English — traits (thorough explanation)
1) What traits are (core idea)
Traits are characteristics or features of an organism.
They describe how an organism looks, behaves, or functions.
Trait = a characteristic of a living thing
Traits can be:
- Inherited (passed down from parents)
- Influenced by the environment
- Or a combination of both
2) Types of traits
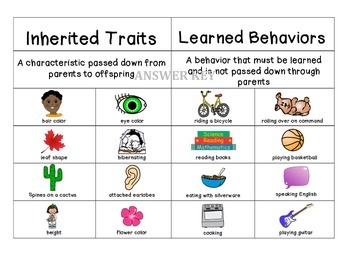
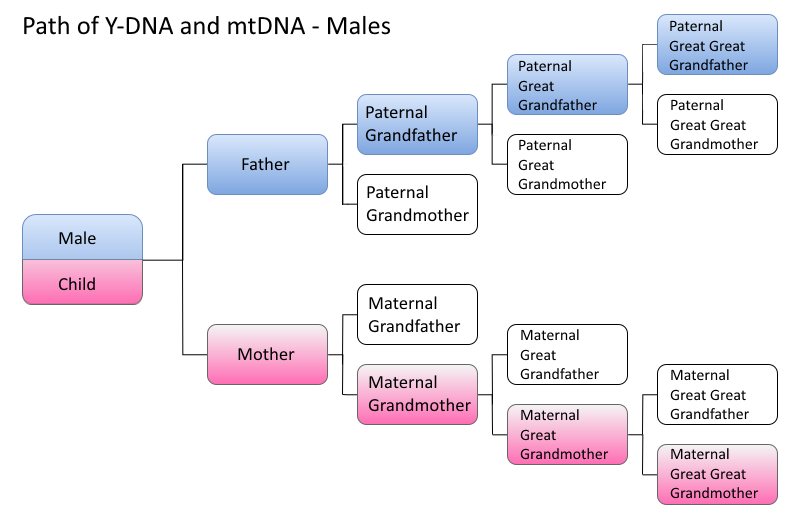
A) Inherited (genetic) traits
These traits are controlled by genes and passed from parents to offspring.
Examples:
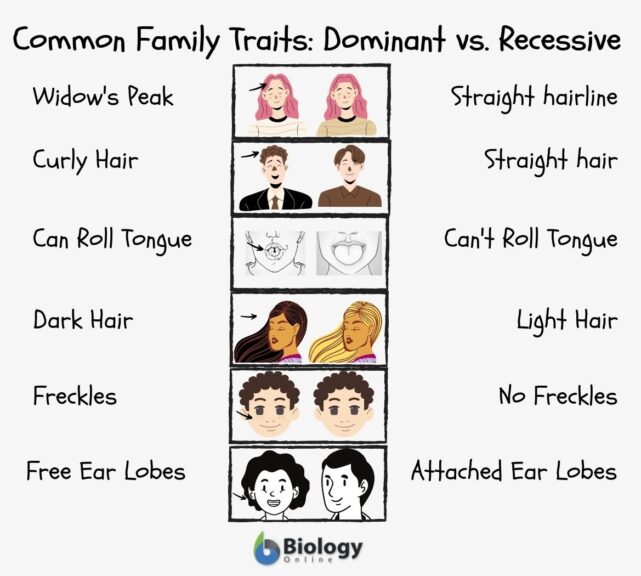
- Eye color
- Hair color and texture
- Blood type
- Natural height potential
📌 These traits are present from birth.
B) Acquired (environmental) traits
These traits develop due to life experiences or environment.
Examples:
- Suntan
- Muscle strength from exercise
- Scars
- Language spoken
📌 These traits are not inherited genetically.
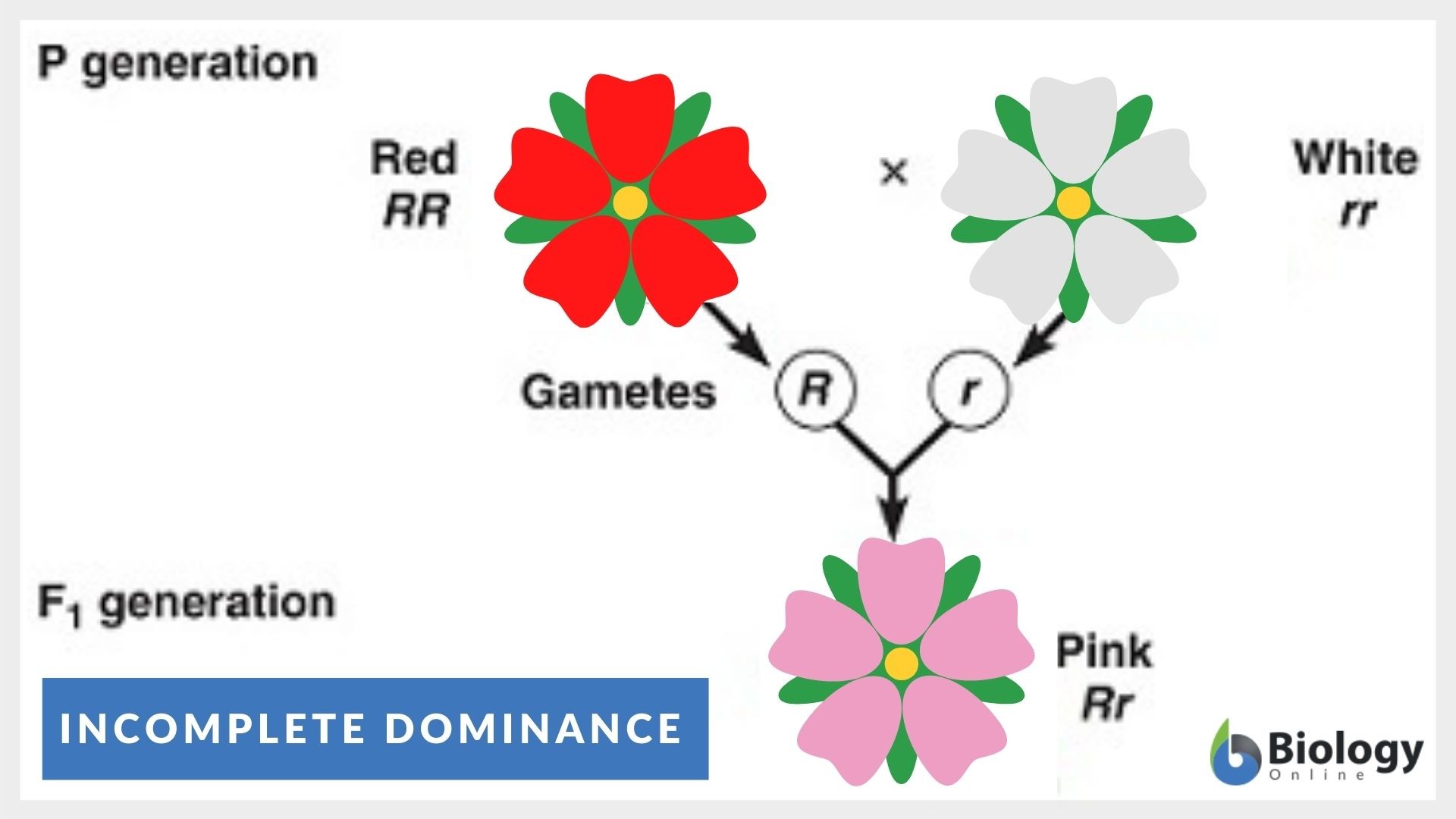
C) Traits influenced by both genes and environment
Many traits result from both heredity and environment.
Examples:
- Height (genes + nutrition)
- Intelligence (genes + education)
- Athletic ability (genes + training)
3) Traits in genetics (Science 10 focus)
In genetics, traits are:
- Controlled by genes
- Each gene may have different alleles
- Alleles can be dominant or recessive
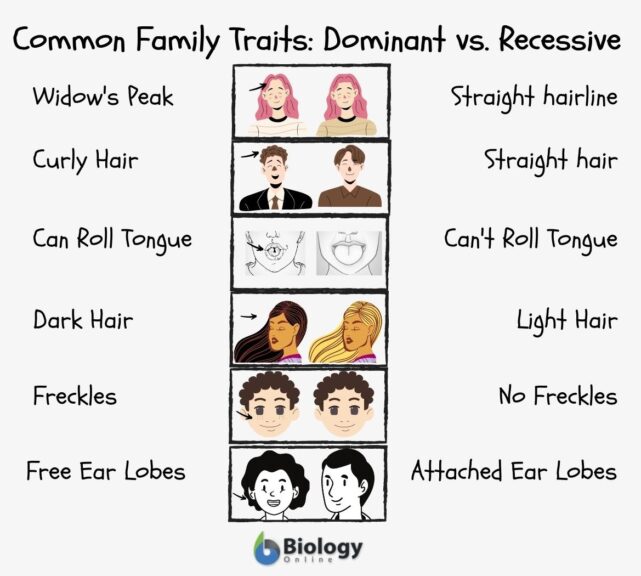
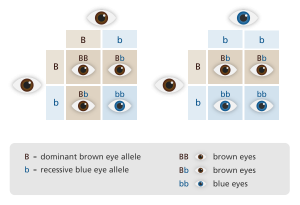
📌 Example:
- Brown eyes (dominant)
- Blue eyes (recessive)
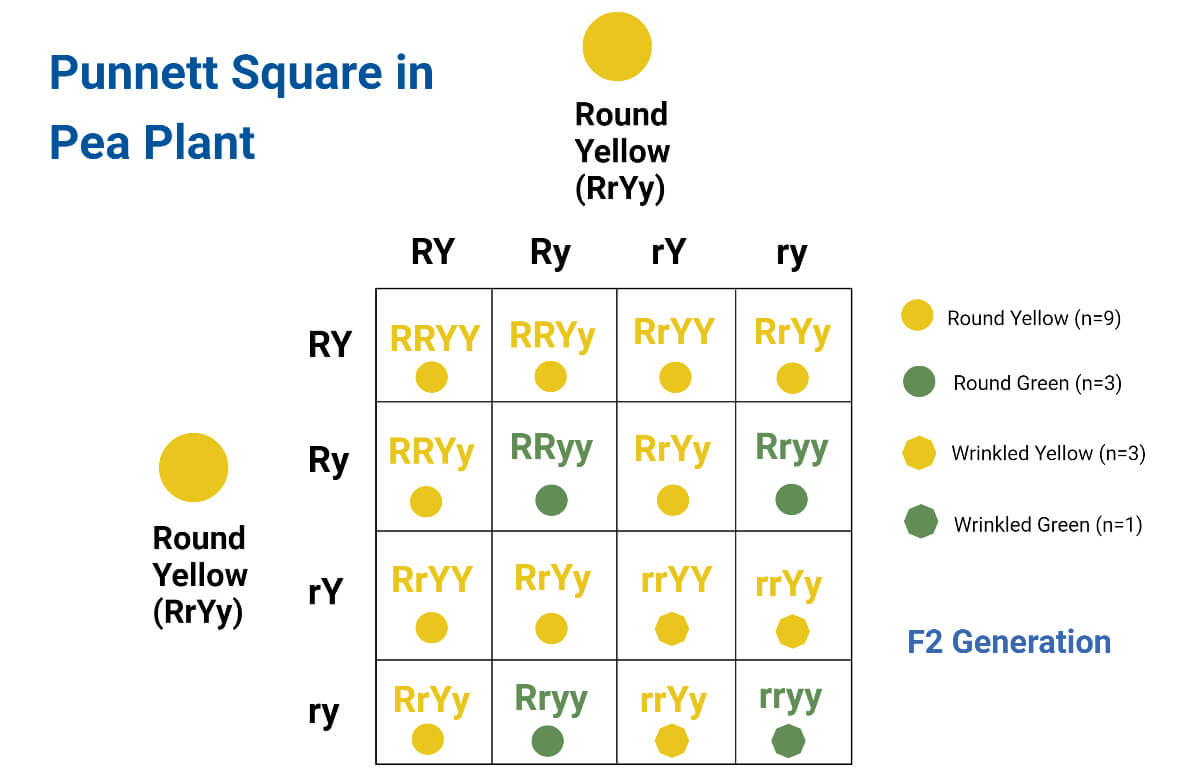
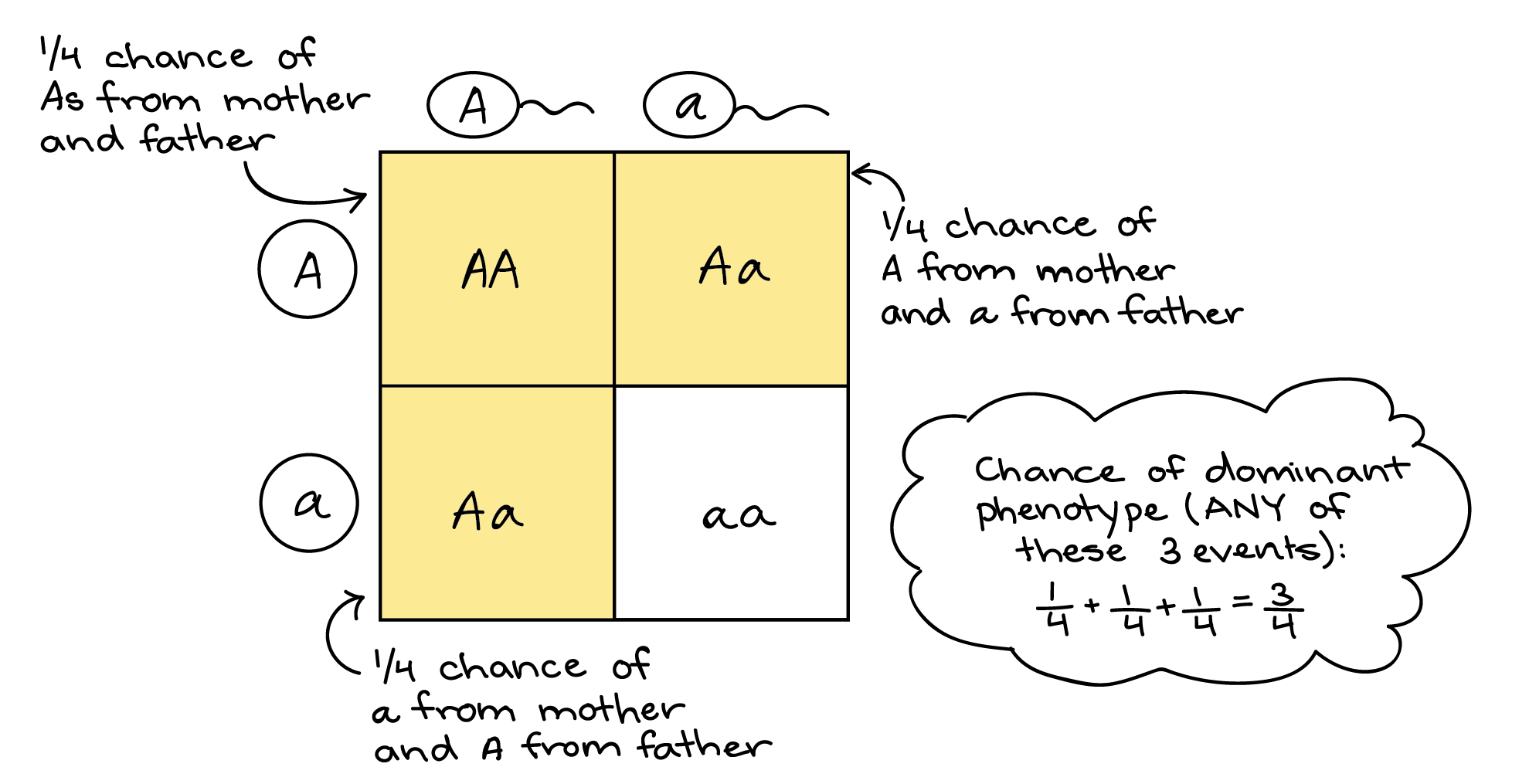
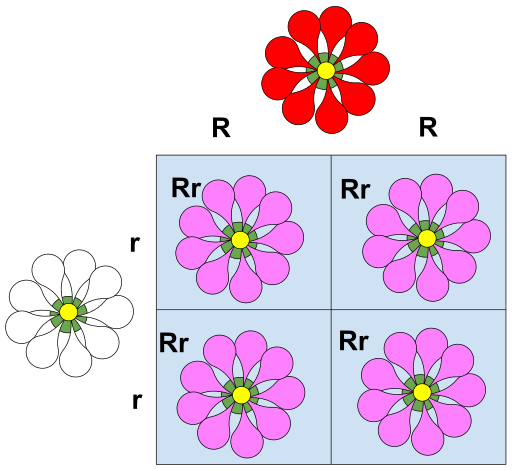
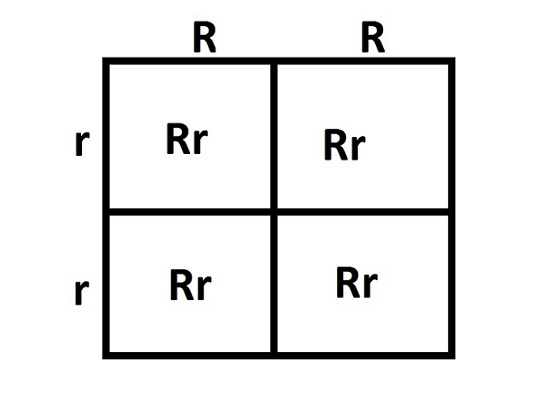
Punnett squares are used to predict traits in offspring.
4) Genotype vs phenotype (important distinction)
| Term | Meaning | Example |
| --------- | ------------------- | ---------- |
| Genotype | Genetic makeup | Bb |
| Phenotype | Physical expression | Brown eyes |
📌 Traits are what you see (phenotype), based on genes (genotype).
5) Why traits matter
Traits:
- Explain similarities and differences between organisms
- Help scientists study inheritance
- Are the basis of natural selection and evolution
One-sentence exam summary
Traits are characteristics of organisms that can be inherited, acquired, or influenced by both genes and environment.
中文 — traits(性状 / 特征) 详细解释
1) 什么是性状(核心概念)
性状(traits)是指生物表现出来的特征或特点。
性状 = 生物的特征
2) 性状的类型
① 遗传性状
由基因决定,从父母传给子女。
例子:
② 获得性状
由环境或经历造成。
例子:
③ 遗传 + 环境共同影响
3) 遗传学中的性状(考试重点)
- 性状由基因控制
- 基因有不同等位基因
- 等位基因有显性和隐性
4) 基因型 vs 表现型
| 概念 | 含义 |
| --- | ---- |
| 基因型 | 基因组合 |
| 表现型 | 外在性状 |
一句话考试版总结
性状是生物的特征,由遗传和环境共同决定。
如果你需要,我可以把 traits / inherited traits / acquired traits / Punnett squares 做成 Science 10 中英对照闪卡或课堂练习题,直接用于教学或复习。
性状(trait)
EN: An inherited characteristic, such as eye colour or hair colour.
Example: Traits like seed colour in pea plants or the ability to taste bitterness are controlled by genes.
中文:一种可遗传的特征,如眼睛颜色、头发颜色等。
例子:豌豆的种子颜色、是否能尝出苦味,都是由基因控制的性状。









 Save this for yourself as a reminder visual or share it with someone you know needs it (and just in case you're not sure who needs it.... erm, that's pretty well everyone)
Save this for yourself as a reminder visual or share it with someone you know needs it (and just in case you're not sure who needs it.... erm, that's pretty well everyone)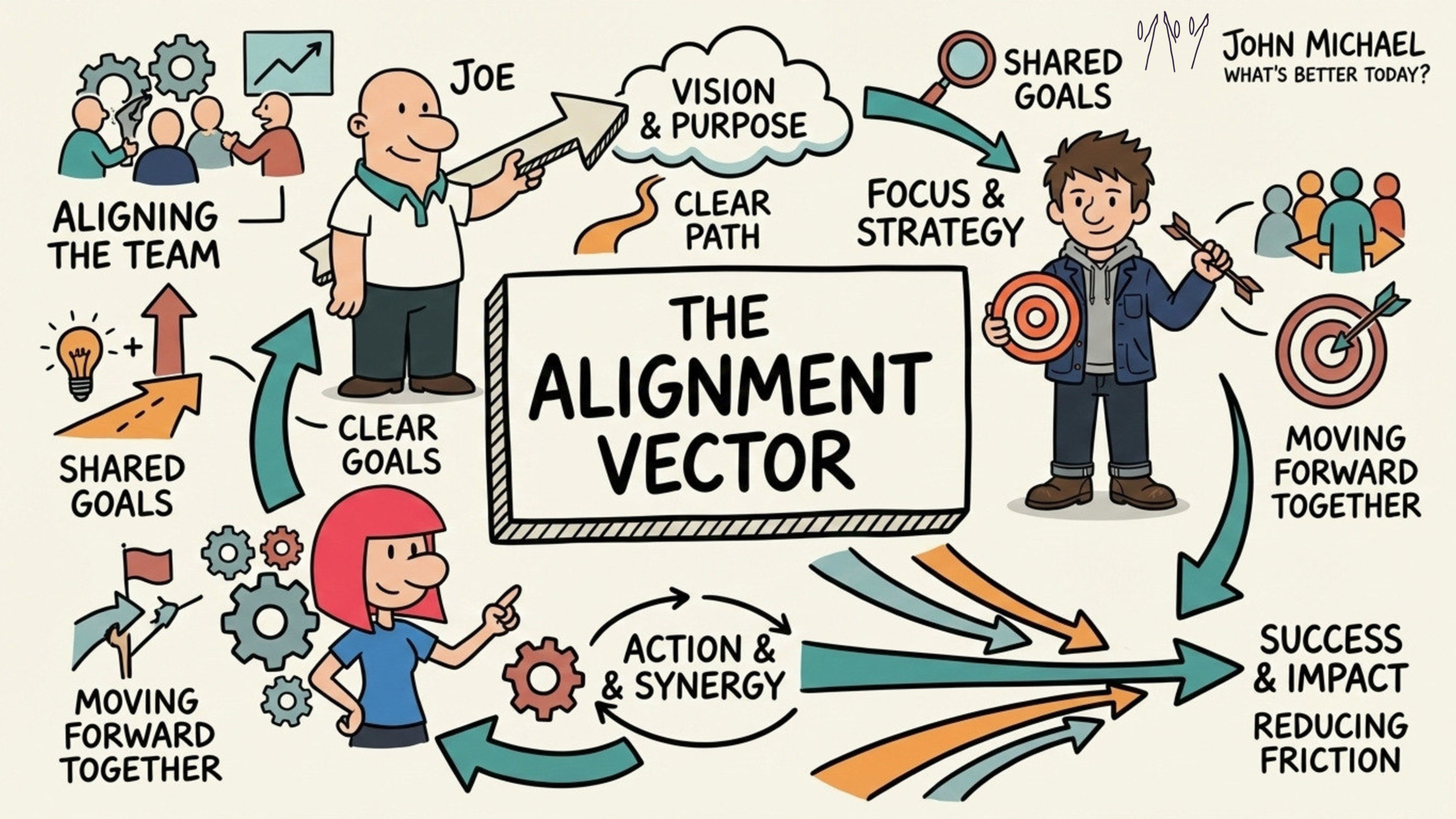 Save this SketchNote about Alignment for yourself or share it with someone you know needs to re-align themselves.
Save this SketchNote about Alignment for yourself or share it with someone you know needs to re-align themselves.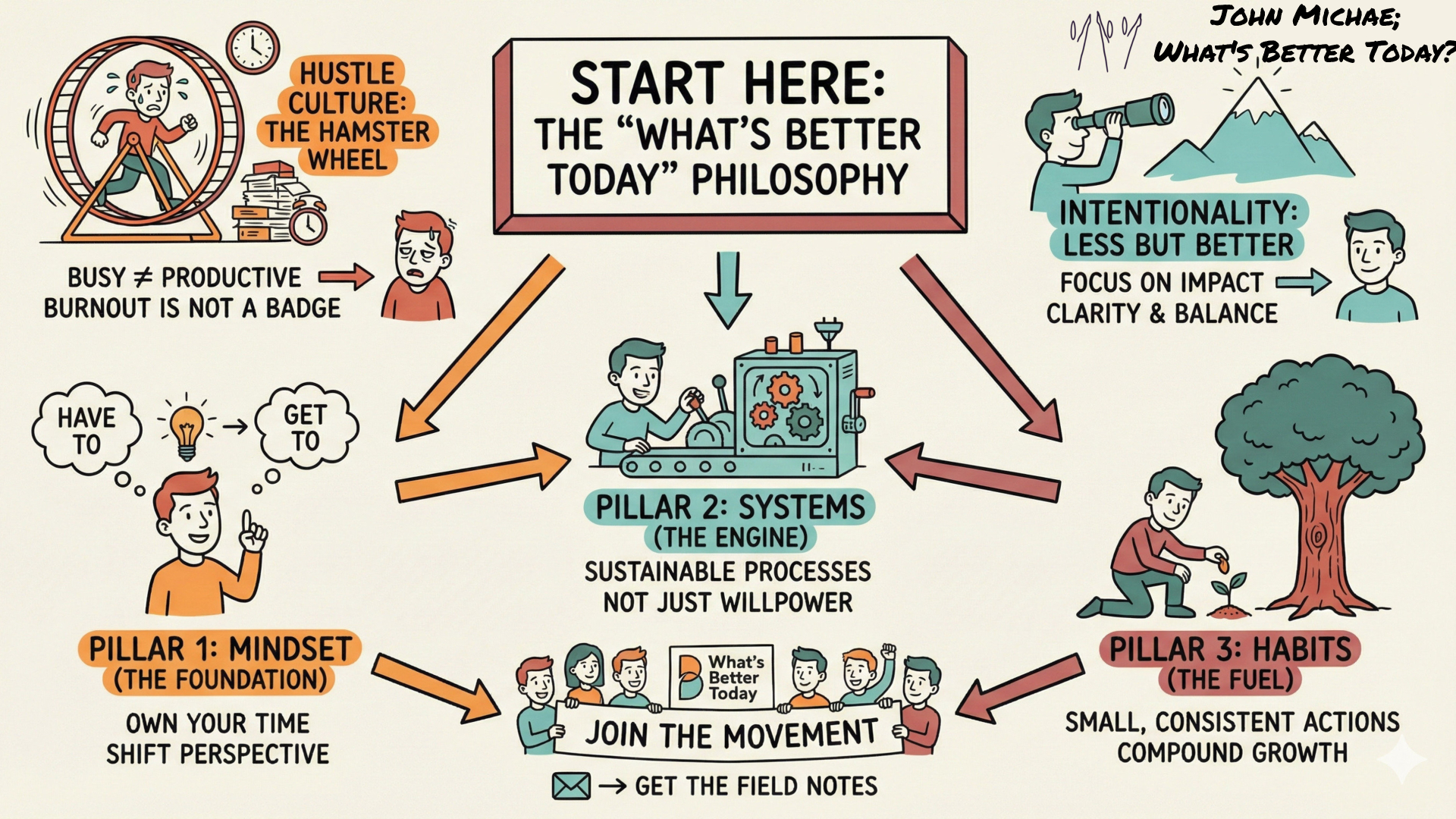 Save a copy of this SketchNote for your reminder or share it with someone you care about.
Save a copy of this SketchNote for your reminder or share it with someone you care about.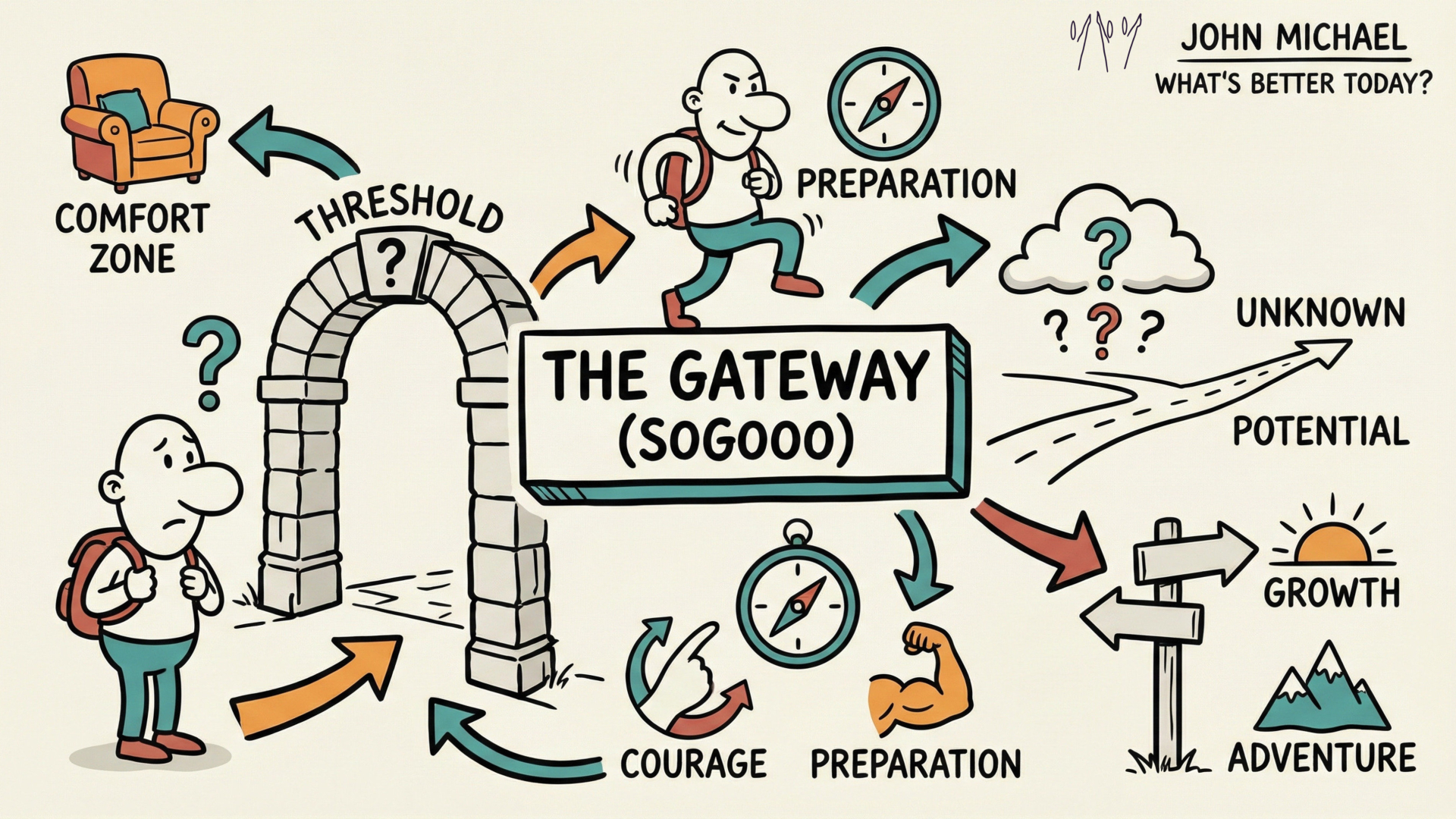 Save this Gateway SketchNote as a quick reminder for yourself or share it with someone you know needs to check this out.
Save this Gateway SketchNote as a quick reminder for yourself or share it with someone you know needs to check this out.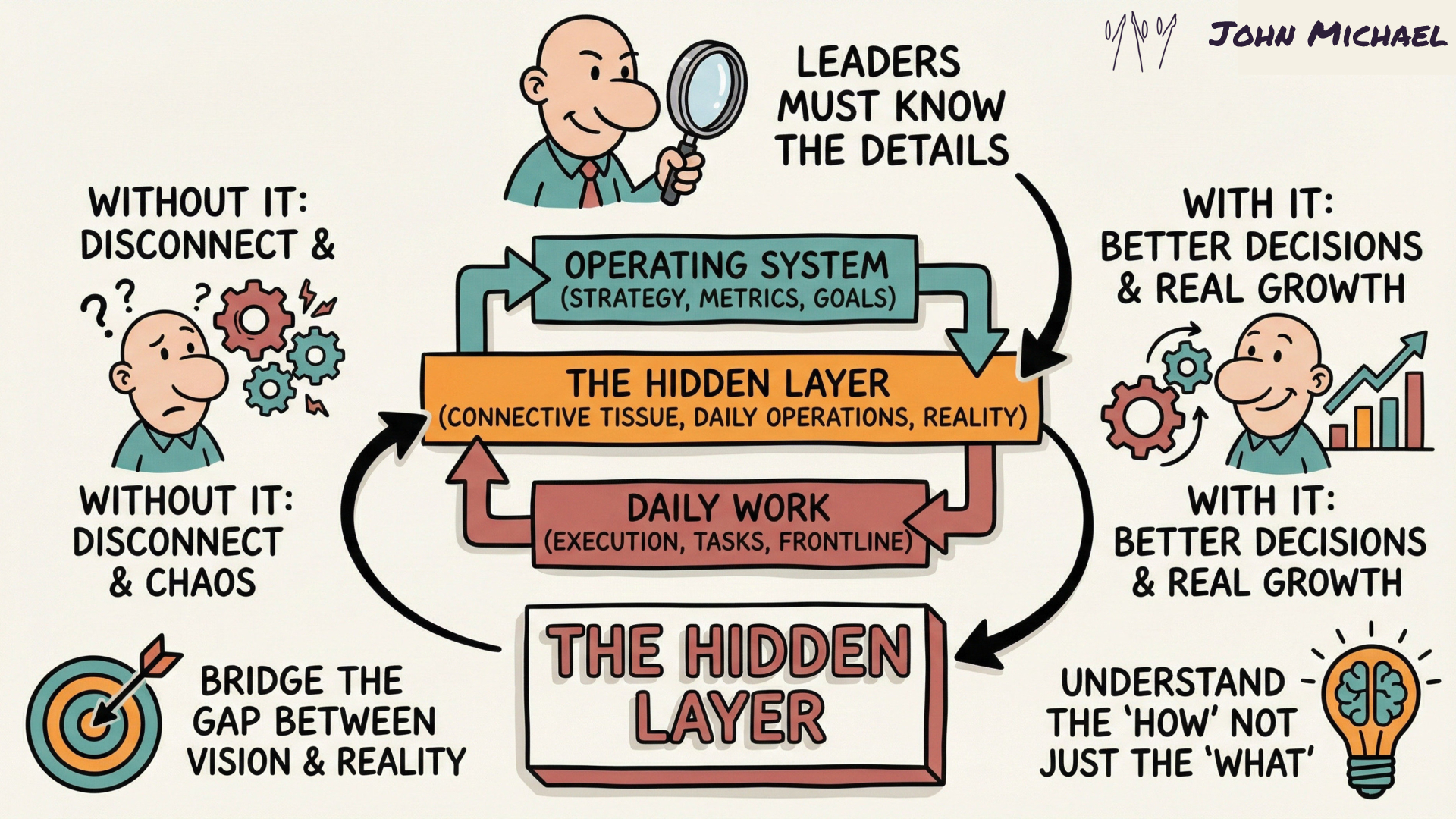 Yes, you're in the "hidden" layer - not so hidden now huh?
Save this for yourself or share it with someone who needs to know that you are one who digs deeper and looks beyond the surface.
Yes, you're in the "hidden" layer - not so hidden now huh?
Save this for yourself or share it with someone who needs to know that you are one who digs deeper and looks beyond the surface.