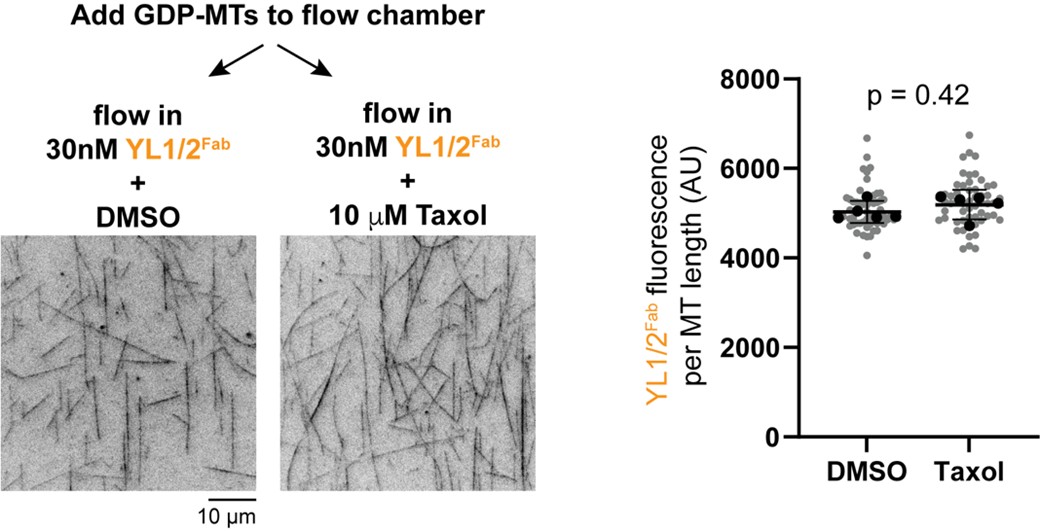
"Normally, an alteration confined to the A cistron (be it a deletion, an acridine mutant, or any other mutant) does not prevent the expression of the B cistron. Conversely, no alteration within the B cistron prevents the function of the A cistron. This implies that there may be a region between the two cistrons which separates them and allows their function to be expressed individually." Are there instances where a mutation on the A cistron would adversely affect the function of the B cistron then? And vice versa? The use of the word "Normally" at the beginning seems to imply that, but I don't know how true that is.
- Jan 2026
-
mssu.blackboard.com mssu.blackboard.comFile7
-
-
"The fact that we can make these changes and can study so large a region probably come about because this part of the protein is not essential for its function." Which part of the protein are they referring to exactly?
-
"The simplest postulate to make is that the shift of the reading frame produces some triplets the reading frame of which is 'unacceptable'; for example, they may be 'nonsense' or stand for 'end of the chain', or be unacceptable in some other way due to the complications of protein structure." Nonsense must mean the resulting codon does not produce a viable protein, 'end of the chain' must result in a stop codon, and the last bit seems pretty self explanatory.
-
"However, when both mutations are present in the same piece of DNA, as in the pseudo-wild double mutant FC (0+1), then although the reading triplets between FC 0 and FC 1 will be altered, the original reading will be restored to the rest of the gene." A mutation that reverses the original mutation as long as everything lines up correct. Once again, a concept that you would think would not be possible to conjure up over 50 years ago.
-
"Thus in all we have about eighty independent r mutants, all suppressors of FC 0, or suppressors of suppressors, or suppressors of suppressors of suppressors" Although I do understand what they authors are trying to convey, the way that this was communicated was not the most efficient. It could have been worded way better. If someone will no knowledge of the sciences were to read this, it would cause great confusion.
-
"Another idea is that certain triplets make 'sense' whereas others make 'nonsense', as in the comma-free codes of Crick, Griffith and Orgel." Nonsense could be interpreted within the literal sense, which I am certain is the author's intention, or it could be used in a more humorous sense.
-
"The code is probably degenerate; that is, in general, one particular amino-acid can coded by one of several triplets of bases." I think it's really interesting that Crick and the other researchers apart of this paper were able to glean such an advanced concept for the 1960s.
-
-
open.library.okstate.edu open.library.okstate.edu
-
With a strong set of ethical values, you will be better prepared to make the right decision and stick to your principles when faced with an ethical dilemma.
This focuses on the importance of having clear personal ethics before encountering workplace dilemmas.
-
This chapter will introduce some of the ethical issues that may arise as technical writers research, write, revise, and produce a technical document.
This explains that ethical decision-making is a core part of technical writing, not just clear communication.
-
-
socialsci.libretexts.org socialsci.libretexts.org
-
In this case, the conflict concerns gender inequality rather than the class inequality emphasized by Marx and Engels
Inequality is a huge issue that both Max & Engels both were inspired by; this inspiration led to them later developing the concept of "conflict theory" .
-
-
open.library.okstate.edu open.library.okstate.edu
-
Culture is part of the very fabric of our thought, and we cannot separate ourselves from it.
This highlights that culture shapes how we think and communicate, making it essential to consider in any rhetorical situation.
-
Another key part of the definition of technical communication is the receiver of the information—the audience.
This supports that understanding the audience is essential for making technical information effective and usable.
-
Technical writing is an audience-centered means of communication that provides a reader with clear and easy access to information
This describes how technical writing and emphasizes that clarity and audience needs are the writer’s main priorities.
-
-
open.ocolearnok.org open.ocolearnok.org
-
A sample plan for responding to off-task behavior might look like the list below. Warning Conference with Teacher Call Home Referral to Office
Does this change depending on the students normal behavior. At what point do we go to the next steps.
-
For example, the rule “Bring all materials to class” covers bringing pencils, paper, textbooks, homework papers, and permission slips—depending on the situation.
What is the best way to communicate what all students need to have, depending on the situation. A handout?
-
“The number one problem in the classroom is not discipline; it is the lack of procedures and routines.” -Harry Wong
Why do so many classrooms lack procedures and routines. How do you know if you have enough procedures and routines in place?
-
Either way, investing the time in planning routines will go a long way in maximizing your instructional time and managing behavior.
Planning out a lesson and routines for the year seems hard for me to fathom. It feels like there is too many unplanned events that could occur that would derail the routine for the day. Also, I still need to learn instructional methods to help me develop a good routine.
-
Classroom rules, also referred to as norms, express standards of behavior for which individual students need to take responsibility.
I really like this definition of classroom rules. To me, when it says standards it emphasizes the importance of structure and clear communication when creating a classroom environment. My concern is as a new teacher, how will I know what the best classroom rules are and how to enforce them? I know I will learn as I go but I also want to maintain some consistency throughout the year.
-
-
www.americanyawp.com www.americanyawp.com
-
I would prefer to live by ourselves, for there is a prejudice against us in the South that will take years to get over; but I do not know that I can answer for my brethren. [Mr. Lynch says he thinks they should not be separated, but live together. All the other persons present, being questioned one by one, answer that they agree with Brother Frazier.]
This had me thinking about what the “freedmen” living by themselves in their own colonies would have looked like. Especially since they would have either had to have their colonies on land “unclaimed” or for the States to remove their own people from parts of the land in order to allow them the space. I had also wondered if it would end up looking far more segregated than not living separated at all. If these new established colonies would be placed on a specific area of land, would it be fit to live on, or fair? Would their colonies get constantly moved and pushed to different areas just to satisfy White land owners—similarly to Native Americans?
-
-
docdrop.org docdrop.org
-
If you ask before the due date I will almost certainly say yes, so just ask! If the due date has passed, the answer will be no.
I greatly appreciate the way this is set up. I feel that it gives enough flexibility that if we are responsible and looking ahead to our schedule and feel as though we might not be able to do the best quality work, we can ask for more time. This does a good job of holding us accountable while at the same time lending us some help when we may need it.
-
-
nmoer.pressbooks.pub nmoer.pressbooks.pub
-
Review each assignment and think about the writing you’ve done in high school and how these assignments might look different in your college composition classes.
From personal experience, the writing I did in high school was less specific and shorter, but still often contained elements of these writing assignments. Then again, my memory can be unreliable.
-
-
www.bitbybitbook.com www.bitbybitbook.com
-
Although these two approaches produced similar estimates in this case, the approach of Blumenstock and colleagues was about 10 times faster and 50 times cheaper than the traditional Demographic and Health Surveys.
This shows how the same data can help public policy but also enable tracking people. Even if the researchers are careful, someone else could use similar methods for harm. It makes me think ethics should include thinking about misuse, not just the study’s intention.
-
Combining these two sources of data, they used the survey data to train a machine learning model to predict a person’s wealth based on their call records.
The survey gives “real” wealth info, and the call records give scale. But what phone behavior is really measuring?
-
-
nmoer.pressbooks.pub nmoer.pressbooks.pub
-
it helps to take detailed notes both when in class and when you read.)
This may just be a personal issue, but I often allow my note taking to get out of hand, so it could help to occasionally check the time in case it is in short supply.
-
-
www.bitbybitbook.com www.bitbybitbook.com
-
researchers can now observe the behavior of millions of people, and as I’ll describe later, researchers can also enroll millions of people in massive experiments.
This is a helpful reminder that big data isn’t enough by itself. Traditional methods (surveys, interviews, careful sampling) still matter for meaning and validity. The best work is probably a mix, like the Rwanda example.
-
-
nmoer.pressbooks.pub nmoer.pressbooks.pub
-
Linguist John Swales defined discourse communities as “groups that have goals and purposes and use communication to achieve their goals.” Swales uses six criteria to determine whether a specific community is, in fact, a discourse community.
All 6 of these are important to some degree, as despite how it may first seem, it would be quite difficult to shrink the definition down any further. If you take out the expert members, vocabulary on a given subject, 1+ genres, active feedback/information, intercommunication, or common goals, then the discourse community would likely struggle to stay intact.
-
-
www.bitbybitbook.com www.bitbybitbook.com
-
Further, it can easily randomize groups of customers to receive different shopping experiences. This ability to randomize on top of tracking means that online stores can constantly run randomized controlled experiments. In fact, if you’ve ever bought anything from an online store, your behavior has been tracked and you’ve almost certainly been a participant in an experiment, whether you knew it or not.
Even if A/B tests are “normal,” it raises consent and transparency issues: does “implied consent” exist when users don’t know they’re being experimented on? It also makes me think about the boundary between product testing and research. At what point does an A/B test become human subjects research?
-
-
nmoer.pressbooks.pub nmoer.pressbooks.pub
-
Is the forum private so that only your instructor or only a group of classmates or only a specific classmate can see it or is it public so that everyone, all of your classmates and your instructor can see your post?
It would be wise to make sure one's private notes are made in a private group, and that one's public notes are made in a public group, as doing so may save time spent fixing such a mistake, and any confusion potentially caused from that error. Generally, double checking things is a solid habit to get into.
-
-
social-media-ethics-automation.github.io social-media-ethics-automation.github.io
-
2003 saw the launch of several popular social networking services: Friendster, Myspace, and LinkedIn. These were websites where the primary purpose was to build personal profiles and create a network of connections with other people, and communicate with them. Facebook was launched in 2004 and soon put most of its competitors out of business, while YouTube, launched in 2005 became a different sort of social networking site built around video.
This section helped me understand how Web 2.0 fundamentally changed social media from static websites into interactive platforms centered around user profiles and ongoing updates. These early design choices still shape how users interact with feeds today and help explain why engagement and visibility became so important later on.
-
-
social-media-ethics-automation.github.io social-media-ethics-automation.github.io
-
Before this centralization of media in the 1900s, newspapers and pamphlets were full of rumors and conspiracy theories. And now as the internet and social media have taken off in the early 2000s, we are again in a world full of rumors and conspiracy theories.
This section helped me understand how early social media platforms were not originally designed for large-scale influence or automation, but rather for small communities and personal connection. Seeing how features like feeds, likes, and sharing evolved over time makes it clear how design decisions made early on still shape user behavior and power dynamics today.
-
-
newtfire.org newtfire.org
-
I feel as if this map is of big importance and think that this map here tells a vivid image of our social lives we live through either the media within our phones, social media pages and biggest thing I see is "Sea Of Memes" I'm much more interested from whomever made this picture
-
-
pubs.aeaweb.org pubs.aeaweb.org
-
percentage of the population in the 1990–2016 period
Is the overall effect negative because the effect of high-skill immigration is more powerful or is it because there are more high skill migrants in absolute terms and so the compounded effect is large?
-
estimate the impact of the shareof foreign citizens on election outcomes using variation over time across districts inthe city of Hamburg between 1987 and 2000. The authors find evidence of a positivecorrelation between the population share of immigrants in a district and the share ofvotes received by extreme right-wing parties with a clearly anti-immigration stance.
In part the previous studies will not capture the specific nuances of the political economy in America or the nuances if the makeup of immigrants (skill, english proficiency, etc...)
-
A scatter plot of the change in the share of Republican votes against the changein the share of immigrants, between the years 1990–1992 and 2014–2016, shows anegative and significant correlation
They'll say because most of the immigrants are high-skill
-
foreign-born residents with no high-school degree. The high-skilled immigrants Hitare counted as the number of adult foreign-born residents with a high-school degreeor more
Oversimplification but thats ok
-
Relative to these works, our paper is the first tofocus on US elections including the whole country, using variation across countiesand isolating a causal mechanism by the use of skill-specific shift-share instruments.
shocking
-
A simple explanation is that immigrants to theEuropean countries analyzed above (Italy, Austria, and Germany) are on averageless skilled than immigrants to the United States.
The migration of high skilled immigrants into NE specifically may result in a higher democratic vote share there (also histiry of immigrants?)
-
we confirm the negative andsignificant impact of high-skilled immigrants on the vote share to the Republicansand the positive and significant impact of low-skilled immigrants
Consistent with what we learned in migration and migration policy
-
This implies that the vote share of the Republican Party shouldincrease if voting residents perceive immigrants as a net cost rather than a benefit
Gonna depend on the type of immigrant, even ignorant americans know that not all immigrants are equal
-
Political leaders’ positions on the topic of immigration can be an important deter-minant of their electoral success.
OOOOH what a hook, way to be poetic eye roll
-
-
www.energy.gov www.energy.gov
-
Report to U.S. Energy Secretar
Experts' review of this review: https://essopenarchive.org/users/260056/articles/1330312-climate-experts-review-of-the-doe-climate-working-group-report?commit=f2b7646f3573ccc4bd74fcc2e9a1111dfbf078c2
-
-
sites.google.com sites.google.com
-
Our review reveals
Dessler et al. experts' review:
-
-
ecampusontario.pressbooks.pub ecampusontario.pressbooks.pub
-
What comes to mind? Sights, sounds, and scents? Something special that happened the last time you were there? Do you contemplate joining them? Do you start to work out a plan of getting from your present location to the restaurant? Do you send your friends a text asking if they want company? Until the moment when you hit the “send” button, you are communicating with yourself.
When I think about a restaurant , my brain starts a private conservation. You imagine the sights, sounds, and smells of the food. You remember a special moment you had there before. You ask yourself if you want to join your friends. You start to plan your trip from where you are now. You think about sending a text to ask if you can come along. Until you actually hit "send", every thought is just you talking to yourself.
