most important to cross-modal integration in the "hub and spokes" model of semantic representation in the brain. The observation in the paper that the perirhinal cortex seems initially biased toward visual structure while more superior ATL is biased toward auditory structure appears generally consistent with the "graded hub" view expressed, for instance, in our group's 2017 review paper (Lambon Ralph et al., Nature Reviews Neuroscience). The balance of visual- versus auditory-sensitivity in that work appears balanced in the anterior fusiform, just a little lateral to the anterior perirhinal cortex. It would be helpful to know if the same pattern is observed for this area specifically in the current dataset.
We thank the reviewer for this suggestion. After close inspection of Lambon Ralph et al. (2017), we believe that our perirhinal cortex mask appears to be overlapping with the ventral ATL/anterior fusiform region that the reviewer mentions. See Author response image 1 for a visual comparison:
Author response image 1.
The top four figures are sampled from Lambon Ralph et al (2017), whereas the bottom two figures visualize our perirhinal cortex mask (white) and temporal pole mask (dark green) relative to the fusiform cortex. The ROIs visualized were defined from the Harvard-Oxford atlas.
We now mention this area of overlap in our manuscript and link it to the hub and spokes model:
“Notably, our perirhinal cortex mask overlaps with a key region of the ventral anterior temporal lobe thought to be the central locus of crossmodal integration in the “hub and spokes” model of semantic representations.9,50 – pg. 20
(D) While most effects seem robust from the information presented, I'm not so sure about the analysis of the perirhinal cortex shown in Figure 5. This compares (I think) the neural similarity evoked by a unimodal stimulus ("feature") to that evoked by the same stimulus when paired with its congruent stimulus in the other modality ("object"). These similarities show an interaction with modality prior to cross-modal association, but no interaction afterward, leading the authors to suggest that the perirhinal cortex has become less biased toward visual structure following learning. But the plots in Figures 4a and b are shown against different scales on the y-axes, obscuring the fact that all of the similarities are smaller in the after-learning comparison. Since the perirhinal interaction was already the smallest effect in the pre-learning analysis, it isn't really surprising that it drops below significance when all the effects diminish in the second comparison. A more rigorous test would assess the reliability of the interaction of comparison (pre- or post-learning) with modality. The possibility that perirhinal representations become less "visual" following cross-modal learning is potentially important so a post hoc contrast of that kind would be helpful.
We apologize for the lack of clarity. We conducted a linear mixed model to assess the interaction between modality and crossmodal learning day (before and after crossmodal learning) in the perirhinal cortex as described by the reviewer. The critical interaction was significant, which is now clarified in the text as well as in the rescaled figure plots.
“To investigate this effect in perirhinal cortex more specifically, we conducted a linear mixed model to directly compare the change in the visual bias of perirhinal representations from before crossmodal learning to after crossmodal learning (green regions in Figure 5a vs. 5b). Specifically, the linear mixed model included learning day (before vs. after crossmodal learning) and modality (visual feature match to crossmodal object vs. sound feature match to crossmodal object). Results revealed a significant interaction between learning day and modality in the perirhinal cortex (F1,775 = 5.56, p = 0.019, η2 = 0.071), meaning that the baseline visual shape bias observed in perirhinal cortex (green region of Figure 5a) was significantly attenuated with experience (green region of Figure 5b). After crossmodal learning, a given shape no longer invoked significant pattern similarity between objects that had the same shape but differed in terms of what they sounded like. Taken together, these results suggest that prior to learning the crossmodal objects, the perirhinal cortex had a default bias toward representing the visual shape information and was not representing sound information of the crossmodal objects. After crossmodal learning, however, the visual shape bias in perirhinal cortex was no longer present. That is, with crossmodal learning, the representations within perirhinal cortex started to look less like the visual features that comprised the crossmodal objects, providing evidence that the perirhinal representations were no longer predominantly grounded in the visual modality.” – pg. 13
We note that not all effects drop in Figure 5b (even in regions with a similar numerical pattern similarity to PRC, like the hippocampus – also see Supplemental Figure S5 for a comparison for patterns only on Day 4), suggesting that the change in visual bias in PRC is not simply due to noise.
“Importantly, the change in pattern similarity in the perirhinal cortex across learning days (Figure 5) is unlikely to be driven by noise, poor alignment of patterns across sessions, or generally reduced responses. Other regions with numerically similar pattern similarity to perirhinal cortex did not change across learning days (e.g., visual features x crossmodal objects in A1 in Figure 5; the exploratory ROI hippocampus with numerically similar pattern similarity to perirhinal cortex also did not change in Supplemental Figure S4c-d).” – pg. 14
(E) Is there a reason the authors did not look at representation and change in the hippocampus? As a rapid-learning, widely-connected feature-binding mechanism, and given the fairly minimal amount of learning experience, it seems like the hippocampus would be a key area of potential import for the cross-modal association. It also looks as though the hippocampus is implicated in the localizer scan (Figure 3c).
We thank the reviewer for this suggestion and now include additional analyses for the hippocampus. We found no evidence of crossmodal integrative coding different from the unimodal features. Rather, the hippocampus seems to represent the convergence of unimodal features, as evidenced by …[can you give some pithy description for what is meant by “convergence” vs “integration”?]. We provide these results in the Supplemental Information and describe them in the main text:
“Analyses for the hippocampus (HPC) and inferior parietal lobe (IPL). (a) In the visual vs. auditory univariate analysis, there was no visual or sound bias in HPC, but there was a bias towards sounds that increased numerically after crossmodal learning in the IPL. (b) Pattern similarity analyses between unimodal features associated with congruent objects and incongruent objects. Similar to Supplemental Figure S3, there was no main effect of congruency in either region. (c) When we looked at the pattern similarity between Unimodal Feature runs on Day 2 to Crossmodal Object runs on Day 2, we found that there was significant pattern similarity when there was a match between the unimodal feature and the crossmodal object (e.g., pattern similarity > 0). This pattern of results held when (d) correlating the Unimodal Feature runs on Day 2 to Crossmodal Object runs on Day 4, and (e) correlating the Unimodal Feature runs on Day 4 to Crossmodal Object runs on Day 4. Finally, (f) there was no significant pattern similarity between Crossmodal Object runs before learning correlated to Crossmodal Object after learning in HPC, but there was significant pattern similarity in IPL (p < 0.001). Taken together, these results suggest that both HPC and IPL are sensitive to visual and sound content, as the (c, d, e) unimodal feature-level representations were correlated to the crossmodal object representations irrespective of learning day. However, there was no difference between congruent and incongruent pairings in any analysis, suggesting that HPC and IPL did not represent crossmodal objects differently from the component unimodal features. For these reasons, HPC and IPL may represent the convergence of unimodal feature representations (i.e., because HPC and IPL were sensitive to both visual and sound features), but our results do not seem to support these regions in forming crossmodal integrative coding distinct from the unimodal features (i.e., because representations in HPC and IPL did not differentiate the congruent and incongruent conditions and did not change with experience). * p < 0.05, ** p < 0.01, *** p < 0.001. Asterisks above or below bars indicate a significant difference from zero. Horizontal lines within brain regions in (a) reflect an interaction between modality and learning day, whereas horizontal lines within brain regions in reflect main effects of (b) learning day, (c-e) modality, or (f) congruency.” – Supplemental Figure S4.
“Notably, our perirhinal cortex mask overlaps with a key region of the ventral anterior temporal lobe thought to be the central locus of crossmodal integration in the “hub and spokes” model of semantic representations.9,50 However, additional work has also linked other brain regions to the convergence of unimodal representations, such as the hippocampus51,52,53 and inferior parietal lobes.54,55 This past work on the hippocampus and inferior parietal lobe does not necessarily address the crossmodal binding problem that was the main focus of our present study, as previous findings often do not differentiate between crossmodal integrative coding and the convergence of unimodal feature representations per se. Furthermore, previous studies in the literature typically do not control for stimulus-based factors such as experience with unimodal features, subjective similarity, or feature identity that may complicate the interpretation of results when determining regions important for crossmodal integration. Indeed, we found evidence consistent with the convergence of unimodal feature-based representations in both the hippocampus and inferior parietal lobes (Supplemental Figure S4), but no evidence of crossmodal integrative coding different from the unimodal features. The hippocampus and inferior parietal lobes were both sensitive to visual and sound features before and after crossmodal learning (see Supplemental Figure S4c-e). Yet the hippocampus and inferior parietal lobes did not differentiate between the congruent and incongruent conditions or change with experience (see Supplemental Figure S4).” – pg. 20
(F) The direction of the neural effects was difficult to track and understand. I think the key observation is that TP and PRh both show changes related to cross-modal congruency - but still it would be helpful if the authors could articulate, perhaps via a schematic illustration, how they think representations in each key area are changing with the cross-modal association. Why does the temporal pole come to activate less for congruent than incongruent stimuli (Figure 3)? And why do TP responses grow less similar to one another for congruent relative to incongruent stimuli after learning (Figure 4)? Why are incongruent stimulus similarities anticorrelated in their perirhinal responses following cross-modal learning (Figure 6)?
We thank the author for identifying this issue, which was also raised by the other reviewers. The reviewer is correct that the key observation is that the TP and PRC both show changes related to crossmodal congruency (given that the unimodal features were equated in the methodological design). However, the structure of the integrative code is less clear, which we now emphasize in the main text. Our findings provide evidence of a crossmodal integrative code that is different from the unimodal features, and future studies are needed to better understand the structure of how such a code might emerge. We now more clearly highlight this distinction throughout the paper:
“By contrast, perirhinal cortex may be involved in pattern separation following crossmodal experience. In our task, participants had to differentiate congruent and incongruent objects constructed from the same three shape and sound features (Figure 2). An efficient way to solve this task would be to form distinct object-level outputs from the overlapping unimodal feature-level inputs such that congruent objects are made to be orthogonal from the representations before learning (i.e., measured as pattern similarity equal to 0 in the perirhinal cortex; Figure 5b, 6, Supplemental Figure S5), whereas non-learned incongruent objects could be made to be dissimilar from the representations before learning (i.e., anticorrelation, measured as patten similarity less than 0 in the perirhinal cortex; Figure 6). Because our paradigm could decouple neural responses to the learned object representations (on Day 4) from the original component unimodal features at baseline (on Day 2), these results could be taken as evidence of pattern separation in the human perirhinal cortex.11,12 However, our pattern of results could also be explained by other types of crossmodal integrative coding. For example, incongruent object representations may be less stable than congruent object representations, such that incongruent objects representation are warped to a greater extent than congruent objects (Figure 6).” – pg. 18
“As one solution to the crossmodal binding problem, we suggest that the temporal pole and perirhinal cortex form unique crossmodal object representations that are different from the distributed features in sensory cortex (Figure 4, 5, 6, Supplemental Figure S5). However, the nature by which the integrative code is structured and formed in the temporal pole and perirhinal cortex following crossmodal experience – such as through transformations, warping, or other factors – is an open question and an important area for future investigation. Furthermore, these anterior temporal lobe structures may be involved with integrative coding in different ways. For example, the crossmodal object representations measured after learning were found to be related to the component unimodal feature representations measured before learning in the temporal pole but not the perirhinal cortex (Figure 5, 6, Supplemental Figure S5). Moreover, pattern similarity for congruent shape-sound pairs were lower than the pattern similarity for incongruent shape-sound pairs after crossmodal learning in the temporal pole but not the perirhinal cortex (Figure 4b, Supplemental Figure S3a). As one interpretation of this pattern of results, the temporal pole may represent new crossmodal objects by combining previously learned knowledge. 8,9,10,11,13,14,15,33 Specifically, research into conceptual combination has linked the anterior temporal lobes to compound object concepts such as “hummingbird”.34,35,36 For example, participants during our task may have represented the sound-based “humming” concept and visually-based “bird” concept on Day 1, forming the crossmodal “hummingbird” concept on Day 3; Figure 1, 2, which may recruit less activity in temporal pole than an incongruent pairing such as “barking-frog”. For these reasons, the temporal pole may form a crossmodal object code based on pre-existing knowledge, resulting in reduced neural activity (Figure 3d) and pattern similarity towards features associated with learned objects (Figure 4b).” – pg. 18
This work represents a key step in our advancing understanding of object representations in the brain. The experimental design provides a useful template for studying neural change related to the cross-modal association that may prove useful to others in the field. Given the broad variety of open questions and potential alternative analyses, an open dataset from this study would also likely be a considerable contribution to the field.
Author Response
Reviewer #1 (Public Review):
This paper tests the hypothesis that 1/f exponent of LFP power spectrum reflects E-I balance in a rodent model and Parkinson's patients. The authors suggest that their findings fit with this hypothesis, but there are concerns about confirmation bias (elaborated on below) and potential methodological issues, despite the strength of incorporating data from both animal model and neurological patients.
First, the frequency band used to fit the 1/f exponent varies between experiments and analyses, inviting concerns about potentially cherry-picking the data to fit with the prior hypothesis. The frequency band used for fitting the exponent was 30-100 Hz in Experiment 1 (rodent model), 40-90 Hz in Experiment 2 (PD, levodopa), and 10-50 Hz in Experiment 3 (PD, DBS). Ad-hoc reasons were given to justify these choices, such as " to avoid a spectral plateau starting > 50 Hz" in Experiment 3. However, at least in Experiment 3 (Fig. 3), if the frequency range was shifted to 1-10 Hz, the authors would have uncovered the opposite effect, where the exponent is smaller for DBS-on condition.
We agree that parameter choice is crucial, in particular, choice of the fitting range. In addition to the 40-90 Hz range (Figure 2C), we have performed aperiodic fitting for five other frequency ranges to test to what extent the reported results are sensitive to the selected frequency range (Figure S2A). This analysis showed that the results are robust when a broad frequency range from 30 to 95 Hz was chosen, which is consistent with what has been suggested by Gao et al., 2017 to make inferences on the E/I ratio.
Accordingly, we have now repeated the analyses for the animal data with the same fitting range used for the ON-OFF medication comparison in humans. Along with Figure S2A where different frequency ranges were tested for data used in Figure 2, this shows that the results in Figure 1 and 2 hold up with higher aperiodic exponents when STN spiking is low and vice versa. Therefore, a broad fitting range from 30 to 90 Hz (excluding harmonics of mains interference) generates consistent results for both human and animal data.
We opted against a fitting range from 1-10 Hz because of two restraints highlighted in Gerster et al., 2022. First, a fitting range starting at 1 Hz could have a larger y-intercept due to the presence of low-frequency oscillations. This could lead to a larger aperiodic exponent and could be misinterpreted as stronger neural inhibition. Therefore, the lower fitting bound should be chosen to best avoid known oscillations in the delta/theta range (Gerster et al., 2022). Second, frequencies should be chosen to avoid oscillations crossing fitting range limits. In Figure 3A, oscillations in the theta/alpha band both ON and OFF stimulation would complicate parameterisation and would likely result in spurious fits.
We also tested the effect of changing the peak threshold, peak width limits and the aperiodic fitting mode on FOOOF parameterisation. Increasing and decreasing the peak threshold from its default value (at 2 standard deviations) did not change results (Figure S2B). Similarly, adapting the peak width limits did not affect the exponent difference between medication states (Figure S2C). Finally, choosing the ‘knee’ mode instead of ‘fixed’ resulted in fundamentally different aperiodic fits that did not differ anymore with medication (Figure S2D). This is most likely a consequence of the near linear PSD in log-log space from 40 to 90 Hz (Figure 2B). If there is no bend in the PSD, the FOOOF algorithm will be forced to assign a ‘random’ knee and the aperiodic fit will then mostly reflect the slope of the spectrum above the knee point.
Second, there are important, fine-grained features in the spectra that are ignored in the analyses, which confounds the interpretation.
One salient example of this is Fig. 2, where based on the plots in B, one would expect that the power of beta-band oscillations to be higher in the Med-On condition, as the oscillatory peaks rise higher above the 1/f floor and reach the same amplitude level as the Med-OFF condition (in other words, similar total power is subtracted by a smaller 1/f power in the Med-ON condition). But this impression is opposite to the model-fitting results in C, where beta power is lower in the Med-ON condition.
We agree that PSDs over a broad frequency range (e.g. 5-90 Hz) typically do not have a single 1/f property. Instead, there can be multiple oscillatory peaks and ‘knees/bends’ in the aperiodic component. For these cases, fitting should be performed using the knee mode. To extract periodic beta power, we parameterise the PSD between 5 and 90 Hz and select the largest oscillatory component between 8 and 35 Hz (this range was extended to include the large oscillatory peaks in hemispheres 27 and 28 at ~ 10 Hz, see Figure R1). We now use the knee mode, to model the aperiodic component between 5 and 90 Hz when periodic beta power is calculated (see our previous comments). Figure R1 provides an overview of all PSDs ON and OFF medication, the aperiodic fits (5-90 Hz (knee) and 40-90 Hz (fixed)) and the detected beta peaks. In spite of this modification in our pipeline, periodic beta power is still larger OFF medication (Figure 2C), in keeping with previous studies (Kim et al., 2022; Kühn et al., 2006; Neumann et al., 2017; Ray et al., 2008). We acknowledge the reviewer’s point that the average spectra in Figure 2B are misleading in that respect and for clarity provide here all 30 spectra in both conditions. Note that the calculation of aperiodic exponents between 40 and 90 Hz is not affected by this change in our pipeline. Figures 2B, D+E were revised accordingly.
We have repeated the analysis of our animal data using the ‘knee mode’ with a fitting range from 30 to 100 Hz. However, using the knee mode did not improve the goodness of fit or fitting error and, in fact, made them slightly worse (Figure S5). Based on this, we think the fixed mode would provide a more holistic model for the PSDs used in this analysis. We have now added this comparison in Figure S5 to justify the choice of the fixed mode.
Figure R1. PSDs from all 30 hemispheres ON and OFF medication. Aperiodic fits are shown between 5-90 Hz (knee mode), which was used to calculate the power of beta peaks, and between 40-90 Hz (fixed mode), which was used to estimate the aperiodic exponent of the spectrum.
Another example is Fig. 1C, where the spectra for high and low STN spiking epochs are identical between 10 and 20 Hz, and the difference in higher frequency range could be well-explained by an overall increase of broadband gamma power (e.g. as observed in Manning et al., J Neurosci 2012, Ray & Maunsell PLoS Biol 2011). This increase of broadband gamma power is trivially expected, as broadband gamma power is tightly coupled with population spiking rate, which was used to define the two conditions.
We agree with the reviewer that in Figure 1C, high and low STN spiking states could well be separated by average gamma power (Figure 1E), too. However, the difference of aperiodic exponents is more prominent between both conditions (Figure 1D+E, based on p-values). What is more, in human LFP data recorded from clinical macroelectrodes, medication states can be reasonably well distinguished using the aperiodic exponent between 40-90 Hz (Figure 2C), but average gamma power does not separate both states (Figure S3A). This suggests that the aperiodic exponent reflects more than just power differences in the high gamma regions. In addition, power changes do not inevitably change the aperiodic exponent and vice versa as elaborated in (Donoghue et al., 2020).
Manning et al., 2009 show that the power spectrum is shifted to higher power values at all observed frequencies (2-150 Hz) as firing rates increase. As the reviewer points out, power spectra of our data are almost identical between 10-20 Hz (despite the marked spiking differences) and only drift apart from > 20 Hz (Figure 1C). This is a relevant difference between our study and Manning et al., 2009 and suggests that power differences in the gamma range are not solely explained by differences in spiking. This is confirmed when cortical activity at different spikes/sec is modelled (Miller et al., 2009). The entire spectrum is shifted to higher power values if spiking rates increase.
Ray & Maunsell, 2011 reported low (30-80 Hz) and high (> 80 Hz) gamma activity in the macaque visual cortex, with a positive correlation between spiking activity and high gamma activity. However, activities in the low gamma range (30-80 Hz), which largely overlaps with the frequency range in our study, does not necessarily correlate with firing rates.
In conclusion, the link between gamma power and spiking activity is not as strong as alluded. Even if the change in spiking activities can lead to changes of both gamma power and the aperiodic exponent, the aperiodic exponent would still constitute a measure to separate E/I levels and medication states.