-
-
Local file Local file
-
A favorite image that kings chose forthemselves in statues from the Early Dynastic period shows themonarch with a basket of earth for construction on his head. Hemight be a mighty ruler, but he was also a builder, erectingmonuments to his gods and taking care of his people.
-
Surprisingly,though, the Mesopotamians rarely wrote about the afterlife. Literarydescriptions suggest that the netherworld was a gloomy place—dark, with bad food, and no way out—and there was little about itthat suggested either a reward or punishment. It simply existed.And yet, since these kings (and many commoners whose burials alsocontained gifts and food) took their worldly possessions with them,perhaps they believed that they could improve their lot in theafterlife.
-
The king of Kish even sometimes enforced order inSumer. For example, Enannatum’s son, Enmetena, wrote that theborder between Lagash and Umma had been determined by thegreat god Enlil himself and had been confirmed by the king of Kish:“Mesalim, king of Kish, at the command of (the god) Ishtaran,measured the field and set up a (boundary-) stone there.” Theauthority of the king of Kish was therefore acknowledged, at leasttemporarily, by both the king of Umma and the king of Lagash.
There is an interesting example of the mnemonic use of stone here in ancient Sumer. It serves as a boundary/border marker by its physical presence, but apart from any (other local) mnemonic uses, it also carries an inscription as a secondary form of long term written memory.
-
At around the same time that they invented inscriptions by which tocommunicate with gods and with future kings, the kings’ scribesbegan to use writing for addressing those at a different kind ofdistance from themselves—kings of other lands.
Note the shifts in potential audience(s) of early writing.
-
Not far from the Ibgal temple of Inanna where the dedicatory tabletwas found, archaeologists excavated the earliest known breweryanywhere in Mesopotamia (a tablet found there even mentioned thebrewer).
-
all of the 1,700administrative tablets that were found at the later capital of Lagash,called Girsu, came from the queen’s palace.
-
The kings had, however, begun to realizeits potential for extending communication, in an almost magical way,beyond what could be accomplished with the spoken word. Writingcould perpetually and eternally address an audience on a king’sbehalf; the words were always there, even when the king was notthinking about them. Given that the population was almost entirelyilliterate, such an audience was mostly made up of gods. Thestatuette of the king’s personal god (or sometimes of the kinghimself), inscribed with the same text as the tablet, could thereforepray continuously in a way that a real person could not.
Is there stronger evidence that this form of permanent writing to an audience of gods was being done? Sources?
-
The scribes did not, however, try toexpress every part of speech; their script was still a shorthandversion of language.
the shift from proto-cuneiform to early dynastic cuneiform showed greater complexity, but was still a variation of shorthand which didn't attempt to express all parts of speech as converted from spoken language. Verbs weren't always appropriately conjugated.
-
The tablet wasfound by archaeologists in the foundations of the temple of Inannain Lagash, called the Ibgal. This extensive complex was oval inshape, as were many Early Dynastic temples in other cities, with alarge courtyard and a platform on which Inanna’s temple wasconstructed.
What is the general history of oval-shaped architecture? Is there an explicit link between the Oval shape of the complex at Ibgal, the temple (or house) of Inanna in Lagash and the oval office at the White House?
Keep in mind that modern knowledge of large portions of the Ancient Near East only surfaced after the 1800s, so the tradition would have required intermediaries from the ANE into other cultures to be passed down to the building of the White House in 1792.
-
Lagash (modern Al Hiba in Iraq), hometo the god Ningirsu and to a dynasty of kings who squabbled forgenerations with their counterparts in the neighboring city-state ofUmma.
-
Every human alive, the king included, was just a servant to thegods, and those gods could choose to treat him or her however theywanted.
At what point in history did it seem apparent to a larger portion of the populace that "god(s) anointed the king" was no longer a presumed reality? When did it seem that way to the kings themselves? Or have they always believed the myth?
-
And just as professions tended to run in families, the kingmight have trained his son in the arts of leadership, grooming himfor the throne.
It certainly makes some sense that in a diversifying economy familial connections of passing down trades from father to son by dint of closeness, teaching, and familial connection that the job/title of king should be passed down in the same way.
-
The Sumerian term for king, “lugal,” literally meant “big man.”
-
Order was maintained in the universe because the king of the godspossessed an object called the “Tablet of Destinies” on which wereinscribed theme (pronounced “may”). Theseme were never writtendown on any earthly tablet, as far as we know, for humanedification. But they encompassed all that kept chaos at bay.
-
Eventhe gods themselves had a king—the great god Enlil, who lived inthe city of Nippur.
-
ngship seemed so obvious and right to the Mesopotamians thatthey believed that it had been invented by the gods, that it hadcome “down from heaven.” Some later scribes made a grand list ofall the kings from the beginning of time to their own era. Theycalled it “When kingship came down from heaven,” which was itsfirst line. To modern scholars it is the Sumerian King List.
Tags
- diplomacy
- The Oval Office
- orality transition to literacy
- ovals
- history of alcohol
- writing to gods
- Girsu (capital of Lagash)
- praying
- leadership
- Enlil
- writing as memory
- administrative tablets
- Mesopotamian art
- iconography
- alcohol
- chaos
- breweries
- construction
- Nippur
- early writing
- stelae
- Sumerian gods
- ovals in architecture
- gods
- House of Inanna
- cuneiform
- Enmetena (King)
- big men
- Ningirsu
- divine right of kings
- audience
- early writing purposes
- myth
- White House
- Al Hiba, Iraq
- political evolution
- primogeniture
- writing to rulers
- builders
- kings
- Lagash
- Sumerian King List
- standing stones
- afterlife
- queens
- open questions
- Tablet of Destinies
- Mesopotamian religion
- stones for memory
- kingship
- Great Man theory of history
- Mesalim (king of Kish)
- stone inscriptions
- Ibgal
- Umma
Annotators
-
-
drive.google.com drive.google.com
-
In addition to no-cost food items, Swoop's Food Pantry offers a wide selection of no cost, non-food items suchas: school supplies, infant diapers and formula, menstrual health items, sexual health items, household goods,paper products, clothing (including commencement gown rentals), accessories and more! All registered EMUstudents are eligible to shop at the pantry as needed.
Another good resource.
-
The University Writing Center at EMU offers free one-to-one writing consultations by appointment. The UWCcan work with you in-person, on Zoom, or via written feedback.
It is nice that this service is available.
-
We will be working with bacteria isolated from soil and so will follow Biological Safety Level (BSL)-2laboratory procedures.
I saw we will be working with a 3 rated bacteria so I'm wondering how that plays out in the a BSL - 2 lab.
-
Each of the specs listed in Canvas will be graded “satisfactory” or “not yet”. “Satisfactory”means that you have demonstrated acceptable proficiency; minor errors may be present.“Not yet” means that your understanding of the material, the logic, or the vocabulary toexplain the concepts can be improved to meet the standard.
I'm interested to see how this affects my grades when I start doing work.
-
ur course-based undergraduate research experience(CURE) is part of the internationalTiny Earth antibiotic discovery network
I wonder what types of progress have come from this project?
-
-
history.wisc.edu history.wisc.edu
-
Everything we do, everything we use, everything else we study is the product of a complex set of causes, ideas, and practices. Even the material we learn in other courses has important historical elements – whether because our understanding of a topic changed over time or because the discipline takes a historical perspective.
I think this is a very important part of why we should study history because many things we take for granted are the results of innovations, contributions, and stuggles of people in the past.
-
-
Local file Local file
-
By recent estimates, 23.2 million of the people eligible to vote in the 2020 presidentialelection, or one-in-ten eligible voters, were naturalized immigrant citizens – a number that hasmore than doubled since 2000
And they influence the politics of NE?
-
Here, too, people who arrived from the Soviet Bloc at older ages are more likely to reportsupporting right-wing political parties.
We're seeing the same phenomon in isreal even though the political climate is differnt, one question might be how differnt the political climate actually is
-
It’s important to remember that the family fixed effect also accounts for much of thepost-migration experience for children, meaning that the primary vehicles for the assimilation ofchildren (e.g. schools, neighborhoods, social milieu) in the US are also fixed once families arrive
Controlling for the American culture they are documented being exposed to
-
The fact that the effects we measure are comparable for this group of refugeesto the effects we measure for those who leave when repression is coupled with economic crisissuggests that repression is the dominant mechanism behind the effects we document in this paper
Makes sense that it is the pirmary although I would think that if communism made you poor you woukld hate it for that
-
the emergence of an economic crisisthat threatened the welfare of the middle class
But why would that makes people more likely to vote
-
The probability plateaus untilthe mid 40s, when the probability of being registered as a Democrat starts rising again. There isno trend for registering as an independent with age of arrival until the beginning of middle-age,when increasing age appears to make non-partisan registration less likely
Young people are more radically right, ot more likely to register as repuiblican
-
1.9% increase in the probability of voting in a generalpresidential election.
Much higher
-
A and B of Figure 5 present a residualized binned scatter plot of the relationship between age atarrival and voting in 2016 and 2014, respectively
The later you come, the more exposure you had to repression, the more likely you are to vote
-
behaviors and preferences of siblings.
Measuring the increased exposure to regimes
-
family members forall families in our data.
And so captured in the family fixed effect
-
Overall therelationship between age of arrival is positive, with every additional year associated with a 0.18percentage point increase in the probability of voting in 2016
Bad model though, need a quadratic term
-
hose who are civically engaged enough to register
Couldn't you see what proportion of the migrants that was
-
this population as right-wing prior to migration.
Suggesting the emotional component not ideological
-
ven when thoseparties are anti-immigrant (
Talked about this last semester, there is a strong factor beyond the ideology of the party in the host country
-
but it might also motivate survivors toparticipate more post-migration to fulfill pent up demand for political voice, cast votes againstparties similar to oppresive ones in their birth countries, or a number of other possible psycho-logical mechanisms beyond the scope of this study
The question is how this ties back to NE, I suspect a bulk of the population will end up there
-
The extant literature suggests a “back-lash effect” for victims who remain in affected areas; victims are more hostile to the perpetratingregime and persistently less likely to express loyalty unless the regime can credibly threaten themagain
Again, intuitive
-
. Ironically, the liberalization of free expressionrules under Gorbachev’s Perestroika heralded an increase in public anti-semitic demonstrations(Gitelman, 1991), all prompting subsequent waves of emigration out of the Soviet Union thatwould continue after the Soviet Union dissolved in 1991
Government and public was persecuting or at least discriminating against jews
-
children are not typically responsible for migrationdecisions
But will still have some of the generational resentment and trauma from the repression
-
Moreover, we also establish similar patternswithin the same refugee population in a different national context (Israel) using data from a largemulti-election survey
One question is if it generalizes beyond jews
-
more likely to participatein presidential and midterm elections and to register as Republicans in the US. Our findings holdwithin our simplest within-family design,
They stay conservative, presumably in a backlash to the regime they are fleeing, but are liable to activly participate in government
-
-
-
Your second reading should be quite different from the first.
Whenever you read something for the second time, you notice things that you didn't notice the first time. It makes you realize that there is more to the topic, and you can relate everything to each other.
-
Our society defines the academically gifted as intelligent, but perhaps book smart would be a better term.
I have heard the term "book smart" my entire life, especially by the older generation. The terms I most often heard were "book smart" and "street smart." Book smart is being intelligent with school work, and street smart is being intelligent on the streets where you live. It is interesting to see this in an article because I thought it was mostly a saying.
-
From the title, you often discover the writer’s position on an issue or attitude toward the topic.
Title's give a lot away about what the topic is about and how the author feels about it. Titles are supposed to give you an insight into what the author is writing about and what they think about this topic.
-
Always read the selection at least twice, no matter how long it is.
This is something I have tried to utilize in the past especially for act preps and tests. I think sometimes it makes things harder to understand though.
-
o g e t t h e m o s t o u t o f y o u r reading, follow the five steps of the reading process.
I am glad that the article is giving these notes. It will be something I will look forward to working on and applying to my day to day readings.
-
-
docdrop.org docdrop.org
-
I am not expecting perfect work. I get it, you want a good grade,
It is appreciative when professors have an understanding that not every student's work is going to be perfect. It gives a sigh of relief that as long as they do there best, the professor will be pleased.
-
If it is not explicitly stated that use of generative AI is permitted, it is not permitted in any fashion.
I've taken many different writing/English courses, and the increase in AI usage is something mentioned more often. Some professors permitted it, and some don't, so it's helpful when they are clear whether they permit it or not.
-
All assignments for this course must be written and submitted directly in Google Docs.
It is interesting we are using google docs. Almost everything for me in the last 3 semester has been on word.
-
Using generative AI for any part of the writing process, including but not limited
This is interesting, because most of my classes are allowing people to use AI as long as it is not just copy and paste, and the way of usage is stated.
-
Developdigital and interviewresearch skills.
This should be helpful for me because I have a research methods class. So I am hoping to use the skills in both to excel this semester.
-
-
www.biorxiv.org www.biorxiv.org
-
eLife Assessment
This important study uncovers a previously unrecognized light-responsive pathway in C. elegans that depends on live food bacteria and is mediated by the bZIP factors ZIP-2/CEBP-2 and the cytochrome P450 enzyme, CYP-14A5. The authors show that this bacteria-linked pathway modulates long-term memory and can be harnessed as a low-cost light-inducible expression system, opening new directions for sensory biology and genetic engineering in worms. The exact means by which live bacteria modulate light signal that activates ZIP-2/CEBP-2 in the worm remains to be elucidated. The evidence supporting the pathway's role uses multiple genetic, transcriptional, and behavioural assays, and is convincing.
-
Reviewer #1 (Public review):
Summary:
The authors set out to understand how animals respond to visible light in an animal without eyes. To do so they used the C. elegans model, which lacks eyes, but nonetheless exhibits robust responses to visible light at several wavelengths. Here, the authors report a promoter that is activated by visible light and independent of known pathways of light resposnes.
Strengths:
The authors convincingly demonstrate that visible light activates the expression of the cyp-14A5 promoter driven gene expression in a variety of contexts and report the finding that this pathway is activated via the ZIP-2 transcriptionally regulated signaling pathway.
Weaknesses:
Because the ZIP-2 pathway has been reported to activated predominantly by changes in the bacterial food source of C. elegans -- or exposure of animals to pathogens -- it remains unclear if visible light activates a pathway in C. elegans (animals) or if visible light potentially is sensed by the bacteria on the plate which also lack eyes. Specifically, it is possible that the the plates are seeded with excess E. coli, that E. coli is altered by light in some way and in this context alters its behavior in such a way that activates a known bacterially responsive pathway in the animals. Consistent with this possibility the authors found that heat-killed bacteria prevented the reporter activation in animals. This weakness would not affect the ability to use this novel discovery as a tool, which would still be useful to the field.
-
Reviewer #2 (Public review):
Summary:
Ji, Ma and colleagues report the discovery of a mechanism in C. elegans that mediates transcriptional responses to low intensity light stimuli. They find that light-induced transcription requires a pair of bZIP transcription factors and induces expression of a cytochrome P450 effector. This unexpected light-sensing mechanism is required for physiologically relevant gene expression that controls behavioral plasticity. The authors further show that this mechanism can be co-opted to create light-inducible transgenes.
Strengths:
The authors rigorously demonstrate that ambient light stimuli regulate gene expression via a mechanism that requires the bZIP factors ZIP-2 and CEBP-2. Transcriptional responses to light stimuli are measured using transgenes and using measurements of endogenous transcripts. The study shows proper genetic controls for these effects. The study shows that this light-response does not require known photoreceptors, is tuned to specific wavelengths, and is highly unlikely to be an artifact of temperature-sensing. The study further shows that the function of ZIP-2 and CEBP-2 in light-sensing can be distinguished from their previously reporter role in mediating transcriptional responses to pathogenic bacteria. The study includes experiments that demonstrate that regulatory motifs from a known light-response gene can be used to confer light-regulated gene expression, demonstrating sufficiency and suggesting an application of these discoveries in engineering inducible transgenes. Finally, the study shows that ambient light and the transcription factors that transduce it into gene expression changes are required to stabilize a learned olfactory behavior, suggesting a physiological function for this mechanism.
Weaknesses:
The study implies but does not show that the effects of ambient light on stabilizing a learned olfactory behavior are through the described pathway. To show this clearly, the authors should determine whether ambient light has any further effects on learning in mutants lacking CYP-14A5, ZIP-2, or CEBP-2.
-
Reviewer #3 (Public review):
Ji et al. report a novel and interesting light-induced transcriptional response pathway in the eyeless roundworm Caenorhabditis elegans that involves a cytochrome P450 family protein (CYP-14A5) and functions independently from previously established photosensory mechanisms. The authors also demonstrate the potential for this pathway to enable robust light-induced control of gene expression and behavior, albeit with some restrictions. Despite the limitations of this tool, including those presented by the authors, it could prove useful for the community. Overall, the evidence supporting the claims of the authors is convincing, and the authors' work suggests numerous interesting lines of future inquiry.
(1) Although the exact mechanisms underlying photoactivation of this pathway remain unclear, light-dependent induction of CYP-14A5 requires bZIP transcription factors ZIP-2 and CEBP-2 that have been previously implicated in worm responses to pathogens. Notably, this light response requires live food bacteria, suggesting a microbial contribution to this phenomenon. The nature of the microbial contribution to the light response is unknown but very interesting.
(2) The authors suggest that light-induced CYP-14A5 activity in the C. elegans hypoderm can unexpectedly and cell-non-autonomously contribute to retention of an olfactory memory. How retention of the olfactory memory is enhanced by light generally remains unclear. Additional experiments, including verification of light-dependent changes in CYP-14A5 levels in the olfactory memory behavioral setup, appropriate would help further interpret these otherwise interesting results.
-
Author response:
The following is the authors’ response to the original reviews
Public Reviews:
Reviewer #1 (Public review):
Summary:
The authors set out to understand how animals respond to visible light in an animal without eyes. To do so, they used the C. elegans model, which lacks eyes, but nonetheless exhibits robust responses to visible light at several wavelengths. Here, the authors report a promoter that is activated by visible light and independent of known pathways of light responses.
Strengths:
The authors convincingly demonstrate that visible light activates the expression of the cyp-14A5 promoter-driven gene expression in a variety of contexts and report the finding that this pathway is activated via the ZIP-2 transcriptionally regulated signaling pathway.
Weaknesses:
Because the ZIP-2 pathway has been reported to be activated predominantly by changes in the bacterial food source of C. elegans -- or exposure of animals to pathogens -- it remains unclear if visible light activates a pathway in C. elegans (animals) or if visible light potentially is sensed by the bacteria on the plate, which also lack eyes. Specifically, it is possible that the plates are seeded with excess E. coli, that E. coli is altered by light in some way, and in this context, alters its behavior in such a way that activates a known bacterially responsive pathway in the animals. This weakness would not affect the ability to use this novel discovery as a tool, which would still be useful to the field, but it does leave some questions about the applicability to the original question of how animals sense light in the absence of eyes.
Thank you for the insightful questions and suggestions. We have now performed a key experiment requested. Interesting new data (Fig. S1I) show that light induction of cyp-14A5p::GFP requires live bacteria that maintain a non-starved physiological state. Neither plates without food nor plates with heat-killed OP50 support robust induction. We now include this interesting new result in the paper and revised discussion on the bacteria-modulated mechanism but note that this bacterial requirement does not alter the central conclusions of the study. Rather, it reveals an intriguing mechanistic layer, namely, that bacterial metabolic activity likely influences the animal’s sensitivity to environmental light. We are pursuing this host–microbe interaction in a separate study. In the present work, we focus on the intrinsic regulation and functional significance of cyp-14A5 under standard laboratory conditions with live OP50. Accordingly, we have revised the Results and Discussion to reflect the appropriate scope.
Reviewer #2 (Public review):
Summary:
Ji, Ma, and colleagues report the discovery of a mechanism in C. elegans that mediates transcriptional responses to low-intensity light stimuli. They find that light-induced transcription requires a pair of bZIP transcription factors and induces expression of a cytochrome P450 effector. This unexpected light-sensing mechanism is required for physiologically relevant gene expression that controls behavioral plasticity. The authors further show that this mechanism can be co-opted to create light-inducible transgenes.
Strengths:
The authors rigorously demonstrate that ambient light stimuli regulate gene expression via a mechanism that requires the bZIP factors ZIP-2 and CEBP-2. Transcriptional responses to light stimuli are measured using transgenes and using measurements of endogenous transcripts. The study shows proper genetic controls for these effects. The study shows that this light-response does not require known photoreceptors, is tuned to specific wavelengths, and is highly unlikely to be an artifact of temperature-sensing. The study further shows that the function of ZIP-2 and CEBP-2 in light-sensing can be distinguished from their previously reported role in mediating transcriptional responses to pathogenic bacteria. The study includes experiments that demonstrate that regulatory motifs from a known light-response gene can be used to confer light-regulated gene expression, demonstrating sufficiency and suggesting an application of these discoveries in engineering inducible transgenes. Finally, the study shows that ambient light and the transcription factors that transduce it into gene expression changes are required to stabilize a learned olfactory behavior, suggesting a physiological function for this mechanism.
Weaknesses:
The study implies but does not show that the effects of ambient light on stabilizing a learned olfactory behavior are through the described pathway. To show this clearly, the authors should determine whether ambient light has any effect on mutants lacking CYP-14A5, ZIP-2, or CEBP-2. Other minor edits to the text and figures are suggested.
We appreciate the reviewer’s comment. Our study indeed implies that ambient light stabilizes learned olfactory behavior through effects on the described pathway. Importantly, the existing data already address this point. Mutants lacking CYP-14A5, ZIP-2, or CEBP-2 display impaired olfactory memory even when exposed to ambient light, indicating that these genes are required for the behavioral effect of light. Consistent with this, ambient light robustly induces cyp-14A5p::GFP in wild-type animals but fails to do so in zip-2 and cebp-2 mutants, demonstrating that light-dependent transcriptional activation is blocked upstream in these pathway mutants. Together, these results support the conclusion that ambient light acts through the ZIP-2 → CEBP-2 → CYP-14A5 pathway to stabilize memory. Minor textual and figure revisions have been made where helpful to clarify this point.
Reviewer #3 (Public review):
Ji et al. report a novel and interesting light-induced transcriptional response pathway in the eyeless roundworm Caenorhabditis elegans that involves a cytochrome P450 family protein (CYP-14A5) and functions independently from previously established photosensory mechanisms. Although the exact mechanisms underlying photoactivation of this pathway remain unclear, light-dependent induction of CYP-14A5 requires bZIP transcription factors ZIP-2 and CEBP-2 that have been previously implicated in worm responses to pathogens. The authors then suggest that light-induced CYP-14A5 activity in the C. elegans hypoderm can unexpectedly and cell-non-autonomously contribute to retention of an olfactory memory. Finally, the authors demonstrate the potential for this pathway to enable robust light-induced control of gene expression and behavior, albeit with some restrictions. Overall, the evidence supporting the claims of the authors is convincing, and the authors' work suggests numerous interesting lines of future inquiry.
(1) The authors determine that light, but not several other stressors tested (temperature, hypoxia, and food deprivation), can induce transcription of cyp-15A5. The authors use these experiments to suggest the potential specificity of the induction of CYP-14A5 by light. Given the established relationship between light and oxidative stress and the authors' later identification of ZIP-2, testing the effect of an oxidative stressor or pathogen exposure on transcription of cyp-14A5 would further strengthen the validity of this statement and potentially shed some insight into the underlying mechanisms.
We appreciate the reviewer’s thoughtful suggestion. We would like to clarify that the “specificity” we refer to is the strong and preferential induction of cyp-14A5 by light among pathogen or detoxification-related genes, rather than an assertion that cyp-14A5 is exclusively light-responsive. This does not preclude the possibility that cyp-14A5 can also be activated under other conditions. Indeed, prior work from the Troemel laboratory has identified cyp-14A5 as one of many pathogen-inducible genes, consistent with its role in stress physiology. Our data show that classical pathogen-responsive genes (e.g., irg-1) are not induced by light, whereas cyp-14A5 is strongly induced, highlighting the selective engagement of this cytochrome P450 by light under the conditions tested. We have revised the text to clarify this point.
(2) The authors suggest that short-wavelength light more robustly increases transcription of cyp-14A5 compared to equally intense longer wavelengths (Figure 2F and 2G). Here, however, the authors report intensities in lux of wavelengths tested. Measurements of and reporting the specific spectra of the incident lights and their corresponding irradiances (ideally, in some form of mW/mm2 - see Ward et al., 2008, Edwards et al., 2008, Bhatla and Horvitz, 2015, De Magalhaes Filho et al., 2018, Ghosh et al., 2021, among others, for examples) is critical for appropriate comparisons across wavelengths and facilitates cross-checking with previous studies of C. elegans light responses. On a related and more minor note, the authors place an ultraviolet shield in front of a visible light LED to test potential effects of ultraviolet light on transcription of cyp-14A5. A measurement of the spectrum of the visible light LED would help confirm if such an experiment was required. Regardless, the principal conclusions the authors made from these experiments will likely remain unchanged.
Thank you. We have revised the text to clarify this point. “Using controlled light versus dark conditions, we confirmed the finding from an integrated cyp-14A5p::GFP reporter and observed its robust widespread GFP expression in many tissues induced by moderate-intensity (500-3000 Lux, 16-48 hr duration) LED light exposure (Fig. 1A). The photometric Lux range is approximately 0.1–0.60 mW/cm<sup>2</sup> in radiometric (total radiant power) metric given the spectrum of the LED light source.”
(3) The authors report an interesting observation that animals exposed to ambient light (~600 lux) exhibit significantly increased memory retention compared to those maintained in darkness (Figure 4). Furthermore, light deprivation within the first 2-4 hours after learning appears to eliminate the effect of light on memory retention. These processes depend on CYP-14A5, loss of which can be rescued by re-expression of cyp-14A5 in mutant animals using a hypoderm-specific- and non-light-inducible- promoter. Taken together, the authors argue convincingly that hypodermal expression of cyp-14A5 can contribute to the retention of the olfactory memory. More broadly, these experiments suggest that cell-non-autonomous signaling can enhance retention of olfactory memory. How retention of the olfactory memory is enhanced by light generally remains unclear. In addition, the authors' experiments in Figure 1B demonstrate - at least by use of the transcriptional reporter - that light-dependent induction of cyp-14A5 transcription at 500 - 1000 lux is minimal and especially so at short duration exposures. Additional experiments, including verification of light-dependent changes in CYP-14A5 levels in the olfactory memory behavioral setup, would help further interpret these otherwise interesting results.
We thank the reviewer for these thoughtful comments. We agree that understanding how light enhances memory retention at a mechanistic level is an important direction for future work. Regarding the light intensities used in Figure 1B, we would like to clarify that 500–1000 lux does produce a measurable and statistically significant induction of cyp-14A5p::GFP, although the magnitude is lower than that observed at higher intensities. We interpret this modest induction as physiologically relevant: intermediate light levels appear sufficient to engage the CYP-14A5–dependent program required for memory stabilization, whereas stronger light intensities are detrimental to learning and reduce behavioral performance. Thus, the behavioral paradigm uses a light regime that activates the pathway without introducing stress-associated confounders.
(4) The experiments in Figure 4 nicely validate the usage of the cyp-14A5 promoter as a potential tool for light-dependent induction of gene expression. Despite the limitations of this tool, including those presented by the authors, it could prove useful for the community.
Thank you and we agree. In addition, we have included in the revised manuscript the single-copy integration strains based on UAS-GAL4 that produced similar results as transgenic strains and will be even more flexible and useful for the community.
Recommendations for the authors:
Reviewing Editor Comments:
While appreciating the quality and presentation of this important study, we had two major concerns that the authors need to address.
(1) Bacteria-versus-worm origin:
To rule out a bacterially derived stimulus, we suggest testing whether cyp-14A5p::GFP is inducible without bacteria (or killed bacteria). Checking whether the canonical immune reporters irg-5p::GFP and gst-4p::GFP are also light-inducible will further clarify this point.
We have now performed the key experiment requested by the reviewers. Interesting new data (Fig. S1I) show that light induction of cyp-14A5p::GFP requires live bacteria that maintain a non-starved physiological state. Neither plates without food nor plates with heat-killed OP50 support robust induction. Importantly, this requirement does not alter any of the central conclusions of the study. Rather, it reveals an intriguing mechanistic layer, namely, that bacterial metabolic activity influences the animal’s sensitivity to environmental light. We are pursuing this host–microbe interaction in a separate study. In the present work, we focus on the regulation and functional significance of cyp-14A5 under standard laboratory conditions with live OP50.
We included the data (Fig. 2D) to show that the canonical immune reporter irg-1p::GFP is not induced by the light condition that robustly induced cyp-14A5p::GFP, and gst-4p::GFP is only very mildly induced (Fig. S1J).
(2) Pathway-behaviour link:
The behavioural relevance of the newly described pathway is intriguing, but it needs direct support. Ideally, this would require comparing memory in WT, zip-2-/-, cebp-2-/-, and cyp-14A5-/- under both dark and light conditions. But at the very least, it would require testing if constitutive CYP-14A5 rescue in the dark bypasses the requirement of light.
We respectfully submit that additional experiments are not required to support the behavioral conclusions. Our model posits that cyp-14A5 is required but not sufficient for memory stabilization, one component within a broader set of light-induced genes. Thus, constitutive hypodermal expression of cyp-14A5 would not be expected to bypass the requirement for ambient light. The existing data are fully consistent with this framework and conclusions of the paper.
Reviewer #1 (Recommendations for the authors):
Overall, I think this paper is interesting to the field of C. elegans researchers at a minimum, as a light-inducible gene expression system might have a variety of uses throughout the diverse research paradigms that use this model system. With that said, I have a couple of suggestions that I think would substantially impact the ability to interpret these findings, which might be useful for broader implications of the study.
(1) Most importantly, the supplemental table of RNA-seq data should likely be updated and discussed further beyond the cyp-14A5 findings. First, the authors report 7,902 genes are differentially expressed in response to light and then break these into upregulated and downregulated genes. But there are only 1,785 upregulated genes and 3,632 downregulated genes. This adds up to 5417 genes, but doesn't match the 7,902 genes reported to change, and I could not find in the text if some other filters were applied that might explain this not adding up.
Thank you for this helpful comment. We agree that the exact numbers depend on statistical thresholds and are therefore somewhat arbitrary. To avoid implying unwarranted precision, we have revised the text to state that “thousands of genes are differentially regulated by light.”
(2) Among the upregulated genes in response to light are irg-5, irg-4, irg-6, irg-8, and gst-4. Indeed, all of these well-studied genes (or most) show even more induction by light than cyp-14A5. It is my opinion that this result needs further criticism as there are existing GFP reporters for gst-4 and irg-5 that are similarly well studied to irg-1, which is in the paper (and is not upregulated). In my opinion, the authors should test if they see activation of the irg-4 and gst-4 GFP reporters by light as well. This would not only validate their RNA-seq but might provide more important evidence for the field, as these other reporters are not considered light-inducible previously. If they are, several major studies might be impacted by this.
Thank you for the comments. We have irg-1p::GFP and gst-4p::GFP in the lab but did not find other reporters for the genes mentioned from CGC. Neither of the two reporters showed light induction (Figs. 2D and S1J) as strongly as cyp-14A5p::GFP. It is possible that irg-1 and gst-4 RNA levels are up-regulated but not reflected in our transgenic reporters that used their promoters to drive GFP expression. Stronger light induction of cyp-14A5p::GFP is unlikely caused by the multi-copy nature of the transgene since newly generated single-copy integration strains based on the UAS-GAL4 system produced similar robust results for light induction (Fig. S1I and see Method).
(3) Along the same lines, if at least 4 (and likely more) well characterized immune response genes are activated by light and these genes are known to mostly respond to differences in C. elegans bacterial food source/diet, then it stands to reason that maybe in this experimental context the light is not acting on "animals" at all, but rather triggering changes in E. coli (i.e. changing E. coli metabolism or pathogenicity like properties). If true, then perhaps the light affects bacteria in such a way that it activates a previously known bacterial pathogen response mechanism. This should be easy to test by seeing if this reporter is still activated by light in the presence of diverse bacterial diets, which are available from the CGC (CeMBio collection, for example). This is likely very important to the conclusions of the manuscript as it relates to animals sensing light, but might not be as important to the use of this system as a tool.
Thank you for the insightful questions and suggestions. Interesting new data (Fig. S1I) show that light induction of cyp-14A5p::GFP requires live bacteria that maintain a non-starved physiological state. Neither plates without food nor plates with heat-killed OP50 support robust induction. Importantly, this requirement does not alter any of the central conclusions of the study. Rather, it reveals an intriguing mechanistic layer, namely, that bacterial metabolic activity influences the animal’s sensitivity to environmental light. We are pursuing this host–microbe interaction in a separate study. In the present work, we focus on the regulation and functional significance of cyp-14A5 under standard laboratory conditions with live OP50. We have revised the Results and Discussion to reflect the appropriate scope of our study and implications of the new findings.
(4) Lastly, it seems unlikely that nearly half the C. elegans genome is transcriptionally regulated by light (or nearly half of the detected genes in the RNA-seq results). It seems likely that this list of 7,902 genes contains false positives. I would suggest upping some sort of filter, like moving to padj < 0.01 instead of 0.05, or adding a 4-fold change filter (2-fold and 0.01 still results in near 5000+ genes changing, which might explain the difference in up and down genes just being due to different padj filters. Along these lines, it is worth noting that the padj is generated using DESeq2 it appears and one of the first assumptions of DESeq2 is that the median expressed genes do not change, and there is a normalization. However, if MOST genes do change in expression, then one of the fundamental assumptions of DESeq2 is not valid, and thus would mean it might not be an appropriate analysis tool - perhaps there is some other normalization that could be done before running DESeq2 due to some other noise present in the RNA-seq runs?
Thank you for this helpful comment. We agree that the exact numbers depend on statistical thresholds and are therefore somewhat arbitrary. To avoid implying unwarranted precision, we have revised the text to state that “thousands of genes are differentially regulated by light.”
(5) Minor point - I would delete the reference to ER in line 92. While most CYPs do localize to the ER, the images shown are not clearly ER and probably do not have enough resolution to make claims about subcellular localization. To me, it would be easier to just delete this claim as it is not required for the main claims of the manuscript.
Reference deleted.
Reviewer #2 (Recommendations for the authors):
I have one request for clarification that likely requires additional data. Figure 3 shows that ambient light stabilizes learned changes to chemotaxis and further shows that CYP-14A5 has a similar function. The implication is that light promotes CYP-14A5 expression, which somehow promotes memory consolidation. The authors should test whether memory consolidation in cyp-15A5, zip-2, or cebp-2 mutants is no longer affected by ambient light.
It is also possible to test whether forced expression of CYP14A5 can bypass the effect of 'no light' conditions on memory consolidation.
Thank you for the comments. We respectfully submit that additional experiments are not required to support the behavioral conclusions. Our model posits that cyp-14A5 is required but not sufficient for memory stabilization, one component within a broader set of light-induced genes. Thus, constitutive hypodermal expression of cyp-14A5 would not be expected to bypass the requirement for ambient light. The existing data are fully consistent with this framework and conclusions of the paper.
I have several minor suggestions relating to the text and figures.
(1) In the introduction, the authors assert that little is known about non-visual light sensing and then list many examples of molecular mechanisms of non-visual light-sensing. They should emphasize that non-visual light sensing is important and accomplished by diverse molecular mechanisms.
Agree and revised accordingly.
(2) Check spacing between gene names (line 109).
Corrected.
(3) There should be a new paragraph break when the uORF experiments are described (line 146).
Corrected.
(4) 'Phenoptosis' is an esoteric word. Please define it (line 206).
Corrected.
(5) 'p' in the transgene name cyp-14A5p::nlp-22 is in italics, unlike the rest of the manuscript.
Corrected.
(6) 'Acknowledgment' should be 'Acknowledgments' (line 384).
Corrected.
(7) The color map in panel 1B should have units.
It was arbitrary unit (now added) to highlight relative not absolute differences.
(8) In panel 1E, it is confusing to have 'DARK' denoted by reddish bars and 'LIGHT' denoted by bluish bars. Perhaps 'DARK' is black/dark grey and 'LIGHT' is white?
Corrected.
(9) In panel 1D, it takes a minute to find the purple diamond. Please mark up the volcano plot to make it easier.
Corrected.
Reviewer #3 (Recommendations for the authors):
The authors generally present convincing experiments detailing interesting results in a well-written manuscript.
One quick note: the same Bhatla and Horvitz (2015) papers appear to be cited twice [line 52].
Corrected.
-
-
viewer.athenadocs.nl viewer.athenadocs.nl
-
two forms are approximately parallel by checking
the criterion need to be equal or very similar
-
-
moodle-courses2527.wolfware.ncsu.edu moodle-courses2527.wolfware.ncsu.edu
-
Need Help?
Its good to have all the tools for success in a spot easily in reach.
-
Tentative Schedule
Good to know the course schedule outright as base even when it might change through out the semester.
-
I will reach out to you directly if you miss 2 consecutive classes, to make sure you’realright and can make up any missed work.
I like the amount of time you put into your students for them to have the best possible success.
-
I may lower these cutoffs, but I will never raise them.
Can you clarify what you mean by lowering the Cutoffs? Like would it be In a situation where someone received a 96.9 and got a A Letter Grade you might lower the Minimum Cutoff to for A+ to 96.9 to increase the letter grade?
-
-
-
eLife Assessment
This important study presents a methodologically rigorous framework for stability-guided fine-mapping, extending PICS and generalizing to methods such as SuSiE, supported by comprehensive simulations and functional enrichment analyses. The evidence is now convincing, demonstrating improved causal variant recovery and offering a robust alternative for cross-population fine-mapping. The approach will be of particular interest to statistical geneticists, computational biologists, and biomedical researchers who rely on fine-mapping to interpret genetic association signals.
-
Reviewer #1 (Public review):
Aw et al. have proposed that utilizing stability analysis can be useful for fine-mapping of cross populations. In addition, the authors have performed extensive analyses to understand the cases where the top eQTL and stable eQTL are the same or different via functional data.
Comments on revisions:
The authors have answered all my concerns.
-
Reviewer #2 (Public review):
Aw et al presents a new stability-guided fine-mapping method by extending the previously proposed PICS method. They applied their stability-based method to fine-map cis-eQTLs in the GEUVADIS dataset and compared it against residualization-based approaches. They evaluated the performance of the proposed method using publicly available functional annotations and demonstrated that the variants identified by their stability-based method show enrichment for these functional annotations.
The authors have substantially strengthened the manuscript by addressing the major concerns raised in the initial review. I acknowledge that they have conducted comprehensive simulation studies to show the performance of their proposed approach and that they have extended their approach to SuSiE ("Stable SuSiE") to demonstrate the broader applicability of the stability-guided principle beyond PICS.
One remaining question is the interpretation of matching variants with very low stable posterior probabilities (~0), which the authors have analyzed in detail but without fully conclusive findings. I agree with the authors that this event is relatively rare and the current sample size is limited but this might be something to keep in mind for future studies.
-
Author response:
The following is the authors’ response to the latest reviews:
"One remaining question is the interpretation of matching variants with very low stable posterior probabilities (~0), which the authors have analyzed in detail but without fully conclusive findings. I agree with the authors that this event is relatively rare and the current sample size is limited but this might be something to keep in mind for future studies."
Fine-mapping stability – on matching variants with very low stable posterior probability
We thank Reviewer 2 for encouraging us to think more about how low stable posterior probability matching variants can be interpreted. We describe a few plausible interpretations, even though – as Reviewer 2 and we have both acknowledged – our present experiments do not point to a clear and conclusive account.
One explanation is that the locus captured by the variant might not be well-resolved, in the sense that many correlated variants exist around the locus. Thus, the variant itself is unlikely causal, but the set of variants in high LD with it may contain the true causal variant, or it's possible that the causal variant itself was not sequenced but lies in that locus. A comparison of LD patterns across ancestries at the locus would be helpful here.
Another explanation rests on the following observation. For a variant to be matching between top and stable PICS and to also have very small stable PP, it has to have the largest PP after residualization on the ALL slice but also have positive PP with gene expression on many other slices. In other words, failing to control for potential confounders shrinks the PP. If one assumes that the matching variant is truly causal, then our observation points to an example of negative confounding (aka suppressor effect). This can occur when the confounders (PCs) are correlated with allele dosage at the causal variant in a different direction than their correlation with gene expression, so that the crude association between unresidualized gene expression and causal variant allele dosage is biased toward 0.
Although our present study does not allow us to systematically confirm either interpretation – since we found that matching variants were depleted in causal variants in our simulations, violating the second argument, but we also found functional enrichment in analyses of GEUVADIS data though only 17 matching variants with low stable PP were reported – we believe a larger-scale study using larger cohort sizes (at least 1000 individuals per ancestry) and many more simulations (to increase yield of such cases) would be insightful.
———
The following is the authors’ response to the original reviews:
Reviewer #1:
Major comments:
(1) It would be interesting to see how much fine-mapping stability can improve the fine-mapping results in cross-population. One can simulate data using true genotype data and quantify the amount the fine-mapping methods improve utilizing the stability idea.
We agree, and have performed simulation studies where we assume that causal variants are shared across populations. Specifically, by mirroring the simulation approach described in Wang et al. (2020), we generated 2,400 synthetic gene expression phenotypes across 22 autosomes, using GEUVADIS gene expression metadata (i.e., gene transcription start site) to ensure largely cis expression phenotypes were simulated. We additionally generated 1,440 synthetic gene expression phenotypes that incorporate environmental heterogeneity, to motivate our pursuit of fine-mapping stability in the first place (see Response to Reviewer 2, Comment 6). These are described in Results section “Simulation study”:
We evaluated the performance of the PICS algorithm, specifically comparing the approach incorporating stability guidance against the residualization approach that is more commonly used — similar to our application to the real GEUVADIS data. We additionally investigated two ways of “combining” the residualization and stability guidance approaches: (1) running stability-guided PICS on residualized phenotypes; (2) prioritizing matching variants returned by both approaches. See Response to Reviewer 2, Comment 5.
(2) I would be very interested to see how other fine-mapping methods (FINEMAP, SuSiE, and CAVIAR) perform via the stability idea.
Thank you for this valuable comment. We ran SuSiE on the same set of simulated datasets. Specifically, we ran a version that uses residualized phenotypes (supposedly removing the effects of population structure), and also a version that incorporates stability. The second version is similar to how we incorporate stability in PICS. We investigated the performance of Stable SuSiE in a similar manner to our investigation of PICS. First we compared the performance relative to SuSiE that was run on residualized phenotypes. Motivated by our finding in PICS that prioritizing matching variants improves causal variant recovery, we did the same analysis for SuSiE. This analysis is described in Results section “Stability guidance improves causal variant recovery in SuSiE.”
We reported overall matching frequencies and causal variant recovery rates of top and stable variants for SuSiE in Figures 2C&D.
Frequencies with which Stable and Top SuSiE variants match, stratified by the simulation parameters, are summarized in Supplementary File 2C (reproduced for convenience in Response to Reviewer 2, Comment 3). Causal variant recovery rates split by the number of causal variants simulated, and stratified by both signal-to-noise ratio and the number of credible sets included, are reported in Figure 2—figure supplements 16-18. We reproduce Figure 2—figure supplement 18 (three causal variants scenario) below for convenience. Analogous recovery rates for matching versus non-matching top or stable variants are reported in Figure 2—figure supplements 19, 21 and 23.
(3) I am a little bit concerned about the PICS's assumption about one causal variant. The authors mentioned this assumption as one of their method limitations. However, given the utility of existing fine-mapping methods (FINEMAP and SuSiE), it is worth exploring this domain.
Thank you for raising this fair concern. We explored this domain, by considering simulations that include two and three causal variants (see Response to Reviewer 2, Comment 3). We looked at how well PICS recovers causal variants, and found that each potential set largely does not contain more than one causal variant (Figure 2—figure supplements 20 and 22). This can be explained by the fact that PICS potential sets are constructed from variants with a minimum linkage disequilibrium to a focal variant. On the other hand, in SuSiE, we observed multiple causal variants appearing in lower credible sets when applying stability guidance (Figure 2—figure supplements 21 and 23). A more extensive study involving more fine-mapping methods and metrics specific to violation of the one causal variant assumption could be pursued in future work.
Reviewer #2:
Aw et al. presents a new stability-guided fine-mapping method by extending the previously proposed PICS method. They applied their stability-based method to fine-map cis-eQTLs in the GEUVADIS dataset and compared it against what they call residualization-based method. They evaluated the performance of the proposed method using publicly available functional annotations and claimed the variants identified by their proposed stability-based method are more enriched for these functional annotations.
While the reviewer acknowledges the contribution of the present work, there are a couple of major concerns as described below.
Major:
(1) It is critical to evaluate the proposed method in simulation settings, where we know which variants are truly causal. While I acknowledge their empirical approach using the functional annotations, a more unbiased, comprehensive evaluation in simulations would be necessary to assess its performance against the existing methods.
Thank you for this point. We agree. We have performed a simulation study where we assume that causal variants are shared across populations (see response to Reviewer 1, Comment 1). Specifically, by mirroring the simulation approach described in Wang et al. (2020), we generated 2,400 synthetic gene expression phenotypes across 22 autosomes, using GEUVADIS gene expression metadata (i.e., gene transcription start site) to ensure cis expression phenotypes were simulated.
(2) Also, simulations would be required to assess how the method is sensitive to different parameters, e.g., LD threshold, resampling number, or number of potential sets.
Thank you for raising this point. The underlying PICS algorithm was not proposed by us, so we followed the default parameters set (LD threshold, r<sup>2</sup> \= 0.5; see Taylor et al., 2021 Bioinformatics) to focus on how stability considerations will impact the existing fine-mapping algorithm. We attempted to derive the asymptotic joint distribution of the p-values, but it was too difficult. Hence, we used 500 permutations because such a large number would allow large-sample asymptotics to kick in. However, following your critical suggestion we varied the number of potential sets in our analyses of simulated data. We briefly mention this in the Results.
“In the Supplement, we also describe findings from investigations into the impact of including more potential sets on matching frequency and causal variant recovery…”
A detailed write-up is provided in Supplementary File 1 Section S2 (p.2):
“The number of credible or potential sets is a parameter in many fine-mapping algorithms. Focusing on stability-guided approaches, we consider how including more potential sets for stable fine-mapping algorithms affects both causal variant recovery and matching frequency in simulations…
Causal variant recovery. We investigate both Stable PICS and Stable SuSiE. Focusing first on simulations with one causal variant, we observe a modest gain in causal variant recovery for both Stable PICS and Stable SuSiE, most noticeably when the number of sets was increased from 1 to 2 under the lowest signal-to-noise ratio setting…”
We observed that increasing the number of potential sets helps with recovering causal variants for Stable PICS (Figure 2—figure supplements 13-15). This observation also accounts for the comparable power that Stable PICS has with SuSiE in simulations with low signal-to-noise ratio (SNR), when we increase the number of credible sets or potential sets (Figure 2—figure supplements 10-12).
(3) Given the previous studies have identified multiple putative causal variants in both GWAS and eQTL, I think it's better to model multiple causal variants in any modern fine-mapping methods. At least, a simulation to assess its impact would be appreciated.
We agree. In our simulations we considered up to three causal variants in cis, and evaluated how well the top three Potential Sets recovered all causal variants (Figure 2—figure supplements 13-15; Figure 2—figure supplement 15). We also reported the frequency of variant matches between Top and Stable PICS stratified by the number of causal variants simulated in Supplementary File 2B and 2C. Note Supplementary File 2C is for results from SuSiE fine-mapping; see Response to Reviewer 1, Comment 2.
Supplementary File 2B. Frequencies with which Stable and Top PICS have matching variants for the same potential set. For each SNR/ “No. Causal Variants” scenario, the number of matching variants is reported in parentheses.
Supplementary File 2C. Frequencies with which Stable and Top SuSiE have matching variants for the same credible set. For each SNR/ “No. Causal Variants” scenario, the number of matching variants is reported in parentheses.
(4) Relatedly, I wonder what fraction of non-matching variants are due to the lack of multiple causal variant modeling.
PICS handles multiple causal variants by including more potential sets to return, owing to the important caveat that causal variants in high LD cannot be statistically distinguished. For example, if one believes there are three causal variants that are not too tightly linked, one could make PICS return three potential sets rather than just one. To answer the question using our simulation study, we subsetted our results to just scenarios where the top and stable variants do not match. This mimics the exact scenario of having modeled multiple causal variants but still not yielding matching variants, so we can investigate whether these non-matching variants are in fact enriched in the true causal variants.
Because we expect causal variants to appear in some potential set, we specifically considered whether these non-matching causal variants might match along different potential sets across the different methods. In other words, we compared the stable variant with the top variant from another potential set for the other approach (e.g., Stable PICS Potential Set 1 variant vs Top PICS Potential Set 2 variant). First, we computed the frequency with which such pairs of variants match. A high frequency would demonstrate that, even if the corresponding potential sets do not have a variant match, there could still be a match between non-corresponding potential sets across the two approaches, which shows that multiple causal variant modeling boosts identification of matching variants between both approaches — regardless of whether the matching variant is in fact causal.
Low frequencies were observed. For example, when restricting to simulations where Top and Stable PICS Potential Set 1 variants did not match, about 2-3% of variants matched between the Potential Set 1 variant in Stable PICS and Potential Sets 2 and 3 variants in Top PICS; or between the Potential Set 1 variant in Top PICS and Potential Sets 2 and 3 variants in Stable PICS (Supplementary File 2D). When looking at non-matching Potential Set 2 or Potential Set 3 variants, we do see an increase in matching frequencies (between 10-20%) between Potential Set 2 variants and other potential set variants between the different approaches. However, these percentages are still small compared to the matching frequencies we observed between corresponding potential sets (e.g., for simulations with one causal variant this was 70-90% between Top and Stable PICS Potential Set 1, and for simulations with two and three causal variants this was 55-78% and 57-79% respectively).
We next checked whether these “off-diagonal” matching variants corresponded to the true causal variants simulated. Here we find that the causal variant recovery rate is mostly less than the corresponding rate for diagonally matching variants, which together with the low matching frequency suggests that the enrichment of causal variants of “off-diagonal” matching variants is much weaker than in the diagonally matching approach. In other words, the fraction of non-matching (causal) variants due to the lack of multiple causal variant modeling is low.
We discuss these findings in Supplementary File 1 Section S2 (bottom of p.2).
(5) I wonder if you can combine the stability-based and the residualization-based approach, i.e., using the residualized phenotypes for the stability-based approach. Would that further improve the accuracy or not?
This is a good idea, thank you for suggesting it. We pursued this combined approach on simulated gene expression phenotypes, but did not observe significant gains in causal variant recovery (Figure 2B; Figure 2—figure supplements 2, 13 and 15). We reported this Results “Searching for matching variants between Top PICS and Stable PICS improves causal variant Recovery.”
“We thus explore ways to combine the residualization and stability-driven approaches, by considering (i) combining them into a single fine-mapping algorithm (we call the resulting procedure Combined PICS); and (ii) prioritizing matching variants between the two algorithms. Comparing the performance of Combined PICS against both Top and Stable PICS, however, we find no significant difference in its ability to recover causal variants (Figure 2B)...”
However, we also confirmed in our simulations that prioritizing matching variants between the two approaches led to gains in causal variant recovery (Figure 2D; Figure 2—figure supplements 4, 19, 20 and 22). We reported this Results “Searching for matching variants between Top PICS and Stable PICS improves causal variant Recovery.”
“On the other hand, matching variants between Top and Stable PICS are significantly more likely to be causal. Across all simulations, a matching variant in Potential Set 1 is 2.5X as likely to be causal than either a non-matching top or stable variant (Figure 2D) — a result that was qualitatively consistent even when we stratified simulations by SNR and number of causal variants simulated (Figure 2—figure supplements 19, 20 and 22)...”
This finding is consistent with our analysis of real GEUVADIS gene expression data, where we reported larger functional significance of matching variants relative to non-matching variants returned by either Top of Stable PICS.
(6) The authors state that confounding in cohorts with diverse ancestries poses potential difficulties in identifying the correct causal variants. However, I don't see that they directly address whether the stability approach is mitigating this. It is hard to say whether the stability approach is helping beyond what simpler post-hoc QC (e.g., thresholding) can do.
Thank you for raising this fair point. Here is a model we have in mind. Gene expression phenotypes (Y) can be explained by both genotypic effects (G, as in genotypic allelic dosage) and the environment (E): Y = G + E. However, both G and E depend on ancestry (A), so that Y = G|A+E|A. Suppose that the causal variants are shared across ancestries, so that (G|A=a)=G for all ancestries a. Suppose however that environments are heterogeneous by ancestry: (E|A=a) = e(a) for some function e that depends non-trivially on a. This would violate the exchangeability of exogenous E in the full sample, but by performing fine-mapping on each ancestry stratum, the exchangeability of exogenous E is preserved. This provides theoretical justification for the stability approach.
We next turned to simulations, where we investigated 1,440 simulated gene expression phenotypes capturing various ways in which ancestry induces heterogeneity in the exogenous E variable (simulation details in Lines 576-610 of Materials and Methods). We ran Stable PICS, as well as a version of PICS that did not residualize phenotypes or apply the stability principle. We observed that (i) causal variant recovery performance was not significantly different between the two approaches (Figure 2—figure supplements 24-32); but (ii) disagreement between the approaches can be considerable, especially when the signal-to-noise ratio is low (Supplementary File 2A). For example, in a set of simulations with three causal variants, with SNR = 0.11 and E heterogeneous by ancestry by letting E be drawn from N(2σ,σ<sup>2</sup>) for only GBR individuals (rest are N(0,σ<sup>2</sup>)), there was disagreement between Potential Set 1 and 2 variants in 25% of simulations — though recovery rates were similar (Probability of recovering at least one causal variant: 75% for Plain PICS and 80% for Stable PICS). These points suggest that confounding in cohorts can reduce power in methods not adjusting or accounting for ancestral heterogeneity, but can be remedied by approaches that do so. We report this analysis in Results “Simulations justify exploration of stability guidance”
In the current version of our work, we have evaluated, using both simulations and empirical evidence, different ways to combine approaches to boost causal variant recovery. Our simulation study shows that prioritizing matching variants across multiple methods improves causal variant recovery. On GEUVADIS data, where we might not know which variants are causal, we already demonstrated that matching variants are enriched for functional annotations. Therefore, our analyses justify that the adverse consequence of confounding on reducing fine-mapping accuracy can be mitigated by prioritizing matching variants between algorithms including those that account for stability.
(7) For non-matching variants, I wonder what the difference of posterior probabilities is between the stable and top variants in each method. If the difference is small, maybe it is due to noise rather than signal.
We have reported differences in posterior probabilities returned by Stable and Top PICS for GEUVADIS data; see Figure 3—figure supplement 1. For completeness, we compute the differences in posterior probabilities and summarize these differences both as histograms and as numerical summary statistics.
Potential Set 1
- Number of non-matching variants = 9,921
- Table of Summary Statistics of (Stable Posterior Probability – Top Posterior Probability)
Author response table 1.
- Histogram of (Stable Posterior Probability – Top Posterior Probability)
Author response image 1.
Potential Set 2
- Number of non-matching variants = 14,454
- Table of Summary Statistics of (Stable Posterior Probability – Top Posterior Probability)
Author response table 2.
- Histogram of (Stable Posterior Probability – Top Posterior Probability)
Author response image 2.
Potential Set 3
- Number of non-matching variants = 16,814
- Table of Summary Statistics of (Stable Posterior Probability – Top Posterior Probability)
Author response table 3.
- Histogram of (Stable Posterior Probability – Top Posterior Probability)
Author response image 3.
We also compared the difference in posterior probabilities between non-matching variants returned by Stable PICS and Top PICS for our 2,400 simulated gene expression phenotypes. Focusing on just Potential Set 1 variants, we find two equally likely scenarios, as demonstrated by two distinct clusters of points in a “posterior probability-posterior probability” plot. The first is, as pointed out, a small difference in posterior probability (points lying close to y=x). The second, however, reveals stable variants with very small posterior probability (of order 4 x 10<sup>–5</sup> to 0.05) but with a non-matching top variant taking on posterior probability well distributed along [0,1]. Moving down to Potential Sets 2 and 3, the distribution of pairs of posterior probabilities appears less clustered, indicating less tendency for posterior probability differences to be small ( Figure 2—figure supplement 8).
Here are the histograms and numerical summary statistics.
Potential Set 1
- Number of non-matching variants = 663 (out of 2,400)
- Table of Summary Statistics of (Stable Posterior Probability – Top Posterior Probability)
Author response table 4.
- Histogram of (Stable Posterior Probability – Top Posterior Probability)
Author response image 4.
Potential Set 2
Number of non-matching variants = 1,429 (out of 2,400)
- Table of Summary Statistics of (Stable Posterior Probability – Top Posterior Probability)
Author response table 5.
- Histogram of (Stable Posterior Probability – Top Posterior Probability)
Author response image 5.
Potential Set 3
- Number of non-matching variants = 1,810 (out of 2,400)
- Table of Summary Statistics of (Stable Posterior Probability – Top Posterior Probability)
Author response table 6.
- Histogram of (Stable Posterior Probability – Top Posterior Probability)
Author response image 6.
(8) It's a bit surprising that you observed matching variants with (stable) posterior probability ~ 0 (SFig. 1). What are the interpretations for these variants? Do you observe functional enrichment even for low posterior probability matching variants?
Thank you for this question. We have performed a thorough analysis of matching variants with very low stable posterior probability, which we define as having a posterior probability < 0.01 (Supplementary File 1 Section S11). Here, we briefly summarize the analysis and key findings.
Analysis
First, such variants occur very rarely — only 8 across all three potential sets in simulations, and 17 across all three potential sets for GEUVADIS (the latter variants are listed in Supplementary 2E). We begin interpreting these variants by looking at allele frequency heterogeneity by ancestry, support size — defined as the number of variants with positive posterior probability in the ALL slice* — and the number of slices including the stable variant (i.e., the stable variant reported positive posterior probability for the slice).
*Note that the stable variant posterior probability need not be at least 1/(Support Size). This is because the algorithm may have picked a SNP that has a lower posterior probability in the ALL slice (i.e., not the top variant) but happens to appear in the most number of other slices (i.e., a stable variant).
For variants arising from simulations, because we know the true causal variants, we check if these variants are causal. For GEUVADIS fine-mapped variants, we rely on functional annotations to compare their relative enrichment against other matching variants that did not have very low stable posterior probability.
Findings
While we caution against generalizing from observations reported here, which are based on very small sample sizes, we noticed the following. In simulations, matching variants with very low stable posterior probability are largely depleted in causal variants, although factors such as the number of slices including the stable variant may still be useful. In GEUVADIS, however, these variants can still be functionally enriched. We reported three examples in Supplementary File 1 Section S11 (pp. 8-9 of Supplement), where the variants were enriched in either VEP or biologically interpretable functional annotations, and were also reported in earlier studies. We partially reproduce our report below for convenience.
“However, we occasionally found variants that stand out for having large functional annotation scores. We list one below for each potential set.
- Potential Set 1 reported the variant rs12224894 from fine-mapping ENSG00000255284.1 (accession code AP006621.3) in Chromosome 11. This variant stood out for lying in the promoter flanking region of multiple cell types and being relatively enriched for GC content with a 75bp flanking region. This variant has been reported as a cis eQTL for AP006632 (using whole blood gene expression, rather than lymphoblastoid cell line gene expression in this study) in a clinical trial study of patients with systemic lupus erythematosus (Davenport et al., 2018). Its nearest gene is GATD1, a ubiquitously expressed gene that codes for a protein and is predicted to regulate enzymatic and catabolic activity. This variant appeared in all 6 slices, with a moderate support size of 23.
- Potential Set 2 reported the variant rs9912201 from fine-mapping ENSG00000108592.9 (mapped to FTSJ3) in Chromosome 17. Its FIRE score is 0.976, which is close to the maximum FIRE score reported across all Potential Set 2 matching variants. This variant has been reported as a SNP in high LD to a GWAS hit SNP rs7223966 in a pan-cancer study (Gong et al., 2018). This variant appeared in all 6 slices, with a moderate support size of 32.
- Potential Set 3 reported the variant rs625750 from fine-mapping ENSG00000254614.1 (mapped to CAPN1-AS1, an RNA gene) in Chromosome 11. Its FIRE score is 0.971 and its B statistic is 0.405 (region under selection), which lie at the extreme quantiles of the distributions of these scores for Potential Set 3 matching variants with stable posterior probability at least 0.01. Its associated mutation has been predicted to affect transcription factor binding, as computed using several position weight matrices (Kheradpour and Kellis, 2014). This variant appeared in just 3 slices, possibly owing to the considerable allele frequency difference between ancestries (maximum AF difference = 0.22). However, it has a small support size of 4 and a moderately high Top PICS posterior probability of 0.64.
To summarize, our analysis of GEUVADIS fine-mapped variants demonstrates that matching variants with very low stable posterior probability could still be functionally important, even for lower potential sets, conditional on supportive scores in interpretable features such as the number of slices containing the stable variant and the posterior probability support size…”
-
-
bookshelf.vitalsource.com bookshelf.vitalsource.com
-
Essentially, we have a “standards-based” educational system in America and many other countries. Standards-based, using commonly accepted objectives for student learning, is now a ubiquitous buzzword in education, if ever there was one. As we’ll see in detail in Chapter 2, standards frame what students should know and do—they formalize and standardize what gets taught and assessed. Every U.S. state has learning standards, with corresponding pacing guides and curriculum at the district level for implementation.
I chose this part because it helps explain how important standards are in schools today. It also helped me understand that standards decide what teachers teach and what students are expected to learn. This matters because it affects lessons, tests, and how students are measured across different schools and states.
-
-
biz.libretexts.org biz.libretexts.org
-
Revenue is the money a company receives by providing services or selling goods to customers. Costs are expenses for rent, salaries, supplies, transportation, and many other items that a company incurs from creating and selling goods and services. For example, some of the costs incurred by Microsoft in developing its software include expenses for salaries, facilities, and advertising. If Microsoft has money left over after it pays all costs, it has a profit. A company whose costs are greater than revenues shows a loss.
revenue, expenses, profits -- profits belong to ownership -- return on investment.
-
-
chrismarino.substack.com chrismarino.substack.com
-
What is ‘cool’ exactly, and what does it mean for art to be undergoing a crisis of cool? For Tatol, cool emerges out of the modernist injunction, explicit since Baudelaire, that the artist represent their time – not in the narrow sense of realistic representation, but as “sense phenomena and feeling” which enable the viewer to discover their own world anew. While some art, like that of Rembrandt and Leonardo, is the work of a master stylist whose immense skill is immediately apparent to the viewer, other art – Tatol offers the examples of Manet’s “Olympia”, Duchamp’s ‘readymade’ urinal, and Johns’s “Flag” – is not inherently impressive, and its significance emerges only in the context of the time in which it was made. “Olympia”, for instance, is important not for its exquisite rendering of a nude – in fact, in some ways it is rather ugly – but for the way its frank and realistic depiction of its subject scandalized the French bourgeoisie of the 1860s. ‘Cool’ thus designates an artwork’s relevance to its time, its ability to represent contemporary reality in an insightful and provocative manner. And more broadly, coolness is a distinctive feature of modern art, its capacity to generate ‘relevance’ its stock-in-trade, and the battle to define and express ‘the now’ a central organizing principle of art-world competition. A crisis of cool then describes an existential problem for contemporary art: its inability to generate this quality of relevance to the present moment.
This is the kind of paragraph that you expect to be fluffy nonsense but it's actually lucid and crisp
-
-
medium.com medium.com
-
Open practice, critical pedagogy and open resourcesWe are thrilled to welcome Doctor Sadia Habib back to the unit this year. Sadia is a Lecturer in Education at the Manchester Institute of Education, as well as the young people’s programme coordinator at the Manchester Museum. She has much experience in incorporating open practices and open/critical pedagogies in her work, and through this has led many novel projects with aspects of openness, including supporting young people to create blogs and zines — which she argues can be open resources.Sadia took OKHE in 2023/4 and wrote about some of her experience in her OKHE1 and OKHE2 posts. If you are attending our scheduled session, you will hear from Sadia. If you will not attend, we recommend you read her blog posts, and we will share a summary of her talk afterwards with PGCertHE participants.Sadia has also recently published Activism in the arts: Co-researching cultural inequalities with young people during the COVID-19 pandemic (open access and CC BY licensed).Please submit any questions for Sadia below. Comments are public and anonymous; please don’t share personal information.Add a comment (public, anonymous) above. If it doesn’t work, load it separately or comment on this post.We hope that by hearing from Sadia, you will see some ways in which openness intersects with her role/work, which may not be areas you had considered before. We hope too that this helps you to consider how openness intersects/could intersect with your work/role/practice.
Likely to need updating based on new guest speaker/s
-
David Wiley provides one framing of openness in education. a) Why might it be useful for individuals and institutions in higher education? b) Are there any aspects of effective education that the 5R’s don’t address? Comment in the second box below.💬 Contribute: Where does your practice fit with the Rs?Please share examples below of where any of your recent practice fits with any of the 5Rs. All comments are anonymous, don’t share personal info.Add a comment (public, anonymous) above. If it doesn’t work, load it separately or comment on this post.💬 Contribute: What’s good and what’s missing from the Rs?Please comment on why Wiley’s 5Rs might be useful for individuals and institutions in HE, and any aspects of effective education that the 5Rs don’t address. All comments are anonymous, don’t share personal information.
We might consider moving the first box in between instructions 2 and 3 because it would remove the need for participants to identify the right boxes for each instruction, could probably get rid of some repetition in the text as well
-
If you only have time for one thing, read Sadia Habib’s OKHE2 blog post: Open practice, critical pedagogy and zine making. Sadia discusses open principles for critical pedagogy, with a strong focus on social justice and inclusion of underrepresented voices. She also highlights the importance of collaborating with learners to create OERs. You might also find Sadia’s OKHE1 post interesting: Decolonising Education with Open Resources on Identity and Heritage.
Likely to change based on new speaker/s for 2026
-
-
medium.com medium.com
-
Openness at a publisher 📚Publishing is of fundamental importance in many aspects of openness, and in one way, to publish is to make something open. However, when we look below the surface, we see that it’s not quite so simple. What does it mean, therefore, to “make things open” as a publisher?In the session, we will hear from Emma Brennan, Editorial Director at Manchester University Press (MUP). Drawing on experience at one of the key players in the open publishing movement, Emma will share her insights and experience of the practicalities of openness at MUP. After the session, we will add a summary of the discussion here; in the meantime, read about MUP’s Open Access (OA) books and OA journals.Please submit any questions for Emma below. Comments are public and anonymous; please don’t share personal information.What questions do you have about openness at MUP?
May need changing if Emma cannot present - JB to confirm
-
The Soul of Open is In Danger
Check link - page not loading
-
-
localhost:8123 localhost:8123
-
the test set generated using stratified sampling has income category proportions almost identical to those in the full dataset, whereas the test set generated using purely random sampling is skewed.
This is interesting. Having a good grasp of the underlying data informs the strategies involved in picking the right test set. In this case, we started with a random sample but we might want to make sure the most important attribute are represented proportionally. This takes time talking to the right people and gettting the context behind the data.
-
the data has been scaled and capped at 15 (actually, 15.0001) for higher median incomes, and at 0.5 (actually, 0.4999) for lower median incomes
Thinking this might help now skew the model towards the extremes by constraining it to a reasonable range
-
this is a multiple regression problem, since the system will use multiple features to make a prediction (the district’s population, the median income, etc.). It is also a univariate regression problem, since we are only trying to predict a single value for each district.
Key point here is: multiple regression and not multivariate .. the _variate suffix only applies to when we have multiple targets we are trying to predict.
-
-
iowastatedaily.com iowastatedaily.com
-
“Digital badges are a great opportunity for Iowa State students to explore different avenues to build career-readiness skills,” Hageman said. “It’s not just a badge, it’s the work you put into earning it that stands out.”
Student quote
-
“LinkedIn is a tool I highly value as a college student,” Hageman said. “Gaining digital badges to display on my LinkedIn profile and résumé motivated me to pursue the Pathways workshops.”
Student quote
-
Mendee said a recent survey found that 85% of employers look for candidates who have both a degree and micro-credentials. “Micro-credentials give students a competitive edge by highlighting their skills in ways that stand out to employers and graduate schools,” Mendee said.
Transcripts communicate as they are intended to. MCs communicate in a different way, for a different audience.
-
-
medium.com medium.com
-
Open Research practices from the OOR.
Will need updating to ensure relevance to new speaker
-
Michael from the Office for Open Research (OOR), on how research can happen openly at the University of Manchester. Michael is both a graduate and former tutor of this unit! Please submit any questions for Michael below. Comments are public and anonymous; please don’t share personal information.What questions do you have about open research practices at Manchester?
Will need updating with details for new guest speaker
-
Draw/Write/Discuss — press the buttons to change functions. If it doesn’t work, try opening it separately, or use another app/website/device or pen and paper. This tool was developed for OKHE, is open source, open access, openly licensed, uses open frameworks, remixes an open tool, and is hosted on an open platform!
GlitchBlog no longer running? May need alternative
-
-
human.libretexts.org human.libretexts.org
-
A theme is not the plot of the story. It is the underlying truth that is being conveyed in the story. Themes can be universal, meaning they are understood by readers no matter what culture or country the readers are in. Common themes include coming of age, circle of life, prejudice, greed, good vs. evil, beating the odds, etc.
Theme is the underlying meaning of the story
-
First-person point of view means that one of the characters in the story will narrate–give an account–of the story. The narrator may be the protagonist, the main character. Writing in first-person point of view brings the readers closer to the story. They can read it as if they are the character because personal pronouns like I, me, my, we, us, and our are used. Third-person point of view means that the narrator is not in the story. The third-person narrator is not a character. Third-person point of view can be done two ways: Third-person limited Third-person omniscient Third-person limited means that the narrator limits him/herself by being able to be in one character’s thoughts. Whereas, third-person omniscient means the narrator has unlimited ability to be in various character’s thoughts. Writing in third-person point of view removes readers from the story because of the pronouns he, she, it, him, her, his, hers, they, them, and theirs.
First person means one of your characters is telling the story, while third person means the writer is telling the story and the writer is either limited or omniscient and can see the character's thoughts
-
Conflict is the struggle between two entities. In story writing the main character, also known as the protagonist, encounters a conflict with the antagonist, which is an adversary. The conflict may be one of six kinds: Character vs. character Character vs. nature or natural forces Character vs. society or culture Character vs. machine or technology Character vs. God Character vs himself or herself
Conflict pits your characters against other characters, nature, society, technology, god, or the character themselves
-
Denouement or resolution provides closure to the story. It ties up loose ends in the story.
Denouement is about the consequences of the events in the climax
-
Falling action includes the events that unfold after the climax. This usually creates an emotional response from the reader.
Falling action is victory or defeat
-
Climax is the turning point in the story. Usually, it is a single event with the greatest intensity and uncertainty. The main character must contend with the problem at this point.
Climax is the big battle with Thanos at the end
-
Rising action includes the events that the main character encounters. Each event, developed in separate scenes, makes the problem more complex.
Rising action happens when the MC decides to take action and faces trials and tribulations
-
Exposition is an introduction to the characters, time, and the problem. At the point where exposition moves into rising action a problem, sometimes called an inciting incident, occurs for the main character to handle or solve. This creates the beginning of the story.
Exposition introduces all the important facts about your character, typically through their actions as opposed to words.
-
Plot is the order of events in the story. The plot usually follows a particular structure called Freytag’s Pyramid. Gustav Freytag, a German playwright who lived during the 1800s, identified this structure. Freytag’s Pyramid has five parts: exposition, rising action, climax, falling action, and denouement, also known as resolution. See Figure 3.1.
Plot is exposition, rising action, climax, falling action, denouement (resolution)
-
Writers write about places they are familiar with. If they aren’t familiar with the place, then they need to research it in order to be accurate about the place.
Write settings you already know
-
Setting can function as a main force that the characters encounter, such as a tornado or flood, or a setting can play a minor role such as setting the mood. Often times, the setting can reveal something about the main character as he/she functions in that place and time period.
The setting can be the antagonist like man against nature or setting can be simply for setting the mood.
-
Setting is where and when the story takes place. It includes the following: The immediate surroundings of the characters such as props in a scene: trees, furniture, food, inside of a house or car, etc. The time of day such as morning, afternoon, or night. The weather such as cloudy, sunny, windy, snow, or rain, etc. The time of year, particularly the seasons: fall, winter, summer, spring. The historical period such as what century or decade the story takes place. The geographical location including the city, state, country, and possibly even the universe, if the writer is writing science fiction.
Setting is the surroundings and props, period in time. weather, geography and so on
-
If writers write about characters outside their own culture, they need to do research so as not to misrepresent a particular culture. The same is also true of characters, who have illnesses. The writer may need to research the illness and treatment for it in order to be accurate about it.
The more research a writer does, the more authentic the characters will be
-
When discussing stories with other readers and writers or when writing an analysis of a story, fictional characters can be described as static or developing. Static means the character stays the same throughout the story. They do not change. Developing, also called dynamic, means the character changes. The change may impact the character’s beliefs, attitudes, or actions. The change may be small or large. This change occurs because the character experiences an epiphany, an insight about life.
A static character doesn't change their beliefs or attitude while a dynamic one does
-
On the other hand, the round characters play an important role, often the lead roles in stories. They are complex, dimensional, and well-developed. The stories are about them; therefore, pages of writing will be about them. They often change by going through a life-changing experience as the story unfolds.
Round characters are usually main characters and they have complexity and usually change by the end of the story.
-
Characters are the people, animals, or aliens in the story. Readers come to know the characters through what they say, what they think, and how they act. E. M. Forster, an English novelist, identified that characters are either flat or round. Flat characters do not play important roles in the stories. They often have only one or two traits with little description about them. A flat character may even be a stock character, which is a stereotypical figure that is easily recognized by readers, for example, the mad scientist or the evil stepmother.
Some characters are 2d and some are 3d but both are important. A 2d character is a stereotype or background characters
-
Fiction is make-believe, invented stories. They may be short stories, fables, vignettes, plays, novellas, or novels. Although writers may base a character on people they have met in real life, the characters and the experiences that the character faces in the story are not real. So, how does a writer write fiction? Characters, setting, plot, conflict, point of view, and theme are six key elements for writing fiction.
Fiction is made up stories that come in many forms and lengths. They involve characters, setting, plot, conflict, and so on.
-
-
www.biorxiv.org www.biorxiv.org
-
eLife Assessment
This fundamental work reveals that the accessibility of the unstructured C-terminal tails of α- and β-tubulins differs with the state of the microtubule lattice. Their accessibility increases with the expansion of the lattice induced by GTP and certain MAPs, which can then dictate the subsequent interactions between MAPs and microtubules, and post-translational modifications of tubulin tails. The evidence supporting the conclusion is compelling, although the characterisation of the probes does not answer whether they directly affect the lattice or expose the C-terminal tails of tubulin. The probes can be used as tools in the future to study differences in microtubule lattice assembly under different conditions both in vitro and in vivo. This work will be of great interest to the cytoskeleton field.
-
Reviewer #1 (Public review):
Summary:
This is a careful and comprehensive study demonstrating that effector-dependent conformational switching of the MT lattice from compacted to expanded deploys the alpha tubulin C-terminal tails so as to enhance their ability to bind interactors.
Strengths:
The authors use 3 different sensors for the exposure of the alpha CTTs. They show that all 3 sensors report exposure of the alpha CTTs when the lattice is expanded by GMPCPP, or KIF1C, or a hydrolysis-deficient tubulin. They demonstrate that expansion-dependent exposure of the alpha CTTs works in tissue culture cells as well as in vitro.
Appraisal:
The authors have gone to considerable lengths to test their hypothesis that microtubule expansion favours deployment of the alpha tubulin C-terminal tail, allowing its interactors, including detyrosinase enzymes, to bind. There is a real prospect that this will change thinking in the field. One very interesting possibility, touched on by the authors, is that the requirement for MAP7 to engage kinesin with the MT might include a direct effect of MAP7 on lattice expansion.
Impact:
The possibility that the interactions of MAPS and motors with a particular MT or region feed forward to determine its future interaction patterns is made much more real. Genuinely exciting.
-
Reviewer #2 (Public review):
The unstructured α- and β-tubulin C-terminal tails (CTTs), which differ between tubulin isoforms, extend from the surface of the microtubule, are post-translationally modified, and help regulate the function of MAPs and motors. Their dynamics and extent of interactions with the microtubule lattice are not well understood. Hotta et al. explore this using a set of three distinct probes that bind to the CTTs of tyrosinated (native) α-tubulin. Under normal cellular conditions, these probes associate with microtubules only to a limited extent, but this binding can be enhanced by various manipulations thought to alter the tubulin lattice conformation (expanded or compact). These include small-molecule treatment (Taxol), changes in nucleotide state, and the binding of microtubule-associated proteins and motors. Overall, the authors conclude that microtubule lattice "expanders" promote probe binding, suggesting that the CTT is generally more accessible under these conditions. Consistent with this, detyrosination is enhanced. Mechanistically, molecular dynamics simulations indicate that the CTT may interact with the microtubule lattice at several sites, and that these interactions are affected by the tubulin nucleotide state.
Strengths and weaknesses:
Key strengths of the work include the use of three distinct probes that yield broadly consistent findings, and a wide variety of experimental manipulations (drugs, motors, MAPs) that collectively support the authors' conclusions, alongside a careful quantitative approach.
The challenges of studying the dynamics of a short, intrinsically disordered protein region within the complex environment of the cellular microtubule lattice, amid numerous other binders and regulators, should not be understated. While it is very plausible that the probes report on CTT accessibility as proposed, the possibility of confounding factors (e.g., effects on MAP or motor binding) cannot be ruled out. Sensitivity to the expression level clearly introduces additional complications. Likewise, for each individual "expander" or "compactor" manipulation, one must consider indirect consequences (e.g., masking of binding sites) in addition to direct effects on the lattice; however, this risk is mitigated by the collective observations all pointing in the same direction.
The discussion does a good job of placing the findings in context and acknowledging relevant caveats and limitations. Overall, this study introduces an interesting and provocative concept, well supported by experimental data, and provides a strong foundation for future work. This will be a valuable contribution to the field.
-
Reviewer #3 (Public review):
Summary:
In this study, the authors investigate how the structural state of the microtubule lattice influences the accessibility of the α-tubulin C-terminal tail (CTT). By developing and applying new biosensors, they reveal that the tyrosinated CTT is largely inaccessible under normal conditions but becomes more accessible upon changes to the tubulin conformational state induced by taxol treatment, MAP expression, or GTP-hydrolysis-deficient tubulin. The combination of live imaging, biochemical assays, and simulations suggests that the lattice conformation regulates the exposure of the CTT, providing a potential mechanism for modulating interactions with microtubule-associated proteins. The work addresses a highly topical question in the microtubule field and proposes a new conceptual link between lattice spacing and tail accessibility for tubulin post-translational modification. Future work is required to distinguish CTT exposure in the microtubule lattice is sensitive to additional factors present in vivo but not in vitro.
Strengths:
(1) The study targets a highly relevant and emerging topic-the structural plasticity of the microtubule lattice and its regulatory implications.
(2) The biosensor design represents a methodological advance, enabling direct visualization of CTT accessibility in living cells.
(3) Integration of imaging, biochemical assays, and simulations provides a multi-scale perspective on lattice regulation.
(4) The conceptual framework proposed lattice conformation as a determinant of post-translational modification accessibility is novel and potentially impactful for understanding microtubule regulation.
[Editors' note: the authors have responded to the reviewers and this version was assessed by the editors.]
-
Author response:
The following is the authors’ response to the original reviews
Public Reviews:
Reviewer #1 (Public review):
Summary:
This is a careful and comprehensive study demonstrating that effector-dependent conformational switching of the MT lattice from compacted to expanded deploys the alpha tubulin C-terminal tails so as to enhance their ability to bind interactors.
Strengths:
The authors use 3 different sensors for the exposure of the alpha CTTs. They show that all 3 sensors report exposure of the alpha CTTs when the lattice is expanded by GMPCPP, or KIF1C, or a hydrolysis-deficient tubulin. They demonstrate that expansion-dependent exposure of the alpha CTTs works in tissue culture cells as well as in vitro.
Weaknesses:
There is no information on the status of the beta tubulin CTTs. The study is done with mixed isotype microtubules, both in cells and in vitro. It remains unclear whether all the alpha tubulins in a mixed isotype microtubule lattice behave equivalently, or whether the effect is tubulin isotype-dependent. It remains unclear whether local binding of effectors can locally expand the lattice and locally expose the alpha CTTs.
Appraisal:
The authors have gone to considerable lengths to test their hypothesis that microtubule expansion favours deployment of the alpha tubulin C-terminal tail, allowing its interactors, including detyrosinase enzymes, to bind. There is a real prospect that this will change thinking in the field. One very interesting possibility, touched on by the authors, is that the requirement for MAP7 to engage kinesin with the MT might include a direct effect of MAP7 on lattice expansion.
Impact:
The possibility that the interactions of MAPS and motors with a particular MT or region feed forward to determine its future interaction patterns is made much more real. Genuinely exciting.
We thank the reviewer for their positive response to our work. We agree that it will be important to determine if the bCTT is subject to regulation similar to the aCTT. However, this will first require the development of sensors that report on the accessibility of the bCTT, which is a significant undertaking for future work.
We also agree that it will be important to examine whether all tubulin isotypes behave equivalently in terms of exposure of the aCTT in response to conformational switching of the microtubule lattice.
We thank the reviewer for the comment about local expansion of the microtubule lattice. We believe that Figure 3 does show that local binding of effectors can locally expand the lattice and locally expose the alpha-CTTs. We have added text to clarify this.
Reviewer #2 (Public review):
The unstructured α- and β-tubulin C-terminal tails (CTTs), which differ between tubulin isoforms, extend from the surface of the microtubule, are post-translationally modified, and help regulate the function of MAPs and motors. Their dynamics and extent of interactions with the microtubule lattice are not well understood. Hotta et al. explore this using a set of three distinct probes that bind to the CTTs of tyrosinated (native) α-tubulin. Under normal cellular conditions, these probes associate with microtubules only to a limited extent, but this binding can be enhanced by various manipulations thought to alter the tubulin lattice conformation (expanded or compact). These include small-molecule treatment (Taxol), changes in nucleotide state, and the binding of microtubule-associated proteins and motors. Overall, the authors conclude that microtubule lattice "expanders" promote probe binding, suggesting that the CTT is generally more accessible under these conditions. Consistent with this, detyrosination is enhanced. Mechanistically, molecular dynamics simulations indicate that the CTT may interact with the microtubule lattice at several sites, and that these interactions are affected by the tubulin nucleotide state.
Strengths:
Key strengths of the work include the use of three distinct probes that yield broadly consistent findings, and a wide variety of experimental manipulations (drugs, motors, MAPs) that collectively support the authors' conclusions, alongside a careful quantitative approach.
Weaknesses:
The challenges of studying the dynamics of a short, intrinsically disordered protein region within the complex environment of the cellular microtubule lattice, amid numerous other binders and regulators, should not be understated. While it is very plausible that the probes report on CTT accessibility as proposed, the possibility of confounding factors (e.g., effects on MAP or motor binding) cannot be ruled out. Sensitivity to the expression level clearly introduces additional complications. Likewise, for each individual "expander" or "compactor" manipulation, one must consider indirect consequences (e.g., masking of binding sites) in addition to direct effects on the lattice; however, this risk is mitigated by the collective observations all pointing in the same direction.
The discussion does a good job of placing the findings in context and acknowledging relevant caveats and limitations. Overall, this study introduces an interesting and provocative concept, well supported by experimental data, and provides a strong foundation for future work. This will be a valuable contribution to the field.
We thank the reviewer for their positive response to our work. We are encouraged that the reviewer feels that the Discussion section does a good job of putting the findings, challenges, and possibility of confounding factors and indirect effects in context.
Reviewer #3 (Public review):
Summary:
In this study, the authors investigate how the structural state of the microtubule lattice influences the accessibility of the α-tubulin C-terminal tail (CTT). By developing and applying new biosensors, they reveal that the tyrosinated CTT is largely inaccessible under normal conditions but becomes more accessible upon changes to the tubulin conformational state induced by taxol treatment, MAP expression, or GTP-hydrolysis-deficient tubulin. The combination of live imaging, biochemical assays, and simulations suggests that the lattice conformation regulates the exposure of the CTT, providing a potential mechanism for modulating interactions with microtubule-associated proteins. The work addresses a highly topical question in the microtubule field and proposes a new conceptual link between lattice spacing and tail accessibility for tubulin post-translational modification.
Strengths:
(1) The study targets a highly relevant and emerging topic-the structural plasticity of the microtubule lattice and its regulatory implications.
(2) The biosensor design represents a methodological advance, enabling direct visualization of CTT accessibility in living cells.
(3) Integration of imaging, biochemical assays, and simulations provides a multi-scale perspective on lattice regulation.
(4) The conceptual framework proposed lattice conformation as a determinant of post-translational modification accessibility is novel and potentially impactful for understanding microtubule regulation.
Weaknesses:
There are a number of weaknesses in the paper, many of which can be addressed textually. Some of the supporting evidence is preliminary and would benefit from additional experimental validation and clearer presentation before the conclusions can be considered fully supported. In particular, the authors should directly test in vitro whether Taxol addition can induce lattice exchange (see comments below).
We thank the reviewer for their positive response to our work. We have altered the text and provided additional experimental validation as requested (see below).
Recommendations for the authors:
Reviewer #1 (Recommendations for the authors):
(1) The resolution of the figures is insufficient.
(2) The provision of scale bars is inconsistent and insufficient.
(3) Figure 1E, the scale bar looks like an MT.
(4) Figure 2C, what does the grey bar indicate?
(5) Figure 2E, missing scale bar.
(6) Figure 3 C, D, significance brackets misaligned.
(7) Figure 3E, consider using the same alpha-beta tubulin / MT graphic as in Figure 1B.
(8) Figure 5E, show cell boundaries for consistency?
(9) Figure 6D, stray box above the y-axis.
(11) Figure S3A, scale bar wrong unit again.
(12) S3B "fixed" and mount missing scale bar in the inset.
(13) S4 scale bars without scale, inconsistency in scale bars throughout all the figures.
We apologize for issues with the figures. We have corrected all of the issues indicated by the reviewer.
(10) Figure 6F, surprising that 300 mM KCL washes out rigor binding kinesin
We thank the reviewer for this important point. To address the reviewer’s concern, we have added a new supplementary figure (new Figure 6 – Figure Supplement 1) which shows that the washing step removes strongly-bound (apo) KIF5C(1-560)-Halo<sup>554</sup> protein from the microtubules. In addition, we have made a correction to the Materials and Methods section noting that ATP was added in addition to the KCl in the wash buffer. We apologize for omitting this detail in the original submission. We also added text noting that the wash out step was based on Shima et al., 2018 where the observation chamber was washed with either 1 mM ATP and 300 mM K-Pipes or with 10 mM ATP and 500 mM K-Pipes buffer. In our case, the chamber was washed with 3 mM ATP and 300 mM KCl. It is likely that the addition of ATP facilitates the detachment of strongly-bound KIF5C.
(14) Supplementary movie, please identify alpha and beta tubules for clarity. Please identify residues lighting up in interaction sites 1,2 & 3.
Thank you for the suggestions. We have made the requested changes to the movie.
Reviewer #2 (Recommendations for the authors):
There appear to have been some minor issues (perhaps with .pdf conversion) that leave some text and images pixelated in the .pdf provided, alongside some slightly jarring text and image positioning (e.g., Figure 5E panels). The authors should carefully look at the figures to ensure that they are presented in the clearest way possible.
We apologize for these issues with the figures. We have reviewed the figures carefully to ensure that they are presented in the clearest way possible.
The authors might consider providing a more definitive structural description of compact vs expanded lattice, highlighting what specific parameters are generally thought to change and by what magnitude. Do these differ between taxol-mediated expansion or the effects of MAPs?
Thank you for the suggestion. We have added additional information to the Introduction section.
Reviewer #3 (Recommendations for the authors):
(1) Figure 1 should include a schematic overview of all constructs used in the study. A clear illustration showing the probe design, including the origin and function of each component (e.g., tags, domains), would improve clarity.
Thank you for the suggestion. We have added new illustrations to Figure 1 showing the origin and design (including domains and tags) of each probe.
(2) Add Western blot data for the 4×CAP-Gly construct to Figure 1C for completeness.
We thank the reviewer for this suggestion. We carried out a far-western blot using the purified 4xCAPGly-mEGFP protein to probe GST-Y, GST-DY, and GST-DC2 proteins (new Figure 1 – Figure Supplement 1C). We note that some bleed-through signal can be seen in the lanes containing GST-ΔY and GST-ΔC2 protein due to the imaging requirements and exposure needed to visualize the 4xCAPGly-mEGFP protein. Nevertheless, the blot shows that the purified CAPGly sensor specifically recognizes the native (tyrosinated) CTT sequence of TUBA1A.
(3) Essential background information on the CAP-Gly domain, SXIP motif, and EB proteins is missing from the Introduction. These concepts appear abruptly in the Results and should be properly introduced.
Thank you for the suggestion. We have added additional information to the Introduction section about the CAP-Gly domain. However, we feel that introducing the SXIP motif and EB proteins at this point would detract from the flow of the Introduction and we have elected to retain this information in the Results section when we detail development of the 4xCAPGly probe.
(4) In Figure 2E, it remains possible that the CAP-Gly domain displacement simply follows the displacement of EB proteins. An experiment comparing EB protein localization upon Taxol treatment would clarify this relationship.
We thank the reviewer for raising this important point. To address the reviewer’s concern, we utilized HeLa cells stably expressing EB3-GFP. We performed live-cell imaging before and after Taxol addition (new Figure 2 – Figure Supplement 1C). EB3-EGFP was lost from the microtubule plus ends within minutes and did not localize to the now-expanded lattice.
(5) Statements such as "significantly increased" (e.g., line 195) should be replaced with quantitative information (e.g., "1.5-fold increase").
We have made the suggested changes to the text.
(6) Phrases like "became accessible" should be revised to "became more accessible," as the observed changes are relative, not absolute. The current wording implies a binary shift, whereas the data show a modest (~1.5-fold) increase.
We have made the suggested changes to the text.
(7) Similarly, at line 209, the terms "minimally accessible" versus "accessible" should be rephrased to reflect the small relative change observed; saturation of accessibility is not demonstrated.
We have made the suggested changes to the text.
(8) Statements that MAP7 "expands the lattice" (line 222) should be made cautiously; to my knowledge, that has not been clearly established in the literature.
We thank the reviewer for this important comment. We have added text indicating that MAP7’s ability to induce or presence an expanded lattice has not been clearly established.
(9) In Figures 3 and 4, the overexpression of MAP7 results in a strikingly peripheral microtubule network. Why is there this unusual morphology?
The reviewer raises an interesting question. We are not sure why the overexpression of MAP7 results in a strikingly peripheral microtubule network but we suspect this is unique to the HeLa cells we are using. We have observed a more uniform MAP7 localization in other cell types [e.g. COS-7 cells (Tymanskyj et al. 2018), consistent with the literature [e.g. BEAS-2B cells (Shen and Ori-McKenney 2024), HeLa cells (Hooikaas et al. 2019)].
(10) In Supplementary Figure 5C, the Western blot of detyrosination levels is inconsistent with the text. Untreated cells appear to have higher detyrosination than both wild-type and E254A-overexpressing cells. Do you have any explanation?
We thank the reviewer for this important comment. We do not have an explanation at this point but plan to revisit this experiment. Unfortunately, the authors who carried out this work recently moved to a new institution and it will be several months before they are able to get the cell lines going and repeat the experiment. We thus elected to remove what was Supp Fig 5C until we can revisit the results. We believe that the important results are in what is now Figure 5 - Figure Supplement 1A,B which shows that the expression levels of the WT and E254E proteins are similar to each other.
(11) The image analysis method in Figures 5B and 5D requires clarification. It appears that "density" was calculated from skeletonized probe length over total area, potentially using a strict intensity threshold. It looks like low-intensity binding has been excluded; otherwise, the density would be the same from the images. If so, this should be stated explicitly. A more appropriate analysis might skeletonize and integrate total fluorescence intensity relative to the overall microtubule network.
We have added additional information to the Materials and Methods section to clarify the image analysis. We appreciate the reviewer’s valuable feedback and the suggestion to use the integrated total fluorescence intensity, which is a theoretically sound approach. While we agree that integrated intensity is a valid metric for specific applications, its appropriate use depends on two main preconditions:
(1) Consistent microscopy image acquisition conditions.
(2) Consistent probe expression levels across all cells and experiments.
We successfully maintained consistent image acquisition conditions (e.g., exposure time) throughout the experiment. However, despite generating a stably-expressing sensor cell lines to minimize variation, there remains an inherent, biological variability in probe expression levels between individual cells. Integrated intensity is highly susceptible to this cell-to-cell variability. Relying on it would lead to a systematic error where differences in the total amount of expressed probe would be mistaken for differences in Y-aCTT accessibility.
The density metric (skeletonized probe length / total cell area) was deliberately chosen as it serves as a geometric measure rather than an intensity-based normalization. The density metric quantifies the proportion of the microtubule network that is occupied by Y-aCTT-labeled structures, independent of fluorescence intensity. Thus, the density metric provides a more robust and interpretable measure of Y-aCTT accessibility under the variable expression conditions inherent to our experimental system. Therefore, we believe that this geometric approach represents the most appropriate analysis for our image dataset.
(12) In Figure 5D, the fold-change data are difficult to interpret due to the compressed scale. Replotting is recommended. The text should also discuss the relative fold changes between E254A and Taxol conditions, Figure 2H.
We appreciate the reviewer's insightful comment. We agree that the presence of significant outliers led to a compressed Y-axis scale in Figure 5D, obscuring the clear difference between the WT-tubulin and E254A-tubulin groups. As suggested, we have replotted Figure 5D using a broken Y-axis to effectively expand the relevant lower range of the data while still accurately representing all data points, including the outliers. We believe that the revised graph significantly enhances the clarity and interpretability of these results. For Figure 2, we have added the relative fold changes to the text as requested.
(13) Figure 6. The authors should directly test in vitro whether Taxol addition can induce lattice exchange, for example, by adding Taxol to GDP-microtubules and monitoring probe binding. Including such an assay would provide critical mechanistic evidence and substantially strengthen the conclusions. I was waiting for this experiment since Figure 2.
We thank the reviewer for this suggestion. As suggested, we generated GDP-MTs from HeLa tubulin and added it to two flow chambers. We then flowed in the YL1/2<sup>Fab</sup>-EGFP probe into the chambers in the presence of DMSO (vehicle control) or Taxol. Static images were taken and the fluorescence intensity of the probe on microtubules in each chamber was quantified. There was a slight but not statistically significant difference in probe binding between control and Taxol-treated GDP-MTs (Author response image 1). While disappointing, these results underscore our conclusion (Discussion section) that microtubule assembly in vitro may not produce a lattice state resembling that in cells, either due to differences in protofilament number and/or buffer conditions and/or the lack of MAPs during polymerization.
Author response image 1.
References
Hooikaas, P. J., Martin, M., Muhlethaler, T., Kuijntjes, G. J., Peeters, C. A. E., Katrukha, E. A., Ferrari, L., Stucchi, R., Verhagen, D. G. F., van Riel, W. E., Grigoriev, I., Altelaar, A. F. M., Hoogenraad, C. C., Rudiger, S. G. D., Steinmetz, M. O., Kapitein, L. C. and Akhmanova, A. (2019). MAP7 family proteins regulate kinesin-1 recruitment and activation. J Cell Biol, 218, 1298-1318.
Shen, Y. and Ori-McKenney, K. M. (2024). Microtubule-associated protein MAP7 promotes tubulin posttranslational modifications and cargo transport to enable osmotic adaptation. Dev Cell, 59, 1553-1570.
Tymanskyj, S. R., Yang, B. H., Verhey, K. J. and Ma, L. (2018). MAP7 regulates axon morphogenesis by recruiting kinesin-1 to microtubules and modulating organelle transport. Elife, 7.
-
-
cghuisunshine.github.io cghuisunshine.github.io
-
Karyotype
Below is a complete explanation of “Karyotype” with English + Chinese plus appropriate images that clearly show what a karyotype looks like.
🧬 Karyotype(核型)
📸 Images of Karyotypes
1. Human Karyotype (46 chromosomes, 23 pairs)
2. Labeled karyotype showing chromosome pairs

3. Process of making a karyotype

🇬🇧 English Explanation
What is a karyotype?
A karyotype is an organized photograph of all the chromosomes in a cell, arranged in matching pairs from largest to smallest. It shows:
- the number of chromosomes
- their sizes
- their shapes
- the position of the centromeres
- whether they appear in homologous pairs
- the sex chromosomes (XX or XY)
A karyotype is often used to check for genetic abnormalities.
How chromosomes are arranged in a karyotype
-
Chromosomes are photographed during metaphase, when they are condensed and visible.
-
They are cut out (digitally) and arranged by:
-
Pair (homologous chromosomes)
- Size (largest first)
- Centromere position
-
Banding pattern
-
The final chart shows 23 pairs in humans:
-
22 pairs of autosomes
- 1 pair of sex chromosomes (XX or XY)
What a karyotype is used for
Doctors and scientists use karyotypes to detect:
1. Chromosome number abnormalities
- Down syndrome (Trisomy 21) – an extra chromosome 21
- Turner syndrome (XO) – missing one X chromosome
- Klinefelter syndrome (XXY) – extra X in males
2. Structural abnormalities
- deletions
- duplications
- translocations
- inversions
3. Determining biological sex
- XX → typically female
- XY → typically male
🇨🇳 中文解释(Chinese Explanation)
什么是核型(karyotype)?
核型 是把细胞中所有染色体 拍照并按固定顺序排列 所得到的图像。 它可以展示:
- 染色体的 数量
- 染色体的 大小
- 形状和带纹模式
- 着丝点的位置
- 同源染色体是否成对
- 性染色体 XX 或 XY
核型常用于检测 遗传疾病或染色体异常。
染色体如何在核型中排列?
-
染色体在 中期(metaphase) 最清晰,因此在该阶段拍照。
-
然后按以下顺序排列:
-
同源染色体成对
- 按 从大到小 排列
- 根据 着丝点位置
-
根据 染色带纹(banding patterns)
-
人类共有 23 对染色体:
-
22 对常染色体(autosomes)
- 1 对性染色体(XX 或 XY)
核型用于哪些检测?
1. 染色体数目异常
- 唐氏综合征(21 三体) – 多了一条 21 号染色体
- 特纳综合征(XO) – 缺少一条 X 染色体
- 克氏综合征(XXY) – 男性多一条 X
2. 染色体结构异常
- 缺失(deletion)
- 重复(duplication)
- 转位(translocation)
- 倒位(inversion)
3. 判断生物学性别
- XX → 通常为女性
- XY → 通常为男性
🧩 Want More?
I can also provide:
- A comparison chart: karyotype vs genotype
- A step-by-step diagram of how a karyotype is made
- A worksheet-style labeled karyotype practice (English + Chinese)
- Explanation of Down syndrome karyotype (Trisomy 21) with pictures
Just tell me!
-
Homologous Chromosomes
Here’s a complete explanation of Homologous Chromosomes with appropriate labeled images to help you see and understand the concept clearly.
🧬 Homologous Chromosomes — With Images


📌 What the Images Show
- Pair of homologous chromosomes: Two chromosomes of similar shape/size (one from each parent) shown side by side.
- Diagram of homologous pairing and crossing-over: Shows how non-sister chromatids can exchange genetic material in meiosis.
- Detailed structure with sister chromatids: Each duplicated chromosome has two sister chromatids.
🧠 English Explanation
Definition
Homologous chromosomes are paired chromosomes in a diploid cell that:
-
Have the same genes at the same positions (loci) — even if the versions (alleles) may differ. (Wikipedia)
-
Are similar in size and shape They align with each other during meiosis. (khanacademy.org)
-
Come as one from each parent In humans, cells typically have 23 homologous pairs (46 total chromosomes). (ThoughtCo)
How They Appear
- Each homologous chromosome is a single chromosome before DNA replication.
- After DNA replicates, each one consists of two identical sister chromatids joined at a centromere.
Importance in Meiosis
- Homologous pairs pair up (synapsis) in meiosis I. (Biology LibreTexts)
- During this pairing, crossing over can occur — where non-sister chromatids exchange DNA. This creates genetic variation in gametes. (Genomics Education Programme)
🈶 中文解释(Chinese Explanation)
定义:什么是同源染色体
同源染色体 是二倍体细胞中 成对出现 的染色体:
-
在 相同位置(基因座)上有相同的基因, 但这些基因可能是不同形式(等位基因)。 (Wikipedia)
-
它们 大小、形状、带纹很相似。 (khanacademy.org)
-
每对染色体中,一条来自母亲,一条来自父亲。 (ThoughtCo)
它们的外观
- 在 DNA 复制之前,每一条是单一染色体。
- 复制后,每条都变成 两个姐妹染色单体(sister chromatids),通过 着丝点 连接起来。
在减数分裂中的角色
- 在 减数分裂 I 时,同源染色体会 配对联会。 (Biology LibreTexts)
- 在配对过程中,非姐妹染色单体之间可能发生交叉互换(crossing over),促使基因重新组合,从而增加遗传多样性。 (Genomics Education Programme)
🧩 Key Features of Homologous Chromosomes
| Feature | English | 中文 | | ------------------- | -------------------------------- | ---------------- | | Pair of chromosomes | Same genes, same positions | 含有相同基因且位置一致的染色体对 | | Origin | One from mother, one from father | 来自父母双方各一条 | | Replication | Becomes two sister chromatids | 复制后形成两个姐妹染色单体 | | Role in meiosis | Pairing & crossing-over | 在减数分裂中配对并发生交叉互换 | | Genetic variation | Increased by crossing-over | 由交叉互换增加遗传多样性 |
If you want, I can also show you diagrams of meiosis I focusing on homologous chromosome behavior, or a labeled worksheet style diagram in both English and Chinese!
-
-
paperlesspaper.de paperlesspaper.de
-
load a new image
test
-
-
digitalfreedom.substack.com digitalfreedom.substack.com
-
The three dimensional framework can be visualized as a 3D coordinate system. The x-axis represents the Data Plane, with each point a unique hash of a dataset. The y-axis represents the Account Plane, with each point a collision-free public key. The z-axis represents the Time Plane, with each point a timestamped hash recorded on a blockchain or hash-registry.
GraphTopologySenseMaking
Generally graphs are visualised in an abstract relational space. You are describing here a 3 dimensional map (or rather a 2-dimensional map with a time dimension). the image above it does not seem to reflect such a space and the added benefits of working in a concrete mapable space are not immediately clear to me.
-
-
human.libretexts.org human.libretexts.org
-
What stages of the writing process do you use? Which are your strengths? And which are your weaknesses?
I'm terrible at writing plots. Truly bad
-
How do you write your first draft? Are you a think-write writer or a write-write writer?
I do both but when I am writing for purely the hell of it, I just plow forward and write until I get tired.
-
What genres do you prefer to write? Why?
I like Sci-fi and dystopias because it says something about the human condition and it isn't afraid to get deeply political in subversive ways.
-
Where do you like to write?
In an office in complete silence, but most of the time I end up having to write with headphones on while listening to music.
-
-
www.windowscentral.com www.windowscentral.com
-
Cloud computing is essentially local computing with extra, quite pricy steps today for consumer use scenarios. Unless the economics of local hardware truly does fall off a cliff somewhere down the line, I can't see Bezos' vision of a cloud-only future coming true any time soon — even for casual PC users.
so, in short the article is bunk? cloud computing is not cheap with subscriptions for every piece of it. Owning a computer amortised over its years of us willbe cheaper (25 euro/month is 1k laptop every 3 yrs).....although most people hardly use the capabilities of their device.
-
There's a hard cap on the amount of chips the human race can physically produce at any one time, at least as of writing. With nation states effectively printing money to outbid consumer tech companies on basic components, I'm not sure demand will come down any time soon. That is, of course, unless those investors and nation states stop believing AI can deliver anything more impressive than repackaged reddit answers, memeslop, and blog posts for the vast majority ...
the AI fever is creating the shortages. The flp side is that when the hype deflates you can pick up compute for low prices
-
That means DRAM, but also increasingly other components too. SSD storage is the next component expected to hit a shortage, battering consumer prices hard in the process.
Article hinges on the notion that prices of dRAM