The above consideration also speaks to a major weakness of the general approach of considering the 1/f spectrum a monolithic spectrum that can be captured by a single exponent. As the authors' Fig. 1C shows, there are distinct frequency regions within the 1/f spectrum that have different slopes. Indeed, this tripartite shape of the 1/f spectrum, including a "knee" feature around 40-70 Hz which is well visible here, was described in multiple previous papers (Miller et al., PLoS Comput Biol 2009; He et al., Neuron 2010), and have been successfully modeled with a neural network model using biologically plausible mechanisms (Chaudhuri et al., Cereb Cortex, 2017). The neglect of these fine-grained features confounds the authors' model fitting, because an overall increase in the broadband gamma power - which can be explained straightforwardly by the change in population firing rates - can result in the exponent, fit over a larger spectral frequency region, to decrease. However, this is not due to the exponent actually changing, but the overall increase of power in a specific sub-frequency-region of the broadband 1/f activity.
We have now used the knee mode for aperiodic fits between 5 and 90 Hz when periodic beta power is calculated. We agree that this broad frequency range is unlikely to have a single 1/f component.
We have also repeated the analysis of our animal data using the knee mode for aperiodic fits between 30 and 100 Hz (Figure S5). However, the goodness of fits had barely changed. In fact, the R2 and error become slightly worse. In addition, the knee parameter complicates interpretation of the aperiodic exponent and has to be considered along with the knee frequency. What is more, we do not see this bend around 40-70 Hz in all subjects. We show PSDs of representative LFP channels in Figure R2 and need to assert that the knee around 40-70 Hz is not a robust finding in our data set. Therefore, we chose the fixed mode for parameterisation within this frequency band.
Please see our answer to the previous comment regarding the link between broad gamma power and changes in population firing rates.
Figure R2. PSDs of representative PSD channels for each animal (data used in Figure 1C). The knee around 40-70 Hz is not a robust finding in all PSDs.
Author Response
We thank the reviewers for their positive feedback and thoughtful suggestions that will improve our manuscript. Here we summarise our plan for immediate action. We will resubmit our manuscript once additional experiments have been performed to clarify all the major and minor concerns of the reviewers and the manuscript has been revised. At that point, we will respond to all reviewer’s points and highlight the changes made in the text.
Reviewer #1 (Public Review):
The authors have tried to correlate changes in the cellular environment by means of altering temperature, the expression of key cellular factors involved in the viral replication cycle, and small molecules known to affect key viral protein-protein interactions with some physical properties of the liquid condensates of viral origin. The ideas and experiments are extremely interesting as they provide a framework to study viral replication and assembly from a thermodynamic point of view in live cells.
The major strengths of this article are the extremely thoughtful and detailed experimental approach; although this data collection and analysis are most likely extremely time-consuming, the techniques used here are so simple that the main goal and idea of the article become elegant. A second major strength is that in other to understand some of the physicochemical properties of the viral liquid inclusion, they used stimuli that have been very well studied, and thus one can really focus on a relatively easy interpretation of most of the data presented here.
There are three major weaknesses in this article. The way it is written, especially at the beginning, is extremely confusing. First, I would suggest authors should check and review extensively for improvements to the use of English. In particular, the abstract and introduction are extremely hard to understand. Second, in the abstract and introduction, the authors use terms such as "hardening", "perturbing the type/strength of interactions", "stabilization", and "material properties", for just citing some terms. It is clear that the authors do know exactly what they are referring to, but the definitions come so late in the text that it all becomes confusing. The second major weakness is that there is a lack of deep discussion of the physical meaning of some of the measured parameters like "C dense vs inclusion", and "nuclear density and supersaturation". There is a need to explain further the physical consequences of all the graphs. Most of them are discussed in a very superficial manner. The third major weakness is a lack of analysis of phase separations. Some of their data suggest phase transition and/or phase separation, thus, a more in-deep analysis is required. For example, could they calculate the change of entropy and enthalpy of some of these processes? Could they find some boundaries for these transitions between the "hard" (whatever that means) and the liquid?
The authors have achieved almost all their goals, with the caveat of the third weakness I mentioned before. Their work presented in this article is of significant interest and can become extremely important if a more detailed analysis of the thermodynamics parameters is assessed and a better description of the physical phenomenon is provided.
We thank reviewer 1 for the comments and, in particular, for being so positive regarding the strengths of our manuscript and for raising concerns that will surely improve the manuscript. At this point, we propose the following actions to address the concerns of Reviewer 1:
1) We will extensively revise the use of English, particularly, in the abstract and introduction, defining key terms as they come along in the text to make the argument clearer.
2) We acknowledge the importance of discussing our data in more detail and we propose the following. We will discuss the graphs and what they mean as exemplified in the paragraph below.
Regarding Figure 3 - As the concentration of vRNPs increases, we observe an increase in supersaturation until 12hpi. This means that contrary to what is observed in a binary mixture, in which the Cdilute is constant (Klosin et al., 2020), the Cdilute in our system increases with concentration. It has been reported that Cdilute increases in a multi-component system with bulk concentration (Riback et al., 2020). Our findings have important implications for how we think about the condensates formed during influenza infection. As the 8 different genomic vRNPs have a similar overall structure, they could, in theory, behave as a binary system between units of vRNPs and Rab11a. However, a change in Cdilute with concentration shows that our system behaves as a multi-component system. This means that the differences in length, RNA sequence and valency that each vRNP have are key for the integrity of condensates.
3) The reviewer calls our attention to the lack of analysis of phase separations. We think that phase separation (or percolation coupled to phase separation) governs the formation of influenza A virus condensates. However, we think we ought to exert caution at this point as the condensates we are working with are very complex and that the physics of our system in cells may not be sufficient to claim phase separation without an in vitro reconstitution system. In fact, IAV inclusions contain cellular membranes, different vRNPs and Rab11a. So far, we can only speculate that the liquid character of IAV inclusions may arise from a network of interacting vRNPs that bridge several cognate vRNP-Rab11 units on flexible membranes, similarly to what happens in phase separated vesicles in neurological synapses. However, the speculative model for our system, although being supported by correlative light and electron microscopy, currently lacks formal experimental validation.
For this reason, we thought of developing the current work as an alternative to explore the importance of the liquid material properties of IAV inclusions. By finding an efficient method to alter the material properties of IAV inclusions, we provide proof of principle that it is possible to impose controlled phase transitions that reduce the dynamics of vRNPs in cells and negatively impact progeny virion production. Despite having discussed these issues in the limitations of the study, we will make our point clearer.
We are currently establishing an in vitro reconstitution system to formally demonstrate, in an independent publication, that IAV inclusions are formed by phase separation. For this future work, we teamed up with Pablo Sartori, a theorical physicist to derive in- depth analysis of the thermodynamics of the viral liquid condensates. Collectively, we think that cells have too many variables to derive meaningful physics parameters (such as entropy and enthalpy) as well as models and need to be complemented by in vitro systems. For example, increasing the concentration inside a cell is not a simple endeavour as it relies on cellular pathways to deliver material to a specific place. At the same time, the 8 vRNPs, as mentioned above, have different size, valency and RNA sequence and can behave very differently in the formation of condensates and maintenance of their material properties. Ideally, they should be analysed individually or in selected combinations. For the future, we will combine data from in vitro reconstitution systems and cells to address this very important point raised by the reviewer.
From the paper on the section Limitations of the study: “Understanding condensate biology in living cells is physiologically relevant but complex because the systems are heterotypic and away from equilibria. This is especially challenging for influenza A liquid inclusions that are formed by 8 different vRNP complexes, which although sharing the same structure, vary in length, valency, and RNA sequence. In addition, liquid inclusions result from an incompletely understood interactome where vRNPs engage in multiple and distinct intersegment interactions bridging cognate vRNP-Rab11 units on flexible membranes (Chou et al., 2013; Gavazzi et al., 2013; Haralampiev et al., 2020; Le Sage et al., 2020; Shafiuddin & Boon, 2019; Sugita, Sagara, Noda, & Kawaoka, 2013). At present, we lack an in vitro reconstitution system to understand the underlying mechanism governing demixing of vRNP-Rab11a-host membranes from the cytosol. This in vitro system would be useful to explore how the different segments independently modulate the material properties of inclusions, explore if condensates are sites of IAV genome assembly, determine thermodynamic values, thresholds accurately, perform rheological measurements for viscosity and elasticity and validate our findings”.
Reviewer #2 (Public Review):
During Influenza virus infection, newly synthesized viral ribonucleoproteins (vRNPs) form cytosolic condensates, postulated as viral genome assembly sites and having liquid properties. vRNP accumulation in liquid viral inclusions requires its association with the cellular protein Rab11a directly via the viral polymerase subunit PB2. Etibor et al. investigate and compare the contributions of entropy, concentration, and valency/strength/type of interactions, on the properties of the vRNP condensates. For this, they subjected infected cells to the following perturbations: temperature variation (4, 37, and 42{degree sign}C), the concentration of viral inclusion drivers (vRNPs and Rab11a), and the number or strength of interactions between vRNPs using nucleozin a well-characterized vRNP sticker. Lowering the temperature (i.e. decreasing the entropic contribution) leads to a mild growth of condensates that does not significantly impact their stability. Altering the concentration of drivers of IAV inclusions impact their size but not their material properties. The most spectacular effect on condensates was observed using nucleozin. The drug dramatically stabilizes vRNP inclusions acting as a condensate hardener. Using a mouse model of influenza infection, the authors provide evidence that the activity of nucleozin is retained in vivo. Finally, using a mass spectrometry approach, they show that the drug affects vRNP solubility in a Rab11a-dependent manner without altering the host proteome profile.
The data are compelling and support the idea that drugs that affect the material properties of viral condensates could constitute a new family of antiviral molecules as already described for the respiratory syncytial virus (Risso Ballester et al. Nature. 2021).
Nevertheless, there are some limitations in the study. Several of them are mentioned in a dedicated paragraph at the end of a discussion. This includes the heterogeneity of the system (vRNP of different sizes, interactions between viral and cellular partners far from being understood), which is far from equilibrium, and the absence of minimal in vitro systems that would be useful to further characterize the thermodynamic and the material properties of the condensates.
We thank reviewer 2 for highlighting specific details that need improving and raising such interesting questions to validate our findings. We will address all the minor comments of Reviewer 2. To address the comments of Reviewer 2, we propose the actions described in blue below each point raised that is written in italics.
1) The concentrations are mostly evaluated using antibodies. This may be correct for Cdilute. However, measurement of Cdense should be viewed with caution as the antibodies may have some difficulty accessing the inner of the condensates (as already shown in other systems), and this access may depend on some condensate properties (which may evolve along the infection). This might induce artifactual trends in some graphs (as seen in panel 2c), which could, in turn, affect the calculation of some thermodynamic parameters.
The concern of using antibodies to calculate Cdense is valid. We will address this concern by validating our results using a fluorescent tagged virus that has mNeon Green fused to the viral polymerase PA (PA-mNeonGreen PR8 virus). Like NP, PA is a component of vRNPs and labels viral inclusions, colocalising with Rab11 when vRNPs are in the cytosol without the need of using antibodies.
This virus would be the best to evaluate inclusion thermodynamics, where it not an attenuated virus (Figure 1A below) with a delayed infection as demonstrated by the reduced levels of viral proteins (Figure 1B below). Consistently, it shows differences in the accumulation of vRNPs in the cytosol and viral inclusions form later in infection. After their emergence, inclusions behave as in the wild-type virus (PR8-WT), fusing and dividing (Figure 1C below) and displaying liquid properties. The differences in concentration may shift or alter thermodynamic parameters such as time of nucleation, nucleation density, inclusion maturation rate, Cdense, Cdilute. This is the reason why we performed the thermodynamics profiling using antibodies upon PR8-WT infection. For validating our results, and taking into account a possible delayed kinetics, and differenced that may occur because of reduced vRNP accumulation in the cytosol, this virus will be useful and therefore we will repeat the thermodynamics using it.
As a side note, vRNPs are composed of viral RNA coated with several molecules of NP and each vRNP also contains 1 copy of the trimeric RNA dependent RNA polymerase formed by PA, PB1 and PB2. It is well documented that in the cytosol the vast majority of PA (and other components of the polymerase) is in the form of vRNPs (Avilov, Moisy, Munier, et al., 2012; Avilov, Moisy, Naffakh, & Cusack, 2012; Bhagwat et al., 2020; Lakdawala et al., 2014), and thus we can use this virus to label vRNPs on condensates to corroborate our studies using antibodies.
Figure 1 – The PA- mNeonGreen virus is attenuated in comparison to the WT virus. A. Cells (A549) were infected or mock-infected with PR8 WT or PA- mNeonGreen (PA-mNG) viruses, at a multiplicity of infection (MOI) of 3, for the indicated times. Viral production was determined by plaque assay and plotted as plaque forming units (PFU) per milliliter (mL) ± standard error of the mean (SEM). Data are a pool from 2 independent experiments. B. The levels of viral PA, NP and M2 proteins and actin in cell lysates at the indicated time points were determined by western blotting. C. Cells (A549) were transfected with a plasmid encoding mCherry-NP and co-infected with PA-mNeonGreen virus for 16h, at an MOI of 10. Cells were imaged under time-lapse conditions starting at 16 hpi. White boxes highlight vRNPs/viral inclusions in the cytoplasm in the individual frames. The dashed white and yellow lines mark the cell nucleus and the cell periphery, respectively. The yellow arrows indicate the fission/fusion events and movement of vRNPs/ viral inclusions. Bar = 10 µm. Bar in insets = 2 µm.
2) Although the authors have demonstrated that vRNP condensates exhibit several key characteristics of liquid condensates (they fuse and divide, they dissolve upon hypotonic shock or upon incubation with 1,6-hexanediol, FRAP experiments are consistent with a liquid nature), their aspect ratio (with a median above 1.4) is much higher than the aspect ratio observed for other cellular or viral liquid compartments. This is intriguing and might be discussed.
IAV inclusions have been shown to interact with microtubules and the endoplasmic reticulum, that confers movement, and also undergo fusion and fission events. We propose that these interactions and movement impose strength and deform inclusions making them less spherical. To validate this assumption, we compared the aspect ratio of viral inclusions in the absence and presence of nocodazole (that abrogates microtubule-based movement). The data in figure 2 shows that in the presence of nocodazole, the aspect ratio decreases from 1.42±0.36 to 1.26 ±0.17, supporting our assumption.
Figure 2 – Treatment with nocodazole reduces the aspect ratio of influenza A virus inclusions. Cells (A549) were infected PR8 WT and treated with nocodazole (10 µg/mL) for 2h time after which the movement of influenza A virus inclusions was captured by live cell imaging. Viral inclusions were segmented, and the aspect ratio measured by imageJ, analysed and plotted in R.
3) Similarly, the fusion event presented at the bottom of figure 3I is dubious. It might as well be an aggregation of condensates without fusion.
We will change this, thank you for the suggestion.
4) The authors could have more systematically performed FRAP/FLAPh experiments on cells expressing fluorescent versions of both NP and Rab11a to investigate the influence of condensate size, time after infection, or global concentrations of Rab11a in the cell (using the total fluorescence of overexpressed GFP-Rab11a as a proxy) on condensate properties.
We will try our best to be able to comply with this suggestion as we think it is important.
Reviewer #3 (Public Review):
This study aims to define the factors that regulate the material properties of the viral inclusion bodies of influenza A virus (IAV). In a cellular model, it shows that the material properties were not affected by lowering the temperature nor by altering the concentration of the factors that drive their formation. Impressively, the study shows that IAV inclusions may be hardened by targeting vRNP interactions via the known pharmacological modulator (also an IAV antiviral), nucleozin, both in vitro and in vivo. The study employs current state-of-the-art methodology in both influenza virology and condensate biology, and the conclusions are well-supported by data and proper data analysis. This study is an important starting point for understanding how to pharmacologically modulate the material properties of IAV viral inclusion bodies.
We thank this reviewer for all the positive comments. We will address the minor issues brought to our attention entirely, including changing the tittle of the manuscript and we will investigate the formation and material properties of IAV inclusions in the presence and absence of nucleozin for the nucleozin escape mutant NP-Y289H.
References
Avilov, S. V., Moisy, D., Munier, S., Schraidt, O., Naffakh, N., & Cusack, S. (2012). Replication- competent influenza A virus that encodes a split-green fluorescent protein-tagged PB2 polymerase subunit allows live-cell imaging of the virus life cycle. J Virol, 86(3), 1433- 1448. doi:10.1128/JVI.05820-11
Avilov, S. V., Moisy, D., Naffakh, N., & Cusack, S. (2012). Influenza A virus progeny vRNP trafficking in live infected cells studied with the virus-encoded fluorescently tagged PB2 protein. Vaccine, 30(51), 7411-7417. doi:10.1016/j.vaccine.2012.09.077
Bhagwat, A. R., Le Sage, V., Nturibi, E., Kulej, K., Jones, J., Guo, M., . . . Lakdawala, S. S. (2020). Quantitative live cell imaging reveals influenza virus manipulation of Rab11A transport through reduced dynein association. Nat Commun, 11(1), 23. doi:10.1038/s41467-019-13838-3
Chou, Y. Y., Heaton, N. S., Gao, Q., Palese, P., Singer, R. H., & Lionnet, T. (2013). Colocalization of different influenza viral RNA segments in the cytoplasm before viral budding as shown by single-molecule sensitivity FISH analysis. PLoS Pathog, 9(5), e1003358. doi:10.1371/journal.ppat.1003358
Gavazzi, C., Yver, M., Isel, C., Smyth, R. P., Rosa-Calatrava, M., Lina, B., . . . Marquet, R. (2013). A functional sequence-specific interaction between influenza A virus genomic RNA segments. Proc Natl Acad Sci U S A, 110(41), 16604-16609. doi:10.1073/pnas.1314419110
Haralampiev, I., Prisner, S., Nitzan, M., Schade, M., Jolmes, F., Schreiber, M., . . . Herrmann, A. (2020). Selective flexible packaging pathways of the segmented genome of influenza A virus. Nat Commun, 11(1), 4355. doi:10.1038/s41467-020-18108-1
Klosin, A., Oltsch, F., Harmon, T., Honigmann, A., Julicher, F., Hyman, A. A., & Zechner, C. (2020). Phase separation provides a mechanism to reduce noise in cells. Science, 367(6476), 464-468. doi:10.1126/science.aav6691
Lakdawala, S. S., Wu, Y., Wawrzusin, P., Kabat, J., Broadbent, A. J., Lamirande, E. W., . . . Subbarao, K. (2014). Influenza a virus assembly intermediates fuse in the cytoplasm. PLoS Pathog, 10(3), e1003971. doi:10.1371/journal.ppat.1003971
Le Sage, V., Kanarek, J. P., Snyder, D. J., Cooper, V. S., Lakdawala, S. S., & Lee, N. (2020). Mapping of Influenza Virus RNA-RNA Interactions Reveals a Flexible Network. Cell Rep, 31(13), 107823. doi:10.1016/j.celrep.2020.107823
Riback, J. A., Zhu, L., Ferrolino, M. C., Tolbert, M., Mitrea, D. M., Sanders, D. W., . . . Brangwynne, C. P. (2020). Composition-dependent thermodynamics of intracellular phase separation. Nature, 581(7807), 209-214. doi:10.1038/s41586-020-2256-2
Shafiuddin, M., & Boon, A. C. M. (2019). RNA Sequence Features Are at the Core of Influenza a Virus Genome Packaging. J Mol Biol. doi:10.1016/j.jmb.2019.03.018
Sugita, Y., Sagara, H., Noda, T., & Kawaoka, Y. (2013). Configuration of viral ribonucleoprotein complexes within the influenza A virion. J Virol, 87(23), 12879- 12884. doi:10.1128/JVI.02096-13
Author Response
Reviewer #1 (Public Review):
The authors reveal dual regulatory activity of the complex nuclear receptor element (cNRE; contains hexads A+B+C) in cardiac chambers and its evolutionary origin using computational and molecular approaches. Building upon a previous observation that hexads A and B act as ventricular repressor sequences, in this study the authors identify a novel hexad C sequence with preferential atrial expression. The authors also reveal that the cNRE emerged from an endogenous viral element using comparative genomic approaches. The strength of this study is in a combination of in silico evolutionary analyses with in vivo transgenic assays in both zebrafish and mouse models. Rapid, transient expression assays in zebrafish together with assays using stable, transgenic mice demonstrate dual functionality of cNRE depending on the chamber context. This is especially intriguing given that the cNRE is present only in Galliformes and has originated likely through viral infection. Interestingly, there seem to be some species-specific differences between zebrafish and mouse models in expression response to mutations within the cNRE. Taken together, these findings bear significant implications for our understanding of dual regulatory elements in the evolutionary context of organ formation.
We thank reviewer 1 for the thorough review and are very satisfied with his favorable view of our manuscript. We also thank reviewer 1 for suggestions and opportunities to further clarify some relevant issues.
Reviewer #2 (Public Review):
Nunes Santos et al. investigated the gene regulatory activity of the promoter of the quail myosin gene, SMyHC III, that is expressed specifically in the atria of the heart in quails. To do so, they computationally identified a novel 6-bp sequence within the promoter that is putatively bound by a nuclear receptor transcription factor, and hence is a putative regulatory sequence. They tested this sequence for regulatory activity using transgenic assays in zebrafish and mice, and subjected this sequence to mutagenesis to investigate whether gene regulatory effects are abrogated. They define this sequence, together with two additional known 6-bp regulatory sequences, as a novel regulatory sequence (denoted cNRE) necessary and sufficient for driving atrial-specific expression of SMyHC III. This cNRE sequence is shared across several galliform species but appears to be absent in other avian species. The authors find that there is sequence homology between the cNRE and several virus genomes, and they conclude that this regulatory sequence arose in the quail genome by viral integration.
Strengths: The evolutionary origins of gene regulatory sequences and their impact on directing tissue-specific expression are of great interest to geneticists and evolutionary biologists. The authors of this paper attempt to bring this evolutionary perspective to the developmental biology question of how genes are differentially expressed in different chambers of the heart. The authors test for regulatory activity of the putative regulatory sequence they identified computationally in both zebrafish and mouse transgenic assays. The authors disrupt this sequence using deletions and mutagenesis, and introduce a tandem repeat of the sequence to a reporter gene to determine its consequences on chamber activity. These experiments demonstrate that the identified sequence has regulatory activity.
We appreciate the thorough review of our manuscript and are very stimulated by the reviewer’s understanding of the contents we presented. We will take the liberty to comment after the reviewer’s considerations, in the hope to better answer the relevant points.
Weaknesses: There are several decisions and assumptions that have been made by the authors, the reasons for which have not been articulated. Firstly, the rationale for the approach is not clear. The study is a follow-up to work previously performed by the authors which identified two 6-bp sequences important for controlling atrial-specific expression of the quail SMyHC III gene. This study appears to be motivated by the fact that these two sequences, bound by nuclear receptors, do not fully direct chamber-specific expression, and therefore this study aims to find additional regulatory sequences. It is assumed that any additional regulatory sequences should also be bound by nuclear receptors, and be 6-bp in length, and therefore the authors search for 6-bp sequences bound by nuclear receptors. It is not clear what the input sequence for this analysis was.
Thank you for giving us the opportunity to clarify our rational. Our approach is justified by the natural progression in the understanding of the mechanisms involved in preferential atrial expression by the SMyHC III promoter. The groundwork was solidly laid down by Wang and colleagues (see references as below). They mapped potential atrial stimulators and ventricular repressors throughout the SMyHC III promoter using atrial and ventricular cultures, respectively. Wang and colleagues pinned down the relevant regulators. First between -840 and -680 bp upstream from the transcription start site, then inside this nucleotide stretch, then in the 72-bp fragment contained between -840 and -680 bp, then identified the ventricular repressor in Hexads A and B inside the 72-bp sequence (see references below). We, in this manuscript, contributed with the identification of Hexad C (immediately downstream of Hexads A and B) as a potential nuclear receptor binding site and as a bona fide atrial activator. In summary, our work represents a logical conclusion of previous work by Wang and colleagues. We continued the process of narrowing down sequences previously proven to contain atrial activators (that were unknown before our present work) and ventricular repressors (that were already described).
Why did we use nuclear receptors as models for the putative cardiac chamber regulators binding to the cNRE? This is because previous work by Wang et al., 1996, 1998, 2001 and by Bruneau et al., 2001 showed that the 5’ portion of the cNRE (Hexads A and B) is indeed a hub for the integration of signals conveyed by nuclear receptors. Originally, Wang et al., 1996 showed that the VDR response element is a ventricular repressor acting via the 5’ portion of the cNRE. In a subsequent manuscript, Wang et al., 1998 showed that both RAR and VDR bind the 5’ portion of the cNRE. Bruneau et al., 2001 showed, by crossing IRX4 knockout mice with SMyHC III-HAP mice (Xavier-Neto et al., 1999), that IRX4 plays the role of a repressor of SMyHC III-HAP expression. Finally, Wang et al., 2001 showed that IRX4 interacts with RXR bound to the 5’ portion of the cNRE to inhibit ventricular expression.
Why was the 3’ Hexad included as a research subject? Very early on in our work it was noted that 3’ of the original VDR response element (Hexads A and B), described by Wang et al., 1996 and 1998 as a ventricular repressor, there was a sequence (Hexad C) with almost equal binding potential to nuclear receptors as Hexads A and B (as initially judged on the basis of comparisons with canonical nuclear receptor binding sequences, but later on confirmed by in silico profiling of nuclear receptor binding, see below). This discovery prompted us to design point mutants in the 3’ portion of the cNRE to investigate whether Hexad C contained relevant regulators of heart chamber expression. These analyses revealed a strong atrial activator in the mouse (the missing atrial activator from Wang et al., 1996, 1998, 2001).
Wang, G. F., Nikovits, W., Schleinitz, M., and Stockdale, F. E. (1996). Atrial chamber-specific expression of the slow myosin heavy chain 3 gene in the embryonic heart. J. Biol. Chem. 271, 19836-19845.
Wang, G. F., Nikovits, W. Jr., Schleinitz, M., and Stockdale, F. E. (1998). A positive GATA element and a negative vitamin D receptorlike element control atrial chamber-specific expression of a slow myosin heavy-chain gene during cardiac morphogenesis. Mol. Cell Biol. 18, 6023-6034.
Xavier-Neto, J., Neville, C. M., Shapiro, M. D., Houghton, L., Wang, G. F., Nikovits, W. Jr, Stockdale, F. E., and Rosenthal, N. (1999). A retinoic acid-inducible transgenic marker of sino-atrial development in the mouse heart. Development 126, 2677-2687.
Bruneau, B. G., Bao, Z. Z., Fatkin, D., Xavier-Neto, J., Georgakopoulos, D., Maguire, C. T., Berul, C. I., Kass, D. A., Kuroski-de Bold, M. L., de Bold, A. J., Conner, D. A., Rosenthal, N., Cepko, C. L., Seidman, C. E., and Seidman, J. G. (2001). Cardiomyopathy in Irx4-deficient mice is preceded by abnormal ventricular gene expression. Mol. Cell Biol. 21, 1730-1736.
Wang, G. F., Nikovits, W. Jr., Bao, Z.Z., and Stockdale, F.E. (2001). Irx4 forms an inhibitory complex with the vitamin D and retinoic X receptors to regulate cardiac chamber-specific slow MyHC3 expression. J Biol Chem. 276, 28835-28841.
The methods section mentions the cNRE sequence, but this is their newly defined regulatory sequence based on the newly identified 6-bp sequence. It is therefore unclear why Hexad C was identified to be of interest, and not the GATA binding site for example, and whether other sequences in the promoter might have stronger effects on driving atrial-specific expression.
As far as the existence of binding sites other than Hexads A, B, and C, we cannot, formally, exclude the possibility that there may be other relevant regulators of the SMyHC III gene. But we note that the sequences that we utilized were previously mapped through deletion mutant promoter approach by Wang et al., 1996 as the most powerful atrial activator(s) and ventricular repressor(s). We addressed these concerns in a new session entitled “Limitations of our work”.
Concerning GATA regulation, Wang et al., 1996, 1998 characterized a GATA-4 site that drives generalized (atrial and ventricular) cardiac expression in quail cultures. However, we were unable to identify any relevant changes in cardiac expression in mutant GATA SMyHC III-HAP transgenic mouse lines produced with the same mutated promoter sequences described by Wang et al., 1996, 1998.
Finding Hexad C as an atrial activator was an experimental finding. We identified it as such because we had two important inputs. First, in 1997, we consulted with Ralff Ribeiro, a specialist on nuclear receptors and he pointed out that downstream of the Hexad A + Hexad B VDRE/RARE (the ventricular repressor), there was a sequence with good potential for a nuclear receptor binding motif. This was exactly Hexad C. Then, we confirmed its potential for nuclear receptor binding by nuclear receptor profiling. After these two pieces of evidence, we thought that there was enough evidence to justify a mutant construct (Mut C). The experimental results we obtained in transgenic mice and zebrafish are consistent with the hypothesis that Hexad C does contain the long sought atrial activator predicted by Wang et al., 1996 in atrial cultures. This seems to be the most important atrial activator (a seven-fold activator) predicted by a deletion approach to be located between -840 and 680 bp in Wang et al., 1996.
Wang, G. F., Nikovits, W., Schleinitz, M., and Stockdale, F. E. (1996). Atrial chamber-specific expression of the slow myosin heavy chain 3 gene in the embryonic heart. J. Biol. Chem. 271, 19836-19845.
Wang, G. F., Nikovits, W. Jr., Schleinitz, M., and Stockdale, F. E. (1998). A positive GATA element and a negative vitamin D receptorlike element control atrial chamber-specific expression of a slow myosin heavy-chain gene during cardiac morphogenesis. Mol. Cell Biol. 18, 6023-6034.
Indeed, the zebrafish transgenic assays use the 32 bp cNRE, while in the mouse transgenic assays, a 72 bp region is used. This choice of sequence length is not justified.
As stated above, our rational was built as a continuation of the thorough work by Wang and colleagues in progressively narrowing down the location of relevant atrial stimulators and ventricular repressors. Throughout our work, we sought to obtain maximal coherence with previous studies (see references below) and to simultaneously probe cNRE function at an increased resolution. For that, we utilized previously described mutant SMyHC III promoter constructs (Wang et al., 1996) and introduced novel site-directed dinucleotide substitution mutants of individual Hexads in the SMyHC III promoter.
Wang, G. F., Nikovits, W., Schleinitz, M., and Stockdale, F. E. (1996). Atrial chamber-specific expression of the slow myosin heavy chain 3 gene in the embryonic heart. J. Biol. Chem. 271, 19836-19845.
Wang, G. F., Nikovits, W. Jr., Schleinitz, M., and Stockdale, F. E. (1998). A positive GATA element and a negative vitamin D receptorlike element control atrial chamber-specific expression of a slow myosin heavy-chain gene during cardiac morphogenesis. Mol. Cell Biol. 18, 6023-6034.
Xavier-Neto, J., Neville, C. M., Shapiro, M. D., Houghton, L., Wang, G. F., Nikovits, W. Jr, Stockdale, F. E., and Rosenthal, N. (1999). A retinoic acid-inducible transgenic marker of sino-atrial development in the mouse heart. Development 126, 2677-2687.
Bruneau, B. G., Bao, Z. Z., Fatkin, D., Xavier-Neto, J., Georgakopoulos, D., Maguire, C. T., Berul, C. I., Kass, D. A., Kuroski-de Bold, M. L., de Bold, A. J., Conner, D. A., Rosenthal, N., Cepko, C. L., Seidman, C. E., and Seidman, J. G. (2001). Cardiomyopathy in Irx4-deficient mice is preceded by abnormal ventricular gene expression. Mol. Cell Biol. 21, 1730-1736.
Wang, G. F., Nikovits, W. Jr., Bao, Z.Z., and Stockdale, F.E. (2001). Irx4 forms an inhibitory complex with the vitamin D and retinoic X receptors to regulate cardiac chamber-specific slow MyHC3 expression. J Biol Chem. 276, 28835-28841.
The decisions about which bases to mutate in the three hexads are also not clear. Why are the first two bases mutated in Hexad B and C and the whole region mutated in Hexad A? Is there a reason to believe these bases are particularly important?
As for the reasons behind mutation of the first two bases in Hexad B and Hexad C, there were two:
One reason is because these point mutations in Hexads B and C were planned after the publication of Wang et al., 1996, which defined the major role of Hexad A in ventricular repression. After this discovery, we decided that a higher level of resolution in our mutation approach would be a better way to search for additional regulators of SMyHC III expression, including the atrial regulator that was readily apparent from the results shown in Wang et al., 1996, but had not yet been described.
The second reason is because the two first nucleotides (purines) in a nuclear-receptor binding hexad are critical for the interaction between target DNA and transcription factors of the nuclear receptor family. Substituting pyrimidines for purines in the two first positions of an hexad drastically reduces the affinity of a nuclear response element, and that is why we chose to use TT substitutions in our mutant constructs. Please refer to: Umesono et al., Cell, 1991 65: 12551266 for a review; Mader et al., J Biol Chem, 1993 268:591-600 for a mutation study; Rastinejad et al., EMBO J., 2000 19:1045-1054 for a crystallographic study (as well as additional references listed below).
Mader, S., Chen, J. Y., Chen, Z., White, J., Chambon, P., and Gronemeyer, H. (1993). The patterns of binding of RAR, RXR and TR homo- and heterodimers to direct repeats are dictated by the binding specificites of the DNA binding domains. EMBO J. 12, 50295041.
Ribeiro, R. C., Apriletti, J. W., Yen, P.M., Chin, W. W., and Baxter, J. D. (1994). Heterodimerization and deoxyribonucleic acid-binding properties of a retinoid X receptor-related factor. Endocrinology.135, 2076-2085.
Zhao, Q., Chasse, S. A., Devarakonda, S., Sierk, M. L., Ahvazi, B., and Rastinejad, F. (2000). Structural basis of RXR-DNA interactions. J. Mol. Biol. 296, 509-520.
Shaffer, P. L. and Gewirth, D. T. (2002). Structural basis of VDR-DNA interactions on direct repeat response elements. EMBO J. 21, 2242-2252.
The control mutant also has effects on the chamber distribution of GFP expression.
We note that, in the mouse, MutS did not produce any major changes from the typical wild type phenotypes linked to SMyHC III-HAP transgenic hearts. We concluded, based on our data, that the spacing mutant worked reasonably well as a negative mutation control in mice. We agree that it would have been particularly elegant if a spacing mutant designed for the mouse context worked in the exact same way in the zebrafish. However, the fact that there are slight differences in behavior for the mutated “spacing” constructs in species separated by, millions of years of independent evolution is not really surprising, given that the amino acid sequence of transcription factors can diverge and co-evolve with binding nucleotides and end up drifting quite substantially from an ancestral setup. As we reiterate below, we consider more fundamental the fact that the cNRE is actually able to bias cardiac expression towards a model of preferential atrial expression, even in the context of species separated by millions of years of independent evolution.
Two claims in the paper have weak evidence. Firstly, the conclusion that the cNRE is necessary and sufficient for driving preferential expression in the atrium. Deleting the cNRE does reduce the amount of atrial reporter gene expression but there is not a "conversion" from atrial to ventricular expression as mentioned in line 205. Similarly, a fusion of 5 tandem repeats of the cNRE can induce expression of a ventricular gene in the atria (I'm assuming a single copy is insufficient), but does not abolish ventricular expression.
We agree that our labelling of the cNRE is perhaps too strong, and we have toned it down accordingly to incorporate the much more equilibrated concept that the cNRE biases cardiac expression towards a model of preferential atrial expression.
However, after the corrections suggested, we believe our assertion is now justified. We show that in the mouse, removal of the cNRE is followed by a major reduction of atrial expression coupled to the release of a low, but quite clear level of expression in the ventricles, when compared to the transgenic mouse harboring the wild type SMyHC III promoter. Note that, as expected, the relative power of the cNRE to establish preferential atrial expression is higher in the mouse (a mammal) than it is in the zebrafish (a teleost), which is biologically sound, as mammals and avians are closer, phylogenetically, than teleosts and avians. Yet, the direction of change of expression in atria and ventricles was exactly as expected, if a given motif responsible for preferential atrial expression was removed (the cNRE in our case), that is: marked reduction in atrial expression and small (albeit clearly evident) release of ventricular expression. We believe that these directional changes observed in species separated by millions of years of independent evolution constitute very good biological evidence for the role of the cNRE in driving preferential atrial expression.
Concerning the 5x fusion of cNREs, we chose to produce this multimer for safety purposes only, because we did not want to risk performing incomplete experiments and having to repeat them. However, more to the point, we later compared the efficiency of one (1) versus five (5) cNRE copies in a cell culture context and the results were not different.
Secondly, the authors claim that the cNRE regulatory sequence arose from viral integration into the genomes of galliform species. While this is an attractive mechanism for explaining novel regulatory sequences, the evidence for this is based purely on sequence homology to viral genomes. And this single observation is not robust as the significance of the sequence matches does not appear to be adjusted for sequence matches expected by chance. The "evolutionary pathway" leading to the direction of chamber-specific expression in the heart as highlighted in the abstract has therefore not been demonstrated.
We agree with the reviewer. Because of space constraints, we decided to omit a substantial part of our work from the initial submission of the manuscript. We now include the relevant data in the revised version. We thus mapped the phylogenetic origins of the SMyHC III family of slow myosins and then established how and when the cNREs became topologically associated with the SMyHC III gene. To do that, we repeat masked all available sequences from avian SMyHC III orthologs. As it will become clear below, the cNRE is a rare sequence, rather than a low complexity repeat. Our search for cNREs outside of the quail context (Coturnix coturnix) followed two independent lines. First, we took a scaled, evolution-oriented approach. Initially, we looked for cNREs in species close to the quail (i.e., Galliformes) and then progressively farther, to include derived (i.e., Passeriformes) and basal avians (i.e., Paleognaths) as well as external groups such as crocodilians. While pursuing this line of investigation, it became clear that the cNRE was a rare form of repetitive element, which showed a conserved topological relationship with the SMyHC III gene (i.e., cNREs flanked the SMyHC III genes at 5’ and 3’ regions). Using this topological relationship as a character, we determined when it appeared during avian evolution and then set out to establish the likely origins of this rare repetitive motif. This search for the origins of the cNRE entailed comparisons to databases of repetitive genome elements, until the extreme telomeric nature of the SMyHC III gene became evident. This finding directed us to the fact that the hexad nature of the cNRE is reminiscent of the hexameric character of telomeric direct repeats. Because direct telomeric repeats are exactly featured in the genomes of avian DNA viruses that can infect the germline and integrate into the avian genome, we focused our search for the cNRE on the members of the subfamily Alphaherpesvirinae (Morissette & Flamand, 2010). In this search, we utilized the human herpes simplex virus 1 (HSV1) as a general model for herpes viruses, and a set of four (4) members of the Alphaherpesvirinae family that specifically infect Galliformes (i.e., GaHV1, the virus responsible for avian infectious laryngotracheitis in chicken, GaHV2, the Marek’s disease virus, GaHV3, a non-pathogenic virus, and MeHV1, the non-pathogenic Meleagrid herpesvirus 1 capable of infecting chicken and wild turkey) (Waidner et al., 2009). The search for cNREs in Alphaherpesvirinae was successful. We found six (6) cNRE hits in HSV1, one (1) in GaHV1, and none in MeHV1, GaHV2, and GaHV3. Our evolution-directed approach thus led to the direct recognition that cNREs can be found in the genomes of a family of viruses that contain members that infect avians and integrate their double-stranded DNA into the host germline (Morissette & Flamand, 2010). Therefore, as a second independent approach, as pointed out by the reviewer, we set out to further extend this proof of concept by broadening our search to all known sequenced viruses and perform an unbiased, internally consistent, and quantitative analysis of cNRE presence in viral genomes, as already reported in the initial submission of this manuscript.
Reviewer #3 (Public Review):
Summary:
In this manuscript Nunes Santos et al. use a combination of computation and experimental methods to identify and characterize a cis-regulatory element that mediates expression of the quail Slow Myosin Heavy Chain III (SMyHC III) gene in the heart (specifically in the atria). Previous studies had identified a cis-regulatory element that can drive expression of SMyHC III in the heart, but not specifically (solely) in the atria, suggesting additional regulatory elements are responsible for the specific expression of SMyHC III in the atria as opposed to other elements of the heart. To identify these elements Nunes Santos et al. first used a bioinformatic approach to identify potentially functional nuclear receptor binding sites ("Hexads") in the SMyHC III promoter; previous studies had already shown that two of these Hexads are important for SMyHC III promoter function. They identified a previously unknown third Hexad within the promoter, and propose that the combination of these three (called the complex Nuclear Receptor Element or cNRE) is necessary and sufficient for specific atrial expression of SMyHC III. Next, they use experimental methods to functionally characterize the cNRE including showing that the quail SMyHC III promoter can drive green fluorescent protein (GFP) expression the atrium of developing zebrafish embryos and that the cNRE is necessary to drive the expression of the human alkaline phosphatase reporter gene (HAP) in transgenic mouse atria. Additional experiments show that the cNRE is portable regulatory element that can drive atrial expression and demonstrate the importance of the three Hexad parts. These data demonstrating that the cNRE mediates atrial-specific expression is well-done and convincing. The authors also note the possibility that the cNRE might be derived from an endogenous viral element but further data are needed to support the hypothesis that the cNRE is of viral origin.
Strengths:
1) The experimental work demonstrating that the cNRE is a regulatory element that can mediate the atrial-specific expression of SMyHC III.
We thank reviewer 3 for this thorough appreciation of our work and are pleased with the evaluation of our manuscript’s potential.
Weaknesses:
1) Justification for use of different regulatory elements in the zebrafish (32 bp cNRE) and the mouse transgenic assays (72 bp cNRE), and discussion of the impact of this difference on the results/interpretation.
In general, throughout our work, we sought to obtain maximal coherence with previous studies (see references below) and to simultaneously probe cNRE function at an increased resolution. For that, we utilized previously described mutant SMyHC III promoter constructs (Wang et al., 1996, 1998) and introduced novel site-directed dinucleotide substitution mutants of individual Hexads in the SMyHC III promoter. Actually, the 72-bp construct is not a 72-bp construct. It is a 5’ deletion construct that removed 72 bp from the 840 bp wild type SMyHC III construct, transforming it into a 768-bp SMyHC III promoter construct. Any directional changes observed in cardiac expression by the 768 bp as compared to the wild type promoter was interpreted in the context as missing regulators present in this 5’ 72 bp.
Wang et al., 1996 and 1998 had already shown that Hexads A and B contained a functional VDRE/RARE, which acted as a ventricular repressor. Using the 768-bp SMyHC III promoter in mouse transgenic lines was thus a natural investigation step for us to evaluate whether regulation of the SMyHC III promoter in the mouse was similar in mice as compared to quail cardiac cultures. As shown in the manuscript, deletion of the 72 bp resulted in the release of a low level of expression in ventricles, consistent with the removal of a ventricular repressor (already described by Wang et al., 1996). It also showed a marked reduction in atrial transgene stimulation, suggesting the elimination of a very important atrial activator.
In 1996, Wang and colleagues mapped an atrial activator to the sequence interval of 160 bp, between -840 and -680 bp (Wang et al., 1996). In our mouse transgenics, we reduced this interval to a mere 72 bp, between -840 to -768 bp. This was very useful information. Wang et al., 1998 showed that HF-1a, M-CAT, and E-box sites located between -840 and -808 bp did not influence atrial expression, so now we had a potential interval of only 40 bp between -808 and -768 bp. Further, our transgenic mice indicated that the GATA site located 3’ from Hexads A, B, and C (GATA site changed to a Sal I site at positions -749 to -743 bp) did not work as a general activator, as in the quail. Thus, the only good candidate for the atrial activator in mice inside the 40-bp fragment between -808 and -768 bp was the cNRE, with its three Hexads, A, B and the novel Hexad C. Because Hexads A plus B composed a functional VDRE/RARE that played a role in ventricular repression in the quail, we hypothesized that the atrial activator would be present in Hexad C. We then mutated the two first purines in Hexad C (the most important ones for nuclear receptor binding, please refer to Umesono et al., Cell, 1991 65: 1255-1266 for a review; Mader et al., J Biol Chem, 1993 268:591-600 for a mutation study; Rastinejad et al., EMBO J., 2000 19:1045-1054 for a crystallographic study as well as additional references listed below) and performed the experiments that demonstrated a profound reduction in atrial expression in the mouse context, revealing the long-sought atrial activator.
Mader, S., Chen, J. Y., Chen, Z., White, J., Chambon, P., and Gronemeyer, H. (1993). The patterns of binding of RAR, RXR and TR homo- and heterodimers to direct repeats are dictated by the binding specificites of the DNA binding domains. EMBO J. 12, 50295041.
Ribeiro, R. C., Apriletti, J. W., Yen, P.M., Chin, W. W., and Baxter, J. D. (1994). Heterodimerization and deoxyribonucleic acid-binding properties of a retinoid X receptor-related factor. Endocrinology.135, 2076-2085.
Wang, G. F., Nikovits, W., Schleinitz, M., and Stockdale, F. E. (1996). Atrial chamber-specific expression of the slow myosin heavy chain 3 gene in the embryonic heart. J. Biol. Chem. 271, 19836-19845.
Wang, G. F., Nikovits, W. Jr., Schleinitz, M., and Stockdale, F. E. (1998). A positive GATA element and a negative vitamin D receptorlike element control atrial chamber-specific expression of a slow myosin heavy-chain gene during cardiac morphogenesis. Mol. Cell Biol. 18, 6023-6034.
Zhao, Q., Chasse, S. A., Devarakonda, S., Sierk, M. L., Ahvazi, B., and Rastinejad, F. (2000). Structural basis of RXR-DNA interactions. J. Mol. Biol. 296, 509-520.
Shaffer, P. L. and Gewirth, D. T. (2002). Structural basis of VDR-DNA interactions on direct repeat response elements. EMBO J. 21, 2242-2252.
2) Is the cNRE really "necessary and sufficient"? I define necessary and sufficient in this context as a regulatory element that fully recapitulates the expression of the target gene, so if the cNRE was "necessary and sufficient" to direct the appropriate expression of SMyHC III it should be able to drive expression of a reporter gene solely in the atria. While deletion of the cNRE does reduce expression of the reporter gene in atria it is not completely lost nor converted from atrial to ventricular expression (as I understand the study design would suggest should be the effect), similarly fusion of 5 repeats of the cNRE induces expression of a ventricular gene in the atria but also does not convert expression from ventricle to atria. This doesn't seem to satisfy the requirements of a "necessary and sufficient" condition. Perhaps a discussion of why the expectations for "necessary and sufficient" are not met but are still consistent would be beneficial here.
We agree with your reasoning. Our description of the cNRE was perhaps too strong, and we have toned it down accordingly in the revised manuscript to incorporate a much more equilibrated concept that the cNRE biases cardiac expression towards a model of preferential atrial expression. After these corrections, we believe our novel assertion is justified. We show that in the mouse, removal of the cNRE is followed by a major reduction of atrial expression coupled to the release of a low, but quite clear level of expression in the ventricles, when compared to the transgenic mouse harboring the wild type SMyHC III promoter. Note that, as expected, the relative power of the cNRE to establish preferential atrial expression is higher in the mouse (a mammal) than it is in the zebrafish (a teleost), which is biologically sound, as mammals and avians are closer, phylogenetically, than teleosts and avians. Yet, the direction of change of expression in atria and ventricles was exactly as expected, if a given motif responsible for preferential atrial expression was removed (the cNRE in our case), that is: marked reduction in atrial expression and small (albeit evident) release of ventricular expression. We believe that these directional changes observed in species separated by millions of years of independent evolution constitute very good biological evidence for the role of the cNRE in driving preferential atrial expression.
3) The claim that the cNRE is derived from a viral integration is not supported by the data. Specifically, the cNRE has sequence similarity to some viral genomes, but this need not be because of homology and can also be because of chance or convergence. Indeed, the region of the chicken genome with the cNRE does have repetitive elements but these are simple sequence repeats, such as (CTCTATGGGG)n and (ACCCATAGAG)n, and a G-rich low complexity region, rather than viral elements; The same is true for the truly genome. These data indicate that the cNRE is not derived from an endogenous virus but is a repetitive and low complexity region, these regions are expected to occur more frequently than expected for larger and more complex regions which would cause the BLAST E value to decrease and appear "significant”, but this is entirely expected because short alignments can have high E values by chance. (Also note that E values do not indicate statistical significance, rather they are the number of hits one can "expect" to see by chance when searching specific database.)
We do understand the criticism, but we would like to advance another concept, based on a series of results that we obtained using bioinformatics-oriented and evolution-oriented analyses. We performed a cNRE scan in the Gallus gallus genome (galGal5), using varying numbers of nucleotide mismatches. When we searched the galGaL5 genome with coordinates matching the localization of cNREs obtained using matchPattern with up to 8 mismatches, only thirty-one (31) and thirty-four (34) hits were found in the 5’ and 3’ strands, respectively. This indicates that a cNRE match is a rather uncommon finding in the Gallus gallus genome.
A more systematic profiling of genome occurrence versus nucleotide mismatch indicated that a significant upward inflexion in the relationship between number of cNRE hits and divergence from the original cNRE version (Coturnix coturnix) is recorded only at 12 mismatches or greater. At 8 mismatches, the total number of cNREs on each DNA strand varied little among all avian species examined, remaining close to the average (31+/- 2,2 cNREs for the 5’ strand, range 1748; 34 +/- 3,3 for the 3’ strand, range 14-64). Consistent with the idea that the cNRE is a specific regulatory motif, rather than an abundant, low complexity sequence, there are only two cNRE occurrences in chromosome 19, which harbors AMHC1, the Gallus gallus ortholog of the Coturnix coturnix SMyHC III gene.
Figure 1: Number of cNRE hits to galGal5 according to maximum mismatches allowed: the cNRE is not an abundant low complexity sequence, but rather a rare repetitive sequence with a clear cutoff level of mismatches allowed. Consistent with this, there are only two (2) cNRE sequences in chromosome 19, the chromosome that contains the AMHC1 gene (the chicken ortholog of the quail SMyHC III gene). ## [1] chr19 [16510, 16541] * | 5’-CAAGGACAAAGAGGGGACAAAGAGGCGGAGGT-3 ## [2] chr19 [32821, 32852] * ‘5’-CAAGGACAAAGAGTGGACAAAGAGGCAGACGT-3
In the evolutionary strategy, which we now include, we first mapped the phylogenetic origins of the SMyHC III family of slow myosins and then established how and when the cNREs became topologically associated with the SMyHC III gene. To do that we repeat masked all available sequences from avian SMyHC III orthologs. As it will become clear below, the cNRE is a rare sequence, rather than a low complexity repeat. Our search for cNREs outside of the quail context (Coturnix coturnix) followed two independent lines. First, we took a scaled, evolution-oriented approach. Initially, we looked for cNREs in species close to the quail (i.e., Galliformes) and then progressively farther, to include derived (i.e., Passeriformes) and basal avians (i.e., Paleognaths) as well as external groups such as crocodilians. While pursuing this line of investigation, it became clear that the cNRE was a rare form of repetitive element, which showed a conserved topological relationship with the SMyHC III gene (i.e., cNREs flanked the SMyHC III genes at 5’ and 3’ regions). Using this topological relationship as a character, we determined when it appeared during avian evolution, and then set out to establish the likely origins of this rare repetitive motif. This search for the origins of the cNRE entailed comparisons to databases of repetitive genome elements, until the extreme telomeric nature of the SMyHC III gene became evident. This finding directed us to the fact that the hexad nature of the cNRE is reminiscent of the hexameric character of telomeric direct repeats. Because direct telomeric repeats are exactly featured in the genomes of avian DNA viruses that can infect the germline and integrate into the avian genome (Morissette & Flamand, 2010), we focused our search for the cNRE on the members of the subfamily Alphaherpesvirinae. In this search, we utilized the human herpes simplex virus 1 (HSV1) as a general model for herpes viruses and a set of four (4) members of the Alphaherpesvirinae family that specifically infect Galliformes (i.e., GaHV1, the virus responsible for avian infectious laryngotracheitis in chickens, GaHV2, the Marek’s disease virus, GaHV3, a non-pathogenic virus and MeHV1, the non-pathogenic Meleagrid herpesvirus 1 capable of infecting chicken and wild turkey) (Waidner et al., 2009). The search for cNREs in Alphaherpesvirinae was successful. We found six (6) cNRE hits in HSV1 and one (1) cNRE was detected in GaHV1, but none in MeHV1, GaHV2, and GaHV3.
Our evolution-directed approach thus led to the direct recognition that cNREs up to a cutoff mismatch value of 11 can be found in the genomes of a family of viruses that contain members that infect avians and integrate their double-stranded DNA into the host germline. Therefore, as a second independent approach, we set out to extend this proof of concept by broadening our search to all known sequenced viruses to perform an unbiased, internally consistent, and quantitative analysis of cNRE presence in viral genomes, as already reported in the initial submission of this manuscript.
Author Response
Public Evaluation Summary:
The authors re-analyzed a previously published dataset and identify patterns suggestive of increased bacterial biodiversity in the gut may creating new niches that lead to gene loss in a focal species and promote generation of more diversity. Two limitations are (i) that sequencing depth may not be sufficient to analyze strain-level diversity and (ii) that the evidence is exclusively based on correlations, and the observed patterns could also be explained by other eco-evolutionary processes. The claims should be supported by a more detailed analysis, and alternative hypotheses that the results do not fully exclude should be discussed. Understanding drivers of diversity in natural microbial communities is an important question that is of central interest to biomedically oriented microbiome scientists, microbial ecologists and evolutionary biologists.
We agree that understanding the drivers of diversity in natural communities is an important and challenging question to address. We believe that our analysis of metagenomes from the gut microbiomes is complementary to controlled laboratory experiments and modeling studies. While these other studies are better able to establish causal relationships, we rely on correlations – a caveat which we make clear, and offer different mechanistic explanations for the patterns we observe.
We also mention the caveat that we are only able to measure sub-species genetic diversity in relatively abundant species with high sequencing depth in metagenomes. These relatively abundant species include dozens of species in two metagenomic datasets, and we see no reason why they would not generalize to other members of the microbiome. Nonetheless, further work will be required to extend our results to rarer species.
Our revised manuscript includes two major new analyses. First, we extend the analysis of within-species nucleotide diversity to non-synonymous sites, with generally similar results. This suggests that evolutionarily older, less selectively constrained synonymous mutations and more recent non-synonymous mutations that affect protein structure both track similarly with measures of community diversity – with some subtle differences described in the manuscript.
Second, we extend our analysis of dense time series data from one individual stool donor and one deeply covered species (B. vulgatus) to four donors and 15 species. This allowed us to reinforce the pattern of gene loss in more diverse communities with greater statistical support. Our correlational results are broadly consistent with the predictions of DBD from modeling and experimental studies, and they open up new lines of inquiry for microbiome scientists, ecologists, and evolutionary biologists.
Reviewer #1 (Public Review):
This paper makes an important contribution to the current debate on whether the diversity of a microbial community has a positive or negative effect on its own diversity at a later time point. In my view, the main contribution is linking the diversity-begets-diversity patterns, already observed by the same authors and others, to genomic signatures of gene loss that would be expected from the Black Queen Hypothesis, establishing an eco-evolutionary link. In addition, they test this hypothesis at a more fine-grained scale (strain-level variation and SNP) and do so in human microbiome data, which adds relevance from the biomedical standpoint. The paper is a well-written and rigorous analysis using state-of-the-art methods, and the results suggest multiple new experiments and testable hypotheses (see below), which is a very valuable contribution.
We thank the reviewer for their generous comments.
That being said, I do have some concerns that I believe should be addressed. First of all, I am wondering whether gene loss could also occur because of environmental selection that is independent of other organisms or the diversity of the community. An alternative hypothesis to the Black Queen is that there might have been a migration of new species from outside and then loss of genes could have occurred because of the nature of the abiotic environment in the new host, without relationship to the community diversity. Telling the difference between these two hypotheses is hard and would require extensive additional experiments, which I don't think is necessary. But I do think the authors should acknowledge and discuss this alternative possibility and adjust the wording of their claims accordingly.
We concur with the reviewer that the drivers of the correlation between community diversity and gene loss are unclear. Therefore, we have now added the following text to the Discussion:
“Here we report that genome reduction in the gut is higher in more diverse gut communities. This could be due to de novo gene loss, preferential establishment of migrant strains encoding fewer genes, or a combination of the two. The mechanisms underlying this correlation remain unclear and could be due to biotic interactions – including metabolic cross-feeding as posited by some models (Estrela et al., 2022; San Roman and Wagner, 2021, 2018) but not others (Good and Rosenfeld, 2022) – or due to unknown abiotic drivers of both community diversity and gene loss.”
Additionally, we have revised Figure 1 to show that strain invasions/replacements, in addition to evolutionary change, could be an important driver of changes in intra-species diversity in the microbiome.
Another issue is that gene loss is happening in some of the most abundant species in the gut. Under Black Queen though, we would expect these species to be most likely "donors" in cross-feeding interactions. Authors should also discuss the implications, limitations, and possible alternative hypotheses of this result, which I think also stimulates future work and experiments.
We thank the reviewer for raising this point. It is unclear to us whether the more abundant species would be donors in cross-feeding interactions. If we understand correctly, the reviewer is suggesting that more abundant donors will contribute more total biomass of shared metabolites to the community. This idea makes sense under the assumption that the abundant species are involved in cross-feeding interactions in the first place, which may or may not be the case. As our work heavily relies on a dataset that we previously analyzed (HMP), we wish to cite Figure S20 in Garud, Good et al. 2019 PLoS Biology in which we found there are comparable rates of gene changes across the ~30 most abundant species analyzed in the HMP. This suggests that among the most abundant species analyzed, there is no relationship between their abundance and gene change rate.
That being said, we acknowledge that our study is limited to the relatively abundant focal species and state now in the Discussion: “Deeper or more targeted sequencing may permit us to determine whether the same patterns hold for rarer members of the microbiome.”
Regarding Figure 5B, there is a couple of questions I believe the authors should clarify. First, How is it possible that many species have close to 0 pathways? Second, besides the overall negative correlation, the data shows some very conspicuous regularities, e.g. many different "lines" of points with identical linear negative slope but different intercept. My guess is that this is due to some constraints in the pathway detection methods, but I struggle to understand it. I think the authors should discuss these patterns more in detail.
We sincerely thank the reviewer for raising this issue, as it prompted us to investigate more deeply the patterns observed at the pathway level. In short, we decided to remove this analysis from the paper because of a number of bioinformatics issues that we realized were contributing to the signal. However, in support of BQH-like mechanisms at play, we do find evidence for gene loss in more diverse communities across multiple species in both the HMP and Poyet datasets. Below we detail our investigation into Figure 5b and how we arrived at the conclusion that is should be removed:
(1) Regarding data points in Figure 5B where many focal species have “zero pathways”,we firstly clarify how we compute pathway presence and richness. Pathway abundance data per species were downloaded from the HMP1-2 database, and these pathway abundances were computed using HUMAnN (HMP Unified Metabolic Analysis Network). According to HUMAnN documentation, pathway abundance is proportional to the number of complete copies of the pathway in the community; this means that if at least one component reaction in a certain pathway is missing coverage (for a sample-species pair), the pathway abundance may be zero (note that HUMAnN also employs “gap filling” to allow no more than one required reaction to have zero abundance). As such, it is likely that insufficient coverage, especially for low-abundance species, causes many pathways to report zero abundance in many species in many samples. Indeed, 556 of the 649 species considered had zero “present” pathways (i.e. having nonzero abundance) in at least 400 of the 469 samples (see figure below).
(2) We thank the reviewer for pointing out the “conspicuous regularities” in Figure 5B,particularly “parallel lines” of data points that we discovered are an artifact of the flawed way in which we computed “community pathway richness [excluding the focal species].” Each diagonal line of points corresponds to different species in the same sample, and because community pathway richness is computed as the total number of pathways [across all species in the sample] minus the number of pathways in the focal species, the current Figure 5B is really plotting y against X-y for each sample (where X is a sample’s total community pathway richness, and y is the pathway richness of an individual species in that sample). This computation fails to account for the possibility that a pathway in an excluded focal species will still be present in the community due to redundancy, and indeed BQH tests for whether this redundancy is kept low in diverse communities due to mechanisms such as gene loss.
We attempted to instead plot community pathway richness defined as the number of unique pathways covered by all species other than the focal species. This is equivalent to [number of unique pathways across all species in a sample] minus the [number of pathways that are ONLY present in the focal species and not any other species in the sample]. However, when we recomputed community pathway richness this way, it is rare that a pathway is present in only one species in a sample. Moreover, we find that with the exception of E. coli, focal species pathway richness tended to be very similar across the 469 samples, often reaching an upper limit of focal species pathway richness observed. (It is unclear to what extent lower pathway richnesses are due to low species abundance/low sample coverage versus gene loss). This new plot reveals even more regularities and is difficult to interpret with respect to BQH. (Note that points are colored by species; the cluster of black dots with outlying high focal pathway richness corresponds to the “unclassified” stratum which can be considered a group of many different species.)
Overall, because community pathway richness (excluding a focal species) seems to primarily vary with sample rather than focal species in this dataset when using the most simple/strict definition of community pathway richness as described above, it is difficult to probe the Black Queen Hypothesis using a plot like Figure 5B. As pointed out by reviewers, lack of sequencing depth to analyze strain-level diversity and accurately quantify pathway abundance, irrespective of species abundance, seems to be a major barrier to this analysis. As such, we have decided to remove Figure 5B from the paper and rewrite some of our conclusions accordingly.
Finally, I also have some conceptual concerns regarding the genomic analysis. Namely, genes can be used for biosynthesis of e.g. building blocks, but also for consumption of nutrients. Under the Black Queen Hypothesis, we would expect the adaptive loss of biosynthetic genes, as those nutrients become provided by the community. However, for catabolic genes or pathways, I would expect the opposite pattern, i.e. the gain of catabolic genes that would allow taking advantage of a more rich environment resulting from a more diverse community (or at least, the absence of pathway loss). These two opposing forces for catabolic and biosynthetic genes/pathways might obscure the trends if all genes are pooled together for the analysis. I believe this can be easily checked with the data the authors already have, and could allow the authors to discuss more in detail the functional implications of the trends they see and possibly even make a stronger case for their claims.
We thank the reviewer for their suggestion. As explained above, we have removed the pathway analysis from the paper due to technical reasons. However, we did investigate catabolic and biosynthetic pathways separately as suggested by the reviewer as we describe below:
We obtained subsets of biosynthetic pathways and catabolic pathways by searching for keywords (such as “degradation” for catabolic) in the MetaCyc pathway database. After excluding the “unclassified” species stratum, we observe a total of 279 biosynthetic and 167 catabolic pathways present in the HMP1-2 pathway abundance dataset. Using the corrected definition of community pathway richness excluding a focal species, for each pathway type—either biosynthetic or catabolic—we plotted focal species pathway richness against community pathway richness including all pathways regardless of type:
We observe the same problem where, within a sample, community pathway richness excluding the focal species hardly varies no matter which focal species it is, due to nearly all of its detected pathways being present in at least one other species; this makes the plots difficult to interpret.
Reviewer #2 (Public Review):
The authors re-analysed two previously published metagenomic datasets to test how diversity at the community level is associated with diversity at the strain level in the human gut microbiota. The overall idea was to test if the observed patterns would be in agreement with the "diversity begets diversity" (DBD) model, which states that more diversity creates more niches and thereby promotes further increase of diversity (here measured at the strain-level). The authors have previously shown evidence for DBD in microbiomes using a similar approach but focusing on 16S rRNA level diversity (which does not provide strain-level insights) and on microbiomes from diverse environments.
One of the datasets analysed here is a subset of a cross-sectional cohort from the Human Microbiome Project. The other dataset comes from a single individual sampled longitudinally over 18 months. This second dataset allowed the authors to not only assess the links between different levels of diversity at single timepoints, but test if high diversity at a given timepoint is associated with increased strain-level diversity at future timepoints.
Understanding eco-evolutionary dynamics of diversity in natural microbial communities is an important question that remains challenging to address. The paper is well-written and the detailed description of the methodological approaches and statistical analyses is exemplary. Most of the analyses carried out in this study seem to be technically sound.
We thank the reviewer for their kind words, comments, and suggestions.
The major limitation of this study comes with the fact that only correlations are presented, some of which are rather weak, contrast each other, or are based on a small number of data points. In addition, finding that diversity at a given taxonomic rank is associated with diversity within a given taxon is a pattern that can be explained by many different underlying processes, e.g. species-area relationships, nutrient (diet) diversity, stressor diversity, immigration rate, and niche creation by other microbes (i.e. DBD). Without experiments, it remains vague if DBD is the underlying process that acts in these communities based on the observed patterns.
We thank the reviewer for their comments. First, regarding the issue of this being a correlative study, we now more clearly acknowledge that mechanistic studies (perhaps in experimental settings) are required to fully elucidate DBD and BQH dynamics. However, we note that our correlational study from natural communities is complementary to experimental and modeling studies, to test the extent to which their predictions hold in more complex, realistic settings. This is now mentioned throughout the manuscript, most explicitly at the end of the Introduction:
“Although such analyses of natural diversity cannot fully control for unmeasured confounding environmental factors, they are an important complement to controlled experimental and theoretical studies which lack real-world complexity.”
Second, to increase the number of data points analyzed in the Poyet study, we now include 15 species and four different hosts (new Figure 5). The association between community diversity and gene loss is now much more statistically robust, and consistent across the Poyet and HMP time series.
Third, we acknowledge more clearly in the Discussion that other processes, including diet and other environmental factors can generate the DBD pattern. We also now stress more prominently the possibility that strain migration across hosts may be responsible for the patterns observed. For example, in Figure 1, we illustrate the possibility of strain migration generating the patterns we observe.
Below we quote a paragraph that we have now added in the Discussion:
"Second, we cannot establish causal relationships without controlled experiments. We are therefore careful to conclude that positive diversity slopes are consistent with the predictions of DBD, and negative slopes with EC, but unmeasured environmental drivers could be at play. For example, increased dietary diversity could simultaneously select for higher community diversity and also higher intra-species diversity. In our previous study, we found that positive diversity slopes persisted even after controlling for potential abiotic drivers such as pH and temperature (Madi et al., 2020), but a similar analysis was not possible here due to a lack of metadata. Neutral processes can account for several ecological patterns such as species-area relationships (Hubbell, 2001), and must be rejected in favor of niche-centric models like DBD or EC. Using neutral models without DBD or EC, we found generally flat or negative diversity slopes due to sampling processes alone and that positive slopes were hard to explain with a neutral model (Madi et al., 2020). These models were intended mainly for 16S rRNA gene sequence data, but we expect the general conclusions to extend to metagenomic data. Nevertheless, further modeling and experimental work will be required to fully exclude a neutral explanation for the diversity slopes we report in the human gut microbiome.”
Finally, we now put more emphasis on the importance of migration (strain invasion) as a non-exclusive alternative to de novo mutation and gene gain/loss. This is mentioned in the Abstract and is also illustrated in the revised Figure 1.
Another limitation is that the total number of reads (5 mio for the longitudinal dataset and 20 mio for the cross-sectional dataset) is low for assessing strain-level diversity in complex communities such as the human gut microbiota. This is probably the reason why the authors only looked at one species with sufficient coverage in the longitudinal dataset.
Indeed, this is a caveat which means we can only consider sub-species diversity in relatively abundant species. Nevertheless, this allows us to study dozens of species in the HMP and 15 in the more frequent Poyet time series. As more deeply sequenced metagenomes become available, future studies will be able to access the rarer species to test whether the same patterns hold or not. This is now mentioned prominently as a caveat our study in the second Discussion paragraph:
“First, using metagenomic data from human microbiomes allowed us to study genetic diversity, but limited us to considering only relatively abundant species with genomes that were well-covered by short sequence reads. Deeper or more targeted sequencing may permit us to determine whether the same patterns hold for rarer members of the microbiome. However, it is notable that the majority of the dozens of species across the two datasets analyzed support DBD, suggesting that the phenomenon may generalize.”
We also note that rarefaction was only applied to calculate community richness, not to estimate sub-species diversity. We apologize for this confusion, which is now clarified in the Methods as follows:
“SNV and gene content variation within a focal species were ascertained only from the full dataset and not the rarefied dataset.”
Analyzing the effect of diversity at a given timepoint on strain-level diversity at a later timepoint adds an important new dimension to this study which was not assessed in the previous study about the DBD in microbiomes by some of the authors. However, only a single species was analysed in the longitudinal dataset and comparisons of diversity were only done between two consecutive timepoints. This dataset could be further exploited to provide more insights into the prevailing patterns of diversity.
We thank the reviewer for raising this point. We now have considered all 15 species for which there was sufficient coverage from the Poyet dataset, which included four different stool donors. Additionally, in the HMP dataset, we analyze 54 species across 154 hosts, with both datasets showing the same correlation between community diversity and gene loss.
Additionally, we followed the suggestion of the reviewer of examining additional time lags, and in Figure 5 we do observe a dependency on time. This is now described in the Results as follows:
“Using the Poyet dataset, we asked whether community diversity in the gut microbiome at one time point could predict polymorphism change at a future time point by fitting GAMs with the change in polymorphism rate as a function of the interaction between community diversity at the first time point and the number of days between the two time points. Shannon diversity at the earlier time point was correlated with increases in polymorphism (consistent with DBD) up to ~150 days (~4.5 months) into the future (Figure S4), but this relationship became weaker and then inverted (consistent with EC) at longer time lags (Fig 5A, Table S8, GAM, P=0.023, Chi-square test). The diversity slope is approximately flat for time lags between four and six months, which could explain why no significant relationship was found in HMP, where samples were collected every ~6 months. No relationship was observed between community richness and changes in polymorphism (Table S8, GAM, P>0.05).”
Finally, the evidence that gene loss follows increase in diversity is weak, as very few genes were found to be lost between two consecutive timepoints, and the analysis is based on only a single species. Moreover, while positive correlation were found between overall community diversity and gene family diversity in single species, the opposite trend was observed when focusing on pathway diversity. A more detailed analysis (of e.g. the functions of the genes and pathways lost/gained) to explain these seemingly contrasting results and a more critical discussion of the limitations of this study would be desirable.
We agree that our previous analysis of one species in one host provided weak support for gene loss following increases in diversity. As described in the response above, we have now expanded this analysis to 15 focal species and 4 independent hosts with extensive time series. We now analyze this larger dataset and report the more statistically robust results as follows:
“We found that community Shannon diversity predicted future gene loss in a focal species, and this effect became stronger with longer time lags (Fig 5B, Table S9, GLMM, P=0.006, LRT for the effect of the interaction between the initial Shannon diversity and time lag on the number of genes lost). The model predicts that increasing Shannon diversity from its minimum to its maximum would result in the loss of 0.075 genes from a focal species after 250 days. In other words, about one of the 15 focal species considered would be expected to lose a gene in this time frame.
Higher Shannon diversity was also associated with fewer gene gains, and this relationship also became stronger over time (Fig 5C, Table S9, GLMM, P=1.11e-09, LRT). We found a similar relationship between community species richness and gene gains, although the relationship was slightly positive at shorter time lags (Fig 5D, Table S9, GLMM, P=3.41e-04, LRT). No significant relationship was observed between richness and gene loss (Table S9, GLMM, P>0.05). Taken together with the HMP results (Fig 4), these longer time series reveal how the sign of the diversity slope can vary over time and how community diversity is generally predictive of reduced focal species gene content.”
As described in detail in the response to Reviewer 1 above, we found that the HUMAnN2 pathway analyses previously described suffered from technical challenges and we deemed them inconclusive. We have therefore removed the pathway results from the manuscript.
Reviewer #3 (Public Review):
This work provides a series of tests of hypothesis, which are not mutually exclusive, on how genomic diversity is structured within human microbiomes and how community diversity may influence the evolution of a focal species.
Strengths:
The paper leverages on existing metagenomic data to look at many focal species at the same time to test for the importance of broad eco-evolutionary hypothesis, which is a novelty in the field.
Thank you for the succinct summary and recognition of the strengths of our work.
Weaknesses:
It is not very clear if the existing metagenomic data has sufficient power to test these models.
It is not clear, neither in the introduction nor in the analysis what precise mechanisms are expected to lead to DBD.
The conclusion that data support DBD appears to depend on which statistics to measure of community diversity are used. Also, performing a test to reject a null neutral model would have been welcome either in the results or in the discussion.
In our revised manuscript, we emphasize several caveats – including that we only have power to test these hypotheses in focal species with sufficient metagenomic coverage to measure sub-species diversity. We also describe more in the Introduction how the processes of competition and niche construction can lead to DBD. We also acknowledge that unmeasured abiotic drivers of both community diversity and sub-species diversity could also lead to the observed patterns. Throughout the manuscript, we attempt to describe the results and acknowledge multiple possible interpretations, including DBD and EC acting with different strengths on different species and time scales. Our previous manuscript assessing the evidence for DBD using 16S rRNA gene amplicon data from the Earth Microbiome Project (Madi et al., eLife 2020) assessed null models based on neutral ecological theory, and found it difficult to explain the observation of generally positive diversity slopes without invoking a non-neutral mechanism like DBD. While a new null model tailored to metagenomic data might provide additional nuance, we think developing one is beyond the scope of the manuscript – which is in the format of a short ‘Research Advance’ to expand on our previous eLife paper, and we expect that the general results of our previously reported null model provide a reasonable intuition for our new metagenomic analysis. This is now mentioned in the Discussion as follows:
“In our previous study, we found that positive diversity slopes persisted even after controlling for potential abiotic drivers such as pH and temperature (Madi et al., 2020), but a similar analysis was not possible here due to a lack of metadata. Neutral processes can account for several ecological patterns such as species-area relationships (Hubbell, 2001), and must be rejected in favor of niche-centric models like DBD or EC. Using neutral models without DBD or EC, we found generally flat or negative diversity slopes due to sampling processes alone and that positive slopes were hard to explain with a neutral model (Madi et al., 2020). These models were intended mainly for 16S rRNA gene sequence data, but we expect the general conclusions to extend to metagenomic data. Nevertheless, further modeling and experimental work will be required to fully exclude a neutral explanation for the diversity slopes we report in the human gut microbiome.”
Author Response
Reviewer #1 (Public Review):
The study by Akter et al demonstrates that astrocyte-derived L-lactate plays a key role in schema memory formation and promotes mitochondrial biogenesis in the Anterior Cingulate Cortex (ACC).
The main tool used by the authors is the DREADD technology that allows to pharmacologically activate receptors in a cell-specific manner. In the study, the authors used the DREADD technique to activate appropriately transfected astrocytes, a subtype of muscarinic receptor that is not normally present in cells. This receptor being coupled to a Gi-mediated signal transduction pathway inhibiting cAMP formation, the authors could demonstrate cell-(astrocyte) specific decreases in cAMP levels that result in decreased L-lactate production by astrocytes.
Behaviorally this pharmacological manipulation results in impairments of schema memory formation and retrieval in the ACC in flavor-place paired associate paradigms. Such impairments are prevented by co-administration of L-lactate.
The authors also show that activation of Gi signaling resulting in L-lactate decreased release by astrocytes impairs mitochondrial biogenesis in neurons in an L-lactate reversible manner.
By using MCT 2 inhibitors and an NMDAR antagonist the authors conclude that the molecular mechanisms underlying the observed effects are mediated by L-lactate entering neurons through MCT2 transporters and involve NMDAR.
Overall, the article's conclusions are warranted by the experimental evidence, but some weak points could be addressed which would make the conclusions even stronger.
The number of animals in some of the experiments is on the low side (4 to 6).
In the revised manuscript, we have increased the animal numbers in two key experimental groups (hM4Di-CNO and Control groups) of behavioral experiments. Now the animal numbers in different groups are as follows:
• 15 rats in hM4Di-CNO group
o Further divided into two subgroups for probe tests (PT1-4) conducted during flavor-place paired associate training; 8 rats in the hM4Di-CNO (saline) and 7 rats in the hM4Di-CNO (CNO) subgroups receiving I.P. saline or I.P. CNO, respectively, before these PTs.
• 8 rats in the Control group
• 7 rats in the Rescue group (hM4Di-CNO+L-lactate)
• 4 rats in the Control-CNO group. Animal number in this group was not increased as it was apparent from these 4 rats that CNO alone was not impairing the PA learning and memory retrieval in these rats (AAV8-GFAP-mCherry injected). Their result was very similar to the control group. Additionally, in a previous study (Liu et al., 2022), we showed that CNO administration in the rats injected with AAV8-GFAP-mCherry into the hippocampus does not show any impairments in schema.
Also, in the newly added open field test experiments to investigate the locomotor activity as suggested by the Reviewer #2, 8 rats were used in each group.
The use of CIN to inhibit MCT2 is not optimal. Authors may want to decrease MCT2 expression by using antisense oligonucleotides.
In the revised manuscript, we have conducted the experiment using MCT2 antisense oligodeoxynucleotide (ODN) as suggested.
To test whether the L-lactate-induced neuronal mitochondrial biogenesis is dependent on MCT2, we bilaterally injected MCT2 antisense oligodeoxynucleotide (MCT2-ODN, n=8 rats, 2 nmol in 1 μl PBS per ACC) or scrambled ODN (SC-ODN, n=8 rats, 2 nmol in 1 μl PBS per ACC) into the ACC. After 11 hours, bilateral infusion of L-lactate (10 nmol, 1 μl) or ACSF (1 μl) was given into the ACC and the rats were kept in the PA event arena. After 60 mins (12 hours from MCT2-ODN or SC-ODN administration), the rats were sacrificed. As shown in Author response image 1B, SC-ODN+L-lactate group showed significantly increased relative mtDNA copy number compared to the SC-ODN+ACSF group (p<0.001, ANOVA followed by Tukey's multiple comparisons test). However, this effect was completely abolished in MCT2-ODN+L-lactate group, suggesting that MCT2 is required for the L-lactate-induced mitochondrial biogenesis in the ACC.
We have integrated this new data and results in the revised manuscript.
Author response image 1.
Mitochondrial biogenesis by L-lactate is dependent on MCT2 and NMDAR. A. Experimental design to investigate whether MCT2 and NMDAR activity are required for L-lactate-induced mitochondrial biogenesis. B and C. mtDNA copy number abundance in the ACC of different rat groups relative to nDNA. Data shown as mean ± SD (n=4 rats in each group). ***p<0.001, ANOVA followed by Tukey's multiple comparisons test.
The experiment using AVP to block NMDAR only partially supports the conclusions. Indeed, blocking NMDAR will knock down any response that involves these receptors, whether L-lactate is necessary or not.
In the current study we found that Astrocytic Gi activation in the ACC reduced L-lactate level in the ECF of ACC which was also associated with decreased PGC-1α/SIRT3/ATPB/mtDNA abundance suggesting downregulation of mitochondrial biogenesis pathway. We also found that exogenous administration of L-lactate into the ACC of astrocytic Gi-activated rats rescued this downregulation. In line with this, in a recently published study (Akter et al., 2023), we found upregulation of mitochondrial biogenesis pathway in the hippocampus neurons of exogenous L-lactate-treated anesthetized rats. Another recent study has demonstrated that exercise-induced L-lactate release from skeletal muscle or I.P. injection of L-lactate can induce hippocampal PGC-1α (which is a master regulator of mitochondrial biogenesis) expression and mitochondrial biogenesis in mice (Park et al., 2021). Together, these results provide compelling evidence that L-lactate promotes mitochondrial biogenesis.
L-lactate is known to promote expression of synaptic plasticity genes like Arc, c-Fos, and Zif268 in neurons (Yang et al., 2014). After entry into the neuronal cytoplasm, mainly through MCT2, it is converted into pyruvate by lactate dehydrogenase 1 (LDH1). This conversion also produces NADH, affecting the redox state of the neuron. NADH positively modulates the activity of NMDAR resulting in enhanced Ca2+ currents, the activation of intracellular signaling cascades, and the induction of the expression of plasticity-associated genes (Yang et al., 2014; Magistretti & Allaman, 2018). The study demonstrated that L-lactate–induced plasticity gene expression was abolished in the presence of NMDAR antagonists including D-APV (Yang et al., 2014). These results suggested that the MCT2 and NMDAR are key players in the regulation of L-lactate induced plasticity gene expression.
In the current study, we investigated whether similar mechanisms might be involved in L-lactate-induced neuronal mitochondrial biogenesis. We now used MCT2 antisense oligodeoxynucleotide to decrease the expression of MCT2 (as mentioned in the previous response and Author response image 1B) and showed that MCT2 is necessary for L-lactate-induced mitochondrial biogenesis to manifest, indicating that L-lactate’s entry into the neuron is required. As mentioned before, after entry into neuron, L-lactate is converted into pyruvate by LDH, which also produce NADH, which in turn potentiates NMDAR activity. Therefore, we investigated whether NMDAR activity is required for L-lactate-induced mitochondrial biogenesis. We used D-APV to inhibit NMDAR (Author response image 1C) and found that L-lactate does not increase mtDNA copy number abundance if D-APV is given, suggesting that NMDAR activity is required for L-lactate to promote mitochondrial biogenesis.
NMDAR serves diverse functions. Therefore, as mentioned by the reviewer, blocking NMDAR may knock down many such functions. While our current data only suggests the involvement of MCT2 and NMDAR in the upregulation of mitochondrial biogenesis by L-lactate, we have not investigated other mechanisms and pathways modulating mitochondrial biogenesis that are either dependent or independent of MCT2 and NMDAR activity. Further studies are needed in future to dissect and better understand this interesting observation. We have now clarified this in the discussion section of the manuscript.
Is inhibition of glycogenolysis involved in the observed effects mediated by Gi signaling? Indeed, L-lactate is formed both by glycolysis and glycogenolysis. The authors could test whether the glycogen metabolism-inhibiting drug DAB would mimic the effects of Gi activation.
In this study we have shown that astrocytic Gi activation in the ACC leads to a decrease in the cAMP and L-lactate. L-lactate is produced by glycogenolysis and glycolysis. cAMP in astrocytes acts as a trigger for L-lactate production (Choi et al., 2012; Horvat, Muhič, et al., 2021; Horvat, Zorec, et al., 2021; Zhou et al., 2021) by promoting glycogenolysis and glycolysis (Vardjan et al., 2018; Horvat, Muhič, et al., 2021; Horvat, Zorec, et al., 2021). Therefore, one promising explanation of reduced L-lactate level observed in our study is the reduction of L-lactate production in the astrocyte due to decreased glycogen metabolism as a result of decreased cAMP. We have now mentioned this in the discussion.
DAB is an inhibitor of glycogen phosphorylase that suppresses L-lactate production. It was shown to impair memory by decreasing L-lactate (Newman et al., 2011; Suzuki et al., 2011; Iqbal et al., 2023). As we found that the impairment in the schema memory and mitochondrial biogenesis was associated with decreased L-lactate level in the ACC and that the exogenous L-lactate administration can rescue the impairments, it is likely that DAB will mimic the effect of Gi activation in terms of schema memory and mitochondrial biogenesis. However, further study is needed to confirm this.
Reviewer #2 (Public Review):
The manuscript of Akter et al is an important study that investigates the role of astrocytic Gi signaling in the anterior cingulate cortex in the modulation of extracellular L-lactate level and consequently impairment in flavor-place associates (PA) learning. However, whereas some of the behavioral observations and signaling mechanism data are compelling, the conclusions about the effect on memory are inadequate as they rely on an experimental design that does not allow to differentiate acute or learning effect from the effect outlasting pharmacological treatments, i.e. effect on memory retention. With the addition of a few experiments, this paper would be of interest to the larger group of researchers interested in neuron-glia interactions during complex behavior.
• Largely, I agree with the authors' conclusion that activating Gi signaling in astrocytes impairs PA learning, however, the effect on memory retrieval is not that obvious. All behavioral and molecular signaling effects described in this study are obtained with the continuous presence of CNO, therefore it is not possible to exclude the acute effect of Gi pathway activation in astrocytes. What will happen with memory on retrieval test when CNO is omitted selectively during early, middle, or late session blocks of PA learning?
We have now added 8 more rats to the hM4Di-CNO group (i.e., the group with astrocytic Gi activation) to clarify the memory retrieval. These rats underwent flavor-place paired associate (PA) training similar to the previously described rats (n=7) of this group, that is they received CNO 30 minutes before and 30 minutes after the PA training sessions (S1-2, S4-8, S10-17). However, contrasting to the previous rats of this group which received CNO before PTs (PT1, PT2, PT3), we omitted the CNO (instead administered I.P. saline) selectively on these PTs conducted at the early, middle, and late stage of PA training, as suggested by the reviewer. These newly added rats did not show memory retrieval in these PTs, suggesting that the rats were not learning the PAs from the PA training sessions. See Author response image 2C-E, where this subgroup is denoted as hM4Di-CNO (Saline).
We then continued more PA training sessions (S21 onwards, Author response image 2B) for these rats without CNO. They gradually learned the PAs. PTs (PT5, PT6, PT7; Author response image 2G-I) were done during this continuation phase of PA training; once without CNO (i.e., with I.P. saline instead), and another one with CNO. As seen in the Author response image 2H and 2I, they retrieved the memory when PT6 and PT7 were done without CNO. However, if these PTs were done with CNO, they could not retrieve the memory. Together these results suggest that ACC astrocytic Gi activation by CNO during PT can impair memory retrieval in rats which have already learned the PAs.
As shown in the Author response image 2B, we replaced two original PAs with two new PAs (NPA 9 and 10) at S34. This was followed by PT8 (S35). As seen in Author response image 2J, these rats retrieved the NPA memory if the PT is done without CNO. However, they could not retrieve the NPA memory if the PT was done with CNO. This result suggests that ACC astrocytic Gi activation by CNO during PT can impair NPA memory retrieval.
In summary, these data show that astrocytic Gi activation in the ACC can impair PA memory retrieval. We have integrated this new data and results in the revised manuscript.
Author response image 2.
A. PI (mean ± SD) during the acquisition of the six original PAs (OPAs) (S1-2, 4-8, 10-17) and new PAs (NPAs) (S19) of the control (n=8), hM4Di-CNO (n=15), and rescue (hM4Di-CNO+L-lactate) (n=7) groups. From S6 onwards, hM4Di-CNO group consistently showed lower PI compared to control. However, concurrent L-lactate administration into the ACC (rescue group) can rescue this impairment. B. PI (mean ± SD) of hM4Di-CNO group (n=8) from S21 onwards showing gradual increase in PI when CNO was withdrawn. C, D, and E. Non-rewarded PTs (PT1, PT2, and PT3 conducted on S3, S9, and S18, respectively) to test memory retrieval of OPAs for the control, hM4Di-CNO, and rescue groups. The percentage of digging time at the cued location relative to that at the non-cued locations are shown (mean ± SD). In both PT2 and PT3, the control group spent significantly more time digging the cued sand well above the chance level, indicating that the rats learned OPAs and could retrieve it. Contrasting to this, hM4Di-CNO group did not spend more time digging the cued sand well above the chance level irrespective of CNO administration before the PTs. The rescue group showed results similar to the hM4Di-CNO group if CNO is given without L-lactate. On the other hand, they showed results similar to the control group if L-lactate is concurrently given with CNO, indicating that this group learned OPAs and could retrieve it. p < 0.05, p < 0.01, p < 0.001, one-sample t-test comparing the proportion of digging time at the cued sand well with the chance level of 16.67%. F. Non-rewarded PT4 (S20) which was conducted after replacing two OPAs with two NPAs (NPA 7 & 8) in S19 for the control, hM4Di-CNO, and rescue groups. Results show that the control group spent significantly more time digging the new cued sand well above the chance level indicating that the rats learned the NPAs from S19 and could retrieve it in this PT. Contrasting to this, hM4Di-CNO group did not spend more time digging the new-cued sand well above the chance level irrespective of CNO administration before the PT. The rescue group showed results similar to the hM4Di-CNO group if CNO is given without L-lactate. On the other hand, they showed results similar to the control group if L-lactate is concurrently given with CNO indicating that this group learned NPAs from S19 and could retrieve it. p < 0.001, one-sample t-test comparing the proportion of digging time at the new cued sand well with the chance level of 16.67%. G, H, and I. Non-rewarded PTs (PT5, PT6, and PT7 conducted on S23, S27, and S33, respectively) to test memory retrieval of OPAs for the hM4Di-CNO group. In both PT6 and PT7, the rats spent significantly more time digging the cued sand well above the chance level if the tests are done without CNO, indicating that the rats learned the OPAs and could retrieve it. However, CNO prevented memory retrieval during these PTs. p < 0.001, one-sample t-test comparing the proportion of digging time at the cued sand well with the chance level of 16.67%. J. Non-rewarded PT4 (S35) which was conducted after replacing two OPAs with two NPAs (NPA 9 & 10) in S34 for the hM4Di-CNO group. Results show that the rats spent significantly more time digging the new cued sand well above the chance level if CNO was not given before the PT, indicating that the rats learned the NPAs from S34 and could retrieve it in this PT. However, if CNO is given before the PT, the retrieval is impaired. *p < 0.001, one-sample t-test comparing the proportion of digging time at the new cued sand well with the chance level of 16.67%.
• I found it truly exciting that the administration of exogenous L-lactate is capable to rescue CNO-induced PA learning impairment, when co-applied. Would it be possible that this treatment has a sensitivity to a particular stage of learning (acquisition, consolidation, or memory retrieval) when L-lactate administration would be the most efficacious?
The hM4Di-CNO group, when continued with PA training without CNO (S21-S32) (Author response image 2B), was able to learn the six original PAs (OPAs). In the PT7 done at S33 (Author response image 2I), this group of rats was able to retrieve the memory if the test was done without CNO but could not retrieve the memory if CNO was given. Similarly, the Rescue group (hM4Di-CNO+L-lactate) (Author response image 2A), which received both CNO and L-lactate during PA training sessions (S1-S17), they were able to learn the OPAs. And at PT3 done at S18 (Author response image 2E), these rats were able to retrieve the memory when the test was done with CNO+L-lactate but not if the test is done with only CNO. Together, these results clearly show that ACC astrocytic Gi activation with CNO impairs memory retrieval and exogenous L-lactate can rescue the impairment. Therefore, it can be concluded that the memory retrieval is sensitive to L-lactate.
The PA learning is hippocampus-dependent. Over the course of repeated PA training, systems consolidation occurs in the ACC, after which the already learned PA memory (schema) becomes hippocampus-independent (Tse et al., 2007; Tse et al., 2011). A higher activation (indicated by expression of c-Fos) in the hippocampus relative to the ACC during the early period of schema development, and the reverse at the late stage was observed in our previous study (Liu et al., 2022). However, rapid assimilation of new PA into the ACC requires simultaneous activation/retrieval of previous schema from ACC and hippocampus dependent new PA learning (Tse et al., 2007; Tse et al., 2011). During new PA learning, increase of c-Fos neurons in both CA1 and ACC was detected (Liu et al., 2022).
Our hM4Di-CNO group received CNO 30 mins before and after each PA training session in S1-S17 (Author response image 2A). Also, the Rescue group similarly received CNO+L-lactate before and after each PA training session in S1-S17. Therefore, while this study design allowed us to conclude that ACC astrocytic Gi activation impairs PA learning and that exogenous L-lactate can rescue the impairment, it does not allow clear differentiation of the effects of these treatments on memory acquisition and consolidation. Further studies are needed to investigate this.
• The hypothesis that observed learning impairments could be associated with diminished mitochondrial biogenesis caused by decreased l-lactate in the result of astrocytic Gi-DREADDS stimulation is very appealing, but a few key pieces of evidence are missing. So far, the hypothesis is supported by experiments demonstrating reduced expression of several components of mitochondrial membrane ATP synthase and a decrease in relative mtDNA copy numbers in ACC of rats injected with Gi-DREADDs. L-lactate injections into ACC restored and even further increased the expression of the above-mentioned markers. Co-administration of NMDAR antagonist D-APV or MCT-2 (mostly neuronal) blocker 4-CIN with L-lactate, prevented L-lactate-induced increase in relative mtDNA copy. I am wondering how the interference with mitochondrial biogenesis is affecting neuronal physiology and if it would result in impaired PA learning or schema memory.
The observation of diminished mitochondrial biogenesis in the astrocytic Gi-activated rats that showed impaired PA learning is exciting. However, our study does not provide experimental data on how mitochondrial biogenesis could be associated with impaired PA learning and schema memory. Results from several previous studies linked mitochondrial biogenesis and its regulators such as PGC-1α and SIRT3 to diverse neuronal and cognitive functions as described in the discussion section of the manuscript. In the revised manuscript, we have provided further discussion as follows to discuss potential mechanisms:
“In this study, we have demonstrated that ACC astrocytic Gi activation impairs PA learning and schema formation, PA memory retrieval, and NPA learning and retrieval by decreasing L-lactate level in the ACC. Although we have shown that these impairments are associated with diminished expression of proteins of mitochondrial biogenesis, the precise mechanisms of how astrocytic Gi activation affects neuronal functions and schema memory remain to be elucidated. We previously demonstrated that neuronal inhibition in either the hippocampus or the ACC impairs PA learning and schema formation (Hasan et al., 2019). In another recent study (Liu et al., 2022), we showed that astrocytic Gi activation in the CA1 impaired PA training-associated CA1-ACC projecting neuronal activation. Yao et al. recently showed that reduction of astrocytic lactate dehydrogenase A (an enzyme that reversibly catalyze L-lactate production from pyruvate) in the dorsomedial prefrontal cortex reduces L-lactate levels and neuronal firing frequencies, promoting depressive-like behaviors in mice (Yao et al., 2023). These impairments could be rescued by L-lactate infusion. It is possible that the impairment in PA learning and schema observed in our study might have involved a similar functional consequence of reduced neuronal activity in the ACC neurons upon astrocytic Gi activation.
Schema consolidation is associated with synaptic plasticity-related gene expression (such as Zif268, Arc) in the ACC (Tse et al., 2011). L-lactate, after entry into neurons, can be converted to pyruvate during which NADH is also produced, promoting synaptic plasticity-related gene expression by potentiating NMDA signaling in neurons (Yang et al., 2014; Margineanu et al., 2018). Furthermore, L-lactate acts as an energy substrate to fuel learning-induced de novo neuronal translation critical for long-term memory (Descalzi et al., 2019). On the other hand, mitochondria play crucial role in fueling local translation during synaptic plasticity (Rangaraju et al., 2019). Therefore, it could be hypothesized that the rescue of astrocytic Gi activation-mediated impairment of schema by exogenous L-lactate could have been mediated by facilitating synaptic plasticity-related gene expression by directly fueling the protein translation, potentiating NMDA signaling, as well as increasing mitochondrial capacity for ATP production by promoting mitochondrial biogenesis. Furthermore, the potential involvement of HCAR1, a receptor for L-lactate that may regulate neuronal activity (Bozzo et al., 2013; Tang et al., 2014; Herrera-López & Galván, 2018; Abrantes et al., 2019), cannot be excluded. Future research could explore these potential mechanisms, examining the interactions among them, and determining their relative contributions to schema. Our previous study also showed that ACC myelination is necessary for PA learning and schema formation, and that repeated PA training is associated with oligodendrogenesis in the ACC (Hasan et al., 2019). Oligodendrocytes facilitate fast, synchronized, and energy efficient transfer of information by wrapping axons in myelin sheath. Furthermore, they supply axons with glycolysis products, such as L-lactate, to offer metabolic support (Fünfschilling et al., 2012; Lee et al., 2012). The association of oligodendrogenesis and myelination with schema memory may suggest an adaptive response of oligodendrocytes to enhance metabolic support and neuronal energy efficiency during PA learning. Given the impairments in PA learning observed in the ACC astrocytic Gi-activated rats in the current study, it is reasonable to conclude that the direct metabolic support to axons provided by oligodendrocytes is not sufficient to rescue the schema impairments caused by decreased L-lactate levels upon astrocytic Gi activation. On the other hand, L-lactate was shown to be important for oligodendrogenesis and myelination (Sánchez-Abarca et al., 2001; Rinholm et al., 2011; Ichihara et al., 2017). Therefore, it is tempting to speculate that a decrease in L-lactate level may also impede oligodendrogenesis and myelination, consequently preventing the enhanced axonal support provided by oligodendrocytes and myelin during schema learning. Recently, a study has demonstrated that upon demyelination, mitochondria move from the neuronal cell body to the demyelinated axon (Licht-Mayer et al., 2020). Enhancement of this axonal response of mitochondria to demyelination, by targeting mitochondrial biogenesis and mitochondrial transport from the cell body to axon, protects acutely demyelinated axons from degeneration. Given the connection between schema and increased myelination, it remains an open question whether L-lactate-induced mitochondrial biogenesis plays a beneficial role in schema through a similar mechanism. Nevertheless, our results contribute to the mounting evidence of the glial role in cognitive functions and underscores the new paradigm in which glial cells are considered as integral players in cognitive functions alongside neurons. Disruption of neurons, myelin, or astrocytes in the ACC can disrupt PA learning and schema memory.”
Reviewer #3 (Public Review):
Akter et al. investigated how the astroglial Gi signaling pathway in the rat anterior cingulate cortex (ACC) affects cognitive functions, in particular schema memory formation. Using a stereotactic approach they intracranially introduced AAV8 vectors carrying mCherry-tagged hM4Di DREADD (Designer Receptor Exclusively Activated by Designer Drugs) under astrocyte selective GFAP promotor (AAV8-GFAP-hM4Di-mCherry) into the AAC region of the rat brain. hM4Di DREADD is a genetically modified form of the human M4 muscarinic (hM4) receptor insensitive to endogenous acetylcholine but is activated by the inert clozapine metabolite clozapine-N-oxide (CNO), triggering the Gi signaling pathway. The authors confirmed that hM4Di DREADD is selectively expressed in astrocytes after the application of the AAV8 vector by analysing the mCherry signals and immunolabeling of astrocytes and neurons in the ACC region of the rat brain. They activated hM4Di DREADD (Gi signalling) in astrocytes by intraperitoneal administration of CNO and measured cognitive functions in animals after CNO administration. Activation of Gi signaling in astrocytes by CNO application decreased paired-associate (PA) learning, schema formation, and memory retrieval in tested animals. This was associated with a decrease in cAMP in astrocytes and L-lactate in extracellular fluid as measured by immunohistochemistry in situ and in awake rats by microdialysis, respectively. Administration of exogenous L-lactate rescued the astroglial Gi-mediated deficits in PA learning, memory retrieval, and schema formation, suggesting that activation of astroglial Gi signalling downregulates L-lactate production in astrocytes and its transport to neurons affecting memory formation. Authors also show that expression level of proteins involved in mitochondrial biogenesis, which is associated with cognitive functions, is decreased in neurons, when Gi signalling is activated in astrocytes, and rescued when exogenous L-lactate is applied, suggesting the implication of astrocyte-derived L-lactate in the maintenance of mitochondrial biogenesis in neurons. The latter depended on lactate MCT2 transporter activity and glutamate NMDA receptor activity.
The paper is very well written and discussed. The conclusions of this paper are well supported by the data. Although this is a study that uses established and previously published methodologies, it provides new insights into L-lactate signalling in the brain, particularly in AAC, and further confirms the role of astroglial L-lactate in learning and memory formation. It also raises new questions about the molecular mechanisms underlying astrocyte-derived L-lactate-mediated mitochondrial biogenesis in neurons and its contribution to schema memory formation.
• The authors discuss astrocytic L-lactate signalling without considering the recently discovered L-lactate-sensitive Gs and Gi protein-coupled receptors in the brain, which are present in both astrocytes and neurons. The use of nonendogenous L-lactate receptor agonists (Compound 2, 3-chloro-5-hydroxybenzoic acid) would clarify the implication of L-lactate receptor signalling in schema memory formation.
In the revised manuscript, we have included this point in the discussion section to mention the potential role of HCAR1 in schema memory as follows:
“Schema consolidation is associated with synaptic plasticity-related gene expression (such as Zif268, Arc) in the ACC (Tse et al., 2011). L-lactate, after entry into neurons, can be converted to pyruvate during which NADH is also produced, promoting synaptic plasticity-related gene expression by potentiating NMDA signaling in neurons (Yang et al., 2014; Margineanu et al., 2018). Furthermore, L-lactate acts as an energy substrate to fuel learning-induced de novo neuronal translation critical for long-term memory (Descalzi et al., 2019). On the other hand, mitochondria play crucial role in fueling local translation during synaptic plasticity (Rangaraju et al., 2019). Therefore, it could be hypothesized that the rescue of astrocytic Gi activation-mediated impairment of schema by exogenous L-lactate could have been mediated by facilitating synaptic plasticity-related gene expression by directly fueling the protein translation, potentiating NMDA signaling, as well as increasing mitochondrial capacity for ATP production by promoting mitochondrial biogenesis. Furthermore, the potential involvement of HCAR1, a receptor for L-lactate that may regulate neuronal activity (Bozzo et al., 2013; Tang et al., 2014; Herrera-López & Galván, 2018; Abrantes et al., 2019), cannot be excluded. Future research could explore these potential mechanisms, examining the interactions among them, and determining their relative contributions to schema.”
• The use of control animals transduced with an "empty" AAV9 vector (AAV8-GFAP-mCherry) compared with animals transduced with AAV8-GFAP-hM4Di-mCherry throughout the study would strengthen the results of this study, since transfection itself, as well as overexpression of the mCherry protein, may affect cell function.
We thank the reviewer for pointing this. The schema experiment includes a control group (Control-CNO group) of rats injected with AAV8-GFAP-mCherry bilaterally into the ACC. As shown in Author response image 3, after habituation and pretraining, these rats were trained for PA learning similarly to the other groups. Before 30 mins and after 30 mins of each PA training session, they received I.P. CNO. The PA learning, schema formation, memory retrieval, NPA learning and retrieval, and latency (time needed to commence digging at the correct well) were similar to the control group of rats. This result is consistent with our previous study where rats bilaterally injected with AAV8-GFAP-mCherry into CA1 of hippocampus did not show impairments in PA learning and schema formation upon CNO treatment (Liu et al., 2022).
Author response image 3.
A. PI (mean ± SD) during the acquisition of the original six PAs (OPAs) (S1-2, 4-8, 10-17) and new PAs (NPAs) (S19) of the control (n=6) and control-CNO (n=4) groups. B. Non-rewarded PTs (PT1, PT2, and PT3 done on S3, S9, and S18, respectively) to test memory retrieval of OPAs for the control-CNO group. C. Non-rewarded PT4 (S20) which was done after replacing two OPAs with two NPAs (NPA 7 & 8) in S19 for the control-CNO group. D. Latency (in seconds) before commencing digging at the correct well for control and control-CNO groups. Data shown as mean ± SD.
References
Abrantes, H. d. C., Briquet, M., Schmuziger, C., Restivo, L., Puyal, J., Rosenberg, N., Rocher, A.-B., Offermanns, S., & Chatton, J.-Y. (2019). The Lactate Receptor HCAR1 Modulates Neuronal Network Activity through the Activation of Gα and Gβγ Subunits. The Journal of Neuroscience, 39(23), 4422-4433. https://doi.org/10.1523/jneurosci.2092-18.2019
Akter, M., Ma, H., Hasan, M., Karim, A., Zhu, X., Zhang, L., & Li, Y. (2023). Exogenous L-lactate administration in rat hippocampus increases expression of key regulators of mitochondrial biogenesis and antioxidant defense [Original Research]. Frontiers in Molecular Neuroscience, 16. https://doi.org/10.3389/fnmol.2023.1117146
Bozzo, L., Puyal, J., & Chatton, J.-Y. (2013). Lactate Modulates the Activity of Primary Cortical Neurons through a Receptor-Mediated Pathway. PLoS One, 8(8), e71721. https://doi.org/10.1371/journal.pone.0071721
Choi, H. B., Gordon, G. R., Zhou, N., Tai, C., Rungta, R. L., Martinez, J., Milner, T. A., Ryu, J. K., McLarnon, J. G., Tresguerres, M., Levin, L. R., Buck, J., & MacVicar, B. A. (2012). Metabolic communication between astrocytes and neurons via bicarbonate-responsive soluble adenylyl cyclase. Neuron, 75(6), 1094-1104. https://doi.org/10.1016/j.neuron.2012.08.032
Covelo, A., Eraso-Pichot, A., Fernández-Moncada, I., Serrat, R., & Marsicano, G. (2021). CB1R-dependent regulation of astrocyte physiology and astrocyte-neuron interactions. Neuropharmacology, 195, 108678. https://doi.org/https://doi.org/10.1016/j.neuropharm.2021.108678
Descalzi, G., Gao, V., Steinman, M. Q., Suzuki, A., & Alberini, C. M. (2019). Lactate from astrocytes fuels learning-induced mRNA translation in excitatory and inhibitory neurons. Communications Biology, 2(1), 247. https://doi.org/10.1038/s42003-019-0495-2
Endo, F., Kasai, A., Soto, J. S., Yu, X., Qu, Z., Hashimoto, H., Gradinaru, V., Kawaguchi, R., & Khakh, B. S. (2022). Molecular basis of astrocyte diversity and morphology across the CNS in health and disease. Science, 378(6619), eadc9020. https://doi.org/10.1126/science.adc9020
Fünfschilling, U., Supplie, L. M., Mahad, D., Boretius, S., Saab, A. S., Edgar, J., Brinkmann, B. G., Kassmann, C. M., Tzvetanova, I. D., Möbius, W., Diaz, F., Meijer, D., Suter, U., Hamprecht, B., Sereda, M. W., Moraes, C. T., Frahm, J., Goebbels, S., & Nave, K.-A. (2012). Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature, 485(7399), 517-521. https://doi.org/10.1038/nature11007
Harris, R. A., Lone, A., Lim, H., Martinez, F., Frame, A. K., Scholl, T. J., & Cumming, R. C. (2019). Aerobic Glycolysis Is Required for Spatial Memory Acquisition But Not Memory Retrieval in Mice. eNeuro, 6(1). https://doi.org/10.1523/ENEURO.0389-18.2019
Hasan, M., Kanna, M. S., Jun, W., Ramkrishnan, A. S., Iqbal, Z., Lee, Y., & Li, Y. (2019). Schema-like learning and memory consolidation acting through myelination. FASEB J, 33(11), 11758-11775. https://doi.org/10.1096/fj.201900910R
Herrera-López, G., & Galván, E. J. (2018). Modulation of hippocampal excitability via the hydroxycarboxylic acid receptor 1. Hippocampus, 28(8), 557-567. https://doi.org/https://doi.org/10.1002/hipo.22958
Horvat, A., Muhič, M., Smolič, T., Begić, E., Zorec, R., Kreft, M., & Vardjan, N. (2021). Ca2+ as the prime trigger of aerobic glycolysis in astrocytes. Cell Calcium, 95, 102368. https://doi.org/https://doi.org/10.1016/j.ceca.2021.102368
Horvat, A., Zorec, R., & Vardjan, N. (2021). Lactate as an Astroglial Signal Augmenting Aerobic Glycolysis and Lipid Metabolism [Review]. Frontiers in Physiology, 12. https://doi.org/10.3389/fphys.2021.735532
Ichihara, Y., Doi, T., Ryu, Y., Nagao, M., Sawada, Y., & Ogata, T. (2017). Oligodendrocyte Progenitor Cells Directly Utilize Lactate for Promoting Cell Cycling and Differentiation. J Cell Physiol, 232(5), 986-995. https://doi.org/10.1002/jcp.25690
Iqbal, Z., Liu, S., Lei, Z., Ramkrishnan, A. S., Akter, M., & Li, Y. (2023). Astrocyte L-Lactate Signaling in the ACC Regulates Visceral Pain Aversive Memory in Rats. Cells, 12(1), 26. https://www.mdpi.com/2073-4409/12/1/26
Jourdain, P., Rothenfusser, K., Ben-Adiba, C., Allaman, I., Marquet, P., & Magistretti, P. J. (2018). Dual action of L-Lactate on the activity of NR2B-containing NMDA receptors: from potentiation to neuroprotection. Sci Rep, 8(1), 13472. https://doi.org/10.1038/s41598-018-31534-y
Kofuji, P., & Araque, A. (2021). G-Protein-Coupled Receptors in Astrocyte-Neuron Communication. Neuroscience, 456, 71-84. https://doi.org/10.1016/j.neuroscience.2020.03.025
Lee, Y., Morrison, B. M., Li, Y., Lengacher, S., Farah, M. H., Hoffman, P. N., Liu, Y., Tsingalia, A., Jin, L., Zhang, P. W., Pellerin, L., Magistretti, P. J., & Rothstein, J. D. (2012). Oligodendroglia metabolically support axons and contribute to neurodegeneration. Nature, 487(7408), 443-448. https://doi.org/10.1038/nature11314
Licht-Mayer, S., Campbell, G. R., Canizares, M., Mehta, A. R., Gane, A. B., McGill, K., Ghosh, A., Fullerton, A., Menezes, N., Dean, J., Dunham, J., Al-Azki, S., Pryce, G., Zandee, S., Zhao, C., Kipp, M., Smith, K. J., Baker, D., Altmann, D., Anderton, S. M., Kap, Y. S., Laman, J. D., Hart, B. A. t., Rodriguez, M., Watzlawick, R., Schwab, J. M., Carter, R., Morton, N., Zagnoni, M., Franklin, R. J. M., Mitchell, R., Fleetwood-Walker, S., Lyons, D. A., Chandran, S., Lassmann, H., Trapp, B. D., & Mahad, D. J. (2020). Enhanced axonal response of mitochondria to demyelination offers neuroprotection: implications for multiple sclerosis. Acta Neuropathologica, 140(2), 143-167. https://doi.org/10.1007/s00401-020-02179-x
Liu, S., Wong, H. Y., Xie, L., Iqbal, Z., Lei, Z., Fu, Z., Lam, Y. Y., Ramkrishnan, A. S., & Li, Y. (2022). Astrocytes in CA1 modulate schema establishment in the hippocampal-cortical neuron network. BMC Biol, 20(1), 250. https://doi.org/10.1186/s12915-022-01445-6
Magistretti, P. J., & Allaman, I. (2018). Lactate in the brain: from metabolic end-product to signalling molecule. Nat Rev Neurosci, 19(4), 235-249. https://doi.org/10.1038/nrn.2018.19
Margineanu, M. B., Mahmood, H., Fiumelli, H., & Magistretti, P. J. (2018). L-Lactate Regulates the Expression of Synaptic Plasticity and Neuroprotection Genes in Cortical Neurons: A Transcriptome Analysis. Front Mol Neurosci, 11, 375. https://doi.org/10.3389/fnmol.2018.00375
Netzahualcoyotzi, C., & Pellerin, L. (2020). Neuronal and astroglial monocarboxylate transporters play key but distinct roles in hippocampus-dependent learning and memory formation. Progress in Neurobiology, 194, 101888. https://doi.org/https://doi.org/10.1016/j.pneurobio.2020.101888
Newman, L. A., Korol, D. L., & Gold, P. E. (2011). Lactate produced by glycogenolysis in astrocytes regulates memory processing. PLoS One, 6(12), e28427. https://doi.org/10.1371/journal.pone.0028427
Park, J., Kim, J., & Mikami, T. (2021). Exercise-Induced Lactate Release Mediates Mitochondrial Biogenesis in the Hippocampus of Mice via Monocarboxylate Transporters. Front Physiol, 12, 736905. https://doi.org/10.3389/fphys.2021.736905
Peterson, S. M., Pack, T. F., & Caron, M. G. (2015). Receptor, Ligand and Transducer Contributions to Dopamine D2 Receptor Functional Selectivity. PLoS One, 10(10), e0141637. https://doi.org/10.1371/journal.pone.0141637
Rangaraju, V., Lauterbach, M., & Schuman, E. M. (2019). Spatially Stable Mitochondrial Compartments Fuel Local Translation during Plasticity. Cell, 176(1), 73-84.e15. https://doi.org/10.1016/j.cell.2018.12.013
Rinholm, J. E., Hamilton, N. B., Kessaris, N., Richardson, W. D., Bergersen, L. H., & Attwell, D. (2011). Regulation of oligodendrocyte development and myelination by glucose and lactate. J Neurosci, 31(2), 538-548. https://doi.org/10.1523/JNEUROSCI.3516-10.2011
Sánchez-Abarca, L. I., Tabernero, A., & Medina, J. M. (2001). Oligodendrocytes use lactate as a source of energy and as a precursor of lipids. Glia, 36(3), 321-329. https://doi.org/10.1002/glia.1119
Suzuki, A., Stern, S. A., Bozdagi, O., Huntley, G. W., Walker, R. H., Magistretti, P. J., & Alberini, C. M. (2011). Astrocyte-neuron lactate transport is required for long-term memory formation. Cell, 144(5), 810-823.
Tang, F., Lane, S., Korsak, A., Paton, J. F. R., Gourine, A. V., Kasparov, S., & Teschemacher, A. G. (2014). Lactate-mediated glia-neuronal signalling in the mammalian brain. Nature Communications, 5(1), 3284. https://doi.org/10.1038/ncomms4284
Tauffenberger, A., Fiumelli, H., Almustafa, S., & Magistretti, P. J. (2019). Lactate and pyruvate promote oxidative stress resistance through hormetic ROS signaling. Cell Death Dis, 10(9), 653. https://doi.org/10.1038/s41419-019-1877-6
Tse, D., Langston, R. F., Kakeyama, M., Bethus, I., Spooner, P. A., Wood, E. R., Witter, M. P., & Morris, R. G. (2007). Schemas and memory consolidation. Science, 316(5821), 76-82. https://doi.org/10.1126/science.1135935
Tse, D., Takeuchi, T., Kakeyama, M., Kajii, Y., Okuno, H., Tohyama, C., Bito, H., & Morris, R. G. (2011). Schema-dependent gene activation and memory encoding in neocortex. Science, 333(6044), 891-895. https://doi.org/10.1126/science.1205274
Vardjan, N., Chowdhury, H. H., Horvat, A., Velebit, J., Malnar, M., Muhič, M., Kreft, M., Krivec, Š. G., Bobnar, S. T., Miš, K., Pirkmajer, S., Offermanns, S., Henriksen, G., Storm-Mathisen, J., Bergersen, L. H., & Zorec, R. (2018). Enhancement of Astroglial Aerobic Glycolysis by Extracellular Lactate-Mediated Increase in cAMP [Original Research]. Frontiers in Molecular Neuroscience, 11. https://doi.org/10.3389/fnmol.2018.00148
Vezzoli, E., Cali, C., De Roo, M., Ponzoni, L., Sogne, E., Gagnon, N., Francolini, M., Braida, D., Sala, M., Muller, D., Falqui, A., & Magistretti, P. J. (2020). Ultrastructural Evidence for a Role of Astrocytes and Glycogen-Derived Lactate in Learning-Dependent Synaptic Stabilization. Cereb Cortex, 30(4), 2114-2127. https://doi.org/10.1093/cercor/bhz226
Wang, J., Tu, J., Cao, B., Mu, L., Yang, X., Cong, M., Ramkrishnan, A. S., Chan, R. H. M., Wang, L., & Li, Y. (2017). Astrocytic l-Lactate Signaling Facilitates Amygdala-Anterior Cingulate Cortex Synchrony and Decision Making in Rats. Cell Rep, 21(9), 2407-2418. https://doi.org/10.1016/j.celrep.2017.11.012
Yang, J., Ruchti, E., Petit, J. M., Jourdain, P., Grenningloh, G., Allaman, I., & Magistretti, P. J. (2014). Lactate promotes plasticity gene expression by potentiating NMDA signaling in neurons. Proc Natl Acad Sci U S A, 111(33), 12228-12233. https://doi.org/10.1073/pnas.1322912111
Yao, S., Xu, M.-D., Wang, Y., Zhao, S.-T., Wang, J., Chen, G.-F., Chen, W.-B., Liu, J., Huang, G.-B., Sun, W.-J., Zhang, Y.-Y., Hou, H.-L., Li, L., & Sun, X.-D. (2023). Astrocytic lactate dehydrogenase A regulates neuronal excitability and depressive-like behaviors through lactate homeostasis in mice. Nature Communications, 14(1), 729. https://doi.org/10.1038/s41467-023-36209-5
Yu, X., Zhang, R., Wei, C., Gao, Y., Yu, Y., Wang, L., Jiang, J., Zhang, X., Li, J., & Chen, X. (2021). MCT2 overexpression promotes recovery of cognitive function by increasing mitochondrial biogenesis in a rat model of stroke. Anim Cells Syst (Seoul), 25(2), 93-101. https://doi.org/10.1080/19768354.2021.1915379
Zhou, Z., Okamoto, K., Onodera, J., Hiragi, T., Andoh, M., Ikawa, M., Tanaka, K. F., Ikegaya, Y., & Koyama, R. (2021). Astrocytic cAMP modulates memory via synaptic plasticity. Proc Natl Acad Sci U S A, 118(3), e2016584118. https://doi.org/10.1073/pnas.2016584118
Zhu, J., Hu, Z., Han, X., Wang, D., Jiang, Q., Ding, J., Xiao, M., Wang, C., Lu, M., & Hu, G. (2018). Dopamine D2 receptor restricts astrocytic NLRP3 inflammasome activation via enhancing the interaction of β-arrestin2 and NLRP3. Cell Death Differ, 25(11), 2037-2049. https://doi.org/10.1038/s41418-018-0127-2
Author Response
Reviewer #1 (Public Review):
We thank the reviewer for carefully reading of the manuscript and for the insightful criticisms and comments. In the following we address them point by point.
The community assembly process is modelled in a very specific way, and the manuscript would benefit from an expanded ecological motivation of the processes that are being mimicked, and thereby explain more clearly what taxonomic level of organization is being considered.
We follow the more recent trait-based approach that shifts the focus from species (and the many traits by which they differ from one another) to groups of species that share the same values of selected functional traits. Since the general context is ecosystem response to drier climates, we choose the functional traits to include a response trait associated with stress tolerance and an effect trait associated with biomass production. We further assume a tradeoff between the two traits which is well supported by earlier studies (see e.g. Angert et al. 2009, https://doi.org/10.1073/pnas.0904512106). So, indeed, the choice we make in characterizing the community is quite specific, but it is highly relevant to the ecological context considered of dryland plant communities where plants compete primarily for water and light. The taxonomic level we consider is species except that we group them in a manner that is more transparent to questions of ecosystem function, ignoring differences between species that are not significant to these questions.
We expanded considerably the text in the section “Modeling spatial assembly of dryland plant communities” to clarify the ecological motivation of the processes we model.
In addition, it would be useful if the authors could provide further clarification as to what extent the community diversity dynamics can be separated from total biomass dynamics of patterned water-limited ecosystems given the current approach. These points are explained in further detail below.
The model describes the dynamics of all functional groups, which provides the biomass distribution 𝐵 = 𝐵(𝜒) in trait space (in the case of patterned states we first integrate over space). That distribution contains information about various community-level properties, including functional diversity (richness, evenness) as figure 3 in the revised manuscript illustrates, and total biomass, which is the area below the distribution curve. The two types of dynamics are tightly connected and cannot be separated, but in principle the approach can be used to study the relationships between diversity and total biomass by calculating biomass distributions along the rainfall gradient and extracting the two properties from the distributions.
We added in the section “Modeling spatial assembly of dryland plant communities” the information that the biomass distribution also contains information about the total biomass.
First, it was not entirely clear to this reviewer how the reaction parts of the model equations determine the optimal trait value χ, and how this value varies as a function of precipitation.
The ‘optimal’ trait value 𝜒𝑚𝑎𝑥 is determined by the interspecific interactions that the model captures, which divide into ‘direct’ and ‘indirect’ interactions. The direct interactions are captured by the dependence of the growth rate Λ𝑖 of the ith functional group (see Eq. (1a)) on the aboveground biomass values of all functional groups, Λ𝑖 = Λ𝑖(𝐵1,𝐵2,… , 𝐵𝑁) (see Eq. (2)). This dependence represents competition for light (taller plants are better competitors) and includes the effect of self-shading. The indirect interactions are through the water uptake term in the soil-water equation (1b) (2nd term from right) and the water dependence of the biomass growth term in Eq. (1a). These terms represent competition for water. For a given precipitation value 𝑃 the net effect of these interspecific interactions result in a particular functional group 𝜒𝑚𝑎𝑥 which is most abundant. For spatially uniform vegetation, as 𝑃 is increased 𝜒𝑚𝑎𝑥 moves to lower values. The precipitation increases surface water (Eq. (1c)) and consequently the amount of water 𝐼𝐻 infiltrating into the soil. The increased soil water gives competitive advantage to species investing in growth, mainly because they better compete for light as they grow taller, and therefore 𝜒𝑚𝑎𝑥 decreases.
… it is then not immediately clear why the most successful trait class is not outcompeting the other classes.
With the current model and parameters set the most successful trait does eventually outcompete all other traits, when trait diffusion is set to zero, 𝐷𝜒 = 0. This is, however, a very long process because the most successful trait suffers from self-shading at late growth stages, which slows down its growth and allows nearby traits to survive for a long time. Choosing a finite but very small 𝐷𝜒 values that represent mutations occurring on evolutionarily long times counteracts the exclusion process and results in a stationary asymptotic community, as Fig. 3 in the revised manuscript shows (this behavior is reminiscent of optical solitons, where self-focusing instability is balanced by dispersion). We note that modeling stronger growth-inhibiting factors, such as pathogens, by including a factor of the form (1 − 𝐵𝑖/𝐾) to the growth rate, results in an asymptotic stationary community also for 𝐷𝜒 = 0 (see also earlier studies Nathan et al. 2016, Yizhaq et al. 2020).
We revised original Fig. 4 (now Fig. 3) by adding a new part (Fig. 3a) that shows the exclusion process for 𝐷𝜒 = 0, and the effect of the counter-acting process of trait diffusion, which results in an asymptotic distribution of finite width (Fig. 3b) from which community level properties such as functional diversity can be derived. We also extended the text in section “Modeling spatial assembly of dryland plant communities” (last paragraph) to clarify the two counter-acting processes of exclusion because of interspecific competition for water and light, and trait diffusion driven by mutations, which together culminate in an asymptotic biomass distribution along the 𝜒 axis of finite width.
The authors model trait adaptation through a diffusion approximation between trait classes. That is, every timestep, a small amount of biomass flows from the class with higher biomass to the neighboring trait class with lower biomass. From an ecological point of view, it seems that this process is describing adaptation of vegetation that is already present, so this process seems to be limited to intraspecific phenotypic plasticity. From the text, however, it seems that the trait classes correspond to higher taxonomic levels of organization, when describing shifts from fast growing to stress-tolerant species, for example. It is not entirely clear, however, how biomass flows as assumed in the model could occur at these higher levels of organization.
We do not study in this work adaptation through diffusion in trait space. That kind of adaptive dynamics can indeed be studied with the current model, but with different initial conditions, namely, initial conditions corresponding to a single resident trait where the biomass of all other traits is zero. The resulting dynamics of mutations and succession are then very slow, occurring on evolutionarily long time scales set by the small value of 𝐷𝜒 (e.g. 10−6). In this study the initial conditions represent the presence of all traits, even if at very low biomass values that may represent a pool of seeds that germinate once environmental conditions allow. For a given precipitation value 𝑃, the functional traits we consider determine which functional groups (of species) overcome environmental filtering and grow, and which of the growing traits survive the competition for water and light. These are relatively fast processes, occurring on ecological time scales, which determine the emerging community. At longer times this community is further shaped by slow processes of interspecific competition among species of similar traits and by trait diffusion (mutations). A final remark about phenotypic changes: although in general 𝜒 can be interpreted as representing different phenotypes, the choice of very small values for 𝐷𝜒 cannot represent relatively fast phenotypic changes and restricts the context to mutations at the taxonomic level of species.
We added an explanation in the 3rd paragraph of the section “Modeling spatial assembly of dryland plant communities” of the need to consider mutations and the role they play in our study.
Combining the observations from the previous two points, there is a concern that for a given level of precipitation, there is a single trait class with optimal biomass/lowest soil water level that is dominant, with the neighboring trait classes being sustained by the diffusion of biomass from the optimal class to neighboring inferior classes. This would seem a bit problematic, as it would mean that most classes are not a true fit for the environment, and only persist due to the continuous inflow of biomass. Taking a clue from the previous papers of the authors, it seems this may not be the case, though. Specifically, in the paper by Nathan et al. (2016) it seems that all trait classes are started at low initial biomass density, and the resulting steady state (in the absence of biomass flows between classes) seems to show similar biomass profiles as shown in Figs. 4,5 and 7 of the current paper. While the current model formulation seems slightly different, similar results may apply here. Indeed, keeping all trait classes at non-zero (but low) density, and when the (abiotic and biotic) environment permits, let each class increase in biomass seems like the most straightforward approach to model community assembly dynamics. Given the above discussion about these trait classes competing for a single resource (soil water), and one trait class being able to drive this resource availability to the lowest level, it would then be useful to readers to explain why multiple trait classes can coexist here, and how(for spatial uniform solutions) the equilibrium soil water level with multiple trait classes present compares to the equilibrium soil water level when only the optimal trait class is present. Furthermore, if results as presented in Nathan et al. (2016) indeed hold in the current case, perhaps it means that the biomass profile responses as shown in e.g. Fig. 5 would also occur if there was no biomass flow between trait classes included, but that the time needed to adjust the profile would take much longer as compared to when the drift term/second trait derivative is included. In summary, further clarification of what the biomass flows between classes represent, and the role it plays in driving the presented results would be useful for readers.
As explained in the reply to previous comments the asymptotic community is tuned by a balance between two slow counter-acting processes, interspecific competition among similar traits and mutations over evolutionarily long time scales. However, the community structure is largely determined by much faster processes of environmental filtering and interspecific competition among widely distinct traits, as all traits are initially present. Indeed, comparing the biomass distributions in new Fig. 3, with and without trait diffusion indicates that the community composition, as measured by 𝜒𝑚𝑎𝑥, is the same. Trait diffusion, however, does affect functional diversity, along with environmental factors. In that sense the emerging community is a true fit for the environment.
We thank the reviewer for these thoughtful comments, which helped us realize that our presentation of these issues was too concise and unclear. We believe that the new extended section on modeling spatial assembly of dryland plant communities, and the new figure 3a clarify these issues.
In addition, it would be useful for readers to understand to what extent the shifts in average trait values and functional diversity can be decoupled from the biomass and soil water responses to changes in precipitation that would occur in a model with only a single biomass variable. For example, early studies on self-organization in semi-arid ecosystems already showed that the shift toward a patterned state involved the formation of patches with higher biomass, and higher soil water availability, as compared to the preceding spatially uniform state, and that the biomass in these patches remains relatively stable under decreasing rainfall, while their geometry changes (e.g. Rietkerket al. 2002). It has also been observed that for a given environmental condition, biomass in vegetation patches tends to increase with pattern wavelength (e.g. Bastiaansen and Doelman 2018; Bastiaansen et al. 2018). Given the model formulation, one wonders whether higher biomass in the single variable model is not automatically corresponding to higher abundance of faster growing species and a higher functional diversity (as the diffusion of biomass can cover a broader range when starting from higher mass in the optimal trait class). There are some indications in the current work that the linkage is more complicated, for example, the biomass peak in Fig. 7c is lower, but also broader as compared to the distribution of Fig. 7b, but it is currently not entirely clear how this result can be explained (for example, it might be the case that in the spatially patterned states, the biomass profiles also vary in space).
We are not sure we understand what the reviewer means by “decoupled”, but much insight indeed can be gained from a study of a model for a single functional group (trait) and observing the behaviors described by the reviewer. In fact, these behaviors, which some of us are familiar with from numerical studies, motivated parts of the current study. Higher biomass in vegetation patches (compared to uniform vegetation) in the single trait model does not automatically imply a shift to faster growing species; in principle the stress-tolerant species that already reside in the system when uniform vegetation destabilizes to a periodic pattern can simply grow denser. To answer this and additional questions we need to take into account interspecific interactions by studying the full community model. As to Fig. 7b,c, the behavior appears to be opposite to that described by the reviewer: the biomass pick in Fig. 7c is higher and narrower than that in Fig. 7b, not lower and broader. This is because of the much larger domain of the patterned state as compared with that of the uniform state, which increases the abundance of low-𝜒 species, i.e. species investing in growth.
The increase of biomass in vegetation patches with pattern wavelength for given environmental conditions, as observed by Bastiaansen et al. 2018, is actually another mechanism for increasing functional diversity. This is because the water stress at the patch center is higher than that in the outer patch areas and thus forms favorable conditions for stress tolerant species while the outer areas form favorable conditions for fast growing species.
We added a new paragraph in the Discussion and Conclusion section (last paragraph in the subsection Insight III) where we discuss the effect of coexisting periodic patterns of different wavelengths on functional diversity and ecosystem management. We also added citations to the references the reviewer mentioned.
The possibility of hybrid states, where part of the landscape is in a spatially uniform state, while the other part of the landscape is in a patterned state, is quite interesting. To better understand how such states could be leveraged in management strategies, it would be useful if a bit more information could be provided on how these hybrid states emerge, and whether one can anticipate whether a perturbation will grow until a fully patterned state, or whether the expansion will halt at some point, yielding the hybrid state. It seems that being able to distinguish this case would be necessary in the design of planning and management strategies
The hybrid states appear in the bistability range of the uniform and patterned vegetation states, and typically occupy most of this range. Their appearance is related to the behavior of ‘front pinning’ in bistability ranges of uniform and patterned states in general. Front pinning refers to fronts that separate a uniform domain and a periodic-pattern domain, which remain stationary in a range of a control parameter (precipitation in our case). This is unlike fronts that separate two uniform states, which always propagate in one direction or another and can be stationary only at a single parameter value – the Maxwell point. Thus, an indication that a given landscape may have the whole multitude of hybrid states is the presence of a front (ecotones) that separates uniform and patterned vegetation. If that front appears stationary over long period of times (on average), this is a strong indication.
We added a new paragraph in the subsection Insight III of the Discussion and conclusion section to clarify this point.
Also, in Fig. 3a, the region of parameter space in which hybrid states occur is not very large; it is not entirely clear whether the full range of hybrid states is left out here for visual considerations, or whether these states only occur within this narrow range in the vicinity of the Turing instability point.
As pointed out in the reply to the previous comment the hybrid states are limited to the bistability range of uniform and patterned vegetation, which is not wide. However, this should not necessarily restrictma nagement of ecosystem services by nonuniform biomass removal, as such management will have similar effects on community structure also outside the bistability range where front propagate slowly.
The new paragraph we added also addresses this point.
Reviewer #2 (Public Review):
We thank the reviewer for carefully reading the manuscript and for the constructive criticisms and comments. In the following we address them point by point.
1) Model presentation.
It would be better to explain the model in ecological terms first, clarifying parameter biological meaning and justifying their choice. In doing so, creating a specific 'Methods' section, which now is lacking, would be of help too. Authors should clarify whether and how the model follows the conservation of mass principle involving precipitation and evapotranspiration. Are root growth and seed dispersal included for this purpose? Why they are not referred to any further in the analysis and discussion? Why a specific term for plant transpiration is not included, or is to somehow phenomenologically incorporated into the growth-tolerance tradeoff? In doing so, authors should also pay attention to water balance as above (H) and below (W) ground water are not independent from each other.
We added a Methods section, which in eLife is placed at the end of the manuscript. The section includes the model equations and more detailed explanations in ecological terms of various parts of the model. We also added Table 1 with a list of all model parameters, their descriptions, units and numerical values used in the simulations. Presenting the model at the end of the manuscript suits more technical information about the model, but not essential information that is needed for understanding the results. We therefore kept the subsection “A model for spatial assembly of dryland plant communities” in the Results section, where we present that information.
There is no conservation of mass in the model (and all other models of this kind) simply because the system that we consider is open. In particular, it does not include the atmosphere, which constitute part of the system’s environment. Including the atmosphere as additional state variables in the model, capturing the feedback of evapotranspiration on the atmosphere, would make the model too complicated for the kind of analysis we perform. So, although the model contains parts that represent mass conservation such as the terms describing below- and above-ground water transport, water mass is not conserved. The biomass variables represent aboveground biomass of living plants or plant parts and are not conserved either as biomass production involve biochemical reactions that convert inorganic substances coming from the system’s environment (atmosphere and the soil) into organic ones, while plant mortality involves organic matter that leaves the system.
Roots in the model platform we consider are modeled indirectly through their relation to aboveground biomass. That relation constitutes one of the scale-dependent feedbacks that produce a Turing instability to vegetation patterns, the so-called root-augmentation feedback (see Meron 2019, Physics Today), but in this particular study we eliminate this feedback for simplicity. The scale-dependent feedback that we do consider is the so-called infiltration feedback, associated with biomass-dependent infiltration rate that produces overland water flow towards vegetation patches, as explained in the subsection “A model for spatial assembly of dryland plant communities”. It will be interesting indeed to extend the study in the future to include also the root-augmentation feedback.
We assume short-range seed dispersal and take it into account through biomass “diffusion” terms (obtained as approximations of dispersal kernels assuming narrow kernels). These terms play important roles in the scale-dependent feedback that induces the Turing instability, as is explained in earlier papers which we cite. Plant transpiration is modeled through the water uptake term in the equation for the soilwater 𝑊. Indeed above-ground water 𝐻 and below-ground water 𝑊 are not independent; the infiltration term IH in the equations for both state variables account for this dependence in a unidirectional manner (loss of 𝐻 and gain of 𝑊). As we do not include the atmosphere in the model the other direction, namely, evapotranspiration that increases air humidity and affects rainfall, is not accounted for. The neglect of this effect can be justified for sparse dryland vegetation.
These good points have already been discussed in many earlier papers as well as in the book Nonlinear Physics of Ecosystems (Meron 2015), and we cannot address them all in this paper. We did however add several clarifications in the section Modeling spatial assembly of dryland plant communities and in the new Methods section, including the consideration of the atmosphere as the system’s environment quantified by the precipitation parameter 𝑃.
Another unclear point is that growth rates for the same plant functional groups are assumed to be constant among different species within the same group and are confounded by biomass production. Why is that the case? Furthermore, how many different species are characterizing each functional group? How are interspecific interactions accounted for (more specifically, see comment below)?
In the trait-based approach we focus on just two functional traits, related to growth rate and tolerance to water stress, ignoring differences in other traits that distinguish species. That is, a given functional group consists of species that share the same values of the two selected functional traits (to a given precision determined by 𝑁), taking all other traits represented in the model to be equal. In this approach we do not care about how many species belong to each functional group, only their total biomass. We wish to add that simplifying assumptions of this kind are necessary if we want the model to be mathematically tractable and capable of providing deep insights by mathematical analysis.
We expanded the discussion of the trait-based approach in the section Modeling spatial assembly of dryland plant communities and added relevant references (second paragraph).
Finally, stress tolerance is purely phenomenological. There is no actual mechanism/parameter describing it. Rather, it "simply" appears as low/high mortality, which in turn is said to be due to high/low tolerance. This leads to a sort of circularity between mortality and tolerance. Yet, mortality can occur due to other biophysical factors (e.g. disturbance, fire, herbivory, pathogens). A drawback of this assumption is that a mechanism of drought tolerance is often to invest in belowground organs, including roots. However, according to the proposed model, it turns out that fast growing species with low investment in tolerance also have high investment in roots; vice versa, tolerant species have low investment in roots. This is a bit counterintuitive and not well biologically supported.
First, we agree with the reviewer that our approach is purely phenomenological, as we model tolerance to water stress by a single parameter that lumps together the effects of various physiological mechanisms. That parameter can be distinguished from other factors affecting mortality by regarding the constant 𝑀𝑚𝑎𝑥 in Eq. (3) as representing several contributions. Since we do not study the effects of these other factors we can absorb them in 𝑀𝑚𝑎𝑥 for mathematical simplicity. Tolerance to water stress is not necessarily associated with roots. Plants can better tolerate water stress by reducing transpiration through stomatal closure, regulating leaf water potential, or develop hydraulically independent multiple stems that lead to a redundancy of independent conduits and higher resistance to drought (see Schenk et al. 2008 - https://doi.org/10.1073/pnas.0804294105).
We added a discussion in the Methods section (5th paragraph, “Tolerance to water stess …”) of the simple form by which we model tolerance to water stress through the mortality parameter.
2) Parameter choice.
N = 128 is an extremely high number for plant functional groups. It is even quite unrealistic to have 128 species per square meter, so this value is not very reasonable. Please run the model and report results with more realistic N (e.g from 4-64) as well as with different sets of N values keeping all other parameters constant.
We wish to clarify two points: 1) N=128 does not imply 128 functional groups per square meter; the emerging community has much lower functional richness (FR) as the average FR is around 0.25, meaning only 128 × 0.25 = 32 functional groups. 2) The model results, as reflected by the key metrics 𝜒𝑚𝑎𝑥, 𝐹𝑅, and 𝐹𝐸, are independent of the particular value of N (for N values sufficiently large), as Figures IA and IB below show. The biomass 𝐵𝑖 of each functional group, however, does change (Figure IA) because by changing N we change the range of traits Δ𝜒 = 1/𝑁 that belong to a given functional group. But if we look at the biomass density in trait space 𝑏𝑖, related to 𝐵𝑖 through the relation 𝐵𝑖 = 𝑏𝑖Δ𝜒, then also the biomass density is independent of 𝑁 as Figure IB shows. So, even if in practice there are less functional groups and thus species as considered in the model studies, the results are not affected by that. On the other hand, choosing higher 𝑁 values provides smoother curves and nicer presentation of our results.
Figure IA
Figure IB
We added a discussion of this issue in the Methods section after Eq. (2).
Gamma (rate of water uptake by plants' roots): why is it in that unit of m^2/kg * y? Why are you now considering the area (and not the volume) per biomass unit?
The vegetation pattern formation model we study, like most other models of this kind, does not explicitly capture the soil depth dimension. Accordingly, W is interpreted as the soil-water content in the soil volume below a unit ground area within the reach of the plant roots. In practice W has units kg/m2, like B, and since Γ𝑊𝐵 should have the same units as 𝜕𝑊/𝜕𝑡 (see Eq. 1b), Γ must have the units of (𝐵𝑡)−1.
A is not defined in the text.
We now define it in Table 1 (see Methods section).
M min: why 0.5 mortality? Having M max set to 0.9, please consider a lower mortality value set to 0.1, and please report evidence(hopefully) demonstrating the robustness of results to such change.
The results are robust to the particular values of 𝑀𝑚𝑖𝑛 and 𝑀𝑚𝑎𝑥, except that there are combinations of these two parameters for which the biomass distributions are pushed towards the edge of the 𝜒 domain, which make the presentation of the results less clear. Figure II shows results of recalculations of the distribution 𝐵 = 𝐵(𝜒) for 𝑀𝑚𝑖𝑛 = 0.1, as requested (using 𝑀𝑚𝑎𝑥 = 0.15) for 3 different precipitation values. As the reviewer can see there’s no qualitative change in the results: lower precipitation push a uniform community to stress tolerant species (higher 𝜒), while the formation of patterns at yet lower precipitation push the community back to fast growing species (low 𝜒).
Figure II
K_min and K_max are in two different units, and should both be kg/m^2.
Thanks, we fixed this typo in Table 1.
Values of precipitation (P, mean annual precipitation) are not reported.
The precipitation parameter is variable, as is now stated in Table 1, and therefore was not include it in the list of parameters’ values used. Whenever a particular precipitation value has been used our intention was to state it in the caption of the corresponding figure. This was done in Figs. 5,6,7, but indeed not in Fig. 4 (Fig. 3 in revised ms.). The insets on the right side of Fig. 3 (Fig. 4 in revised ms.) where also calculated for particular precipitation values, but that information is not essential as the intention is to show typical forms of the various solution branches, which do not qualitatively change along the branches (i.e. at different P values).
We added the precipitation value (P=180mm/y) at which all the biomass distributions shown in new Fig. 3 (Fig. 4 in original ms) were calculated.
3) Results presentation and interpretation.
Parameter range of precipitation in figure 3 is odd. Why in one case precipitation ranges from 0 to 160 while in another it is only 60-120? Furthermore, in paragraph 198-213 and associated results in fig. 5. the Choice of precipitation values is somehow discordant from the previous model. Please provide motivation for this choice, clarify and uniformize it.
In Fig. 3b (Fig. 4b in revised ms) we restricted the precipitation range to 60-120 as the curves, which are limited to 0 < 𝜒 < 1 (by the definition of 𝜒), do not extend to 𝑃 < 60 and to 𝑃 > 120. Extending the range to 0 < 𝑃 < 160 would make the figure less compact and nice as it will contain blank parts with no information.
We are not sure we understand what the reviewer means by “is somehow discordant from the previous model”. The motivation of the choices we made for the precipitation values P=150, 100 and 80 was to show the shift of a spatially uniform community to a higher 𝜒 value as the precipitation is decreased to a lower value (from 150 to 100), and the shift back to a lower 𝜒 value at yet lower precipitation (80) past the Turing instability.
Finally, authors seem to create confusion around community composition, which is defined as the (taxonomic) identity of all different species inhabiting a community. Notably, it is remarkably different from the x_max parameter used in the model, which as a matter of fact is just the value of the most productive (notably, not necessarily the most abundant) functional group.
We thank the reviewer for this comment. Since all the emerging communities in the model studies are pretty localized around the value of 𝜒𝑚𝑎𝑥, that value does contain information about the identity of other functional groups in the community when complemented by FR (functional richness) and FE (functional evenness). More significantly to our study, shifts in 𝜒𝑚𝑎𝑥 represent the shifts in community composition we focus on in this study, i.e. shifts towards fast growing species or towards stress-tolerant species.
We modified the description of the community-level properties that can be derived from the biomass distribution in trait space (see modified text towards the end of the section “Modeling spatial assembly …” and also the caption of Fig. 3b), explaining that both functional diversity and community composition can be described by several metrics, and clarifying the significance of 𝜒𝑚𝑎𝑥 in describing community-composition shifts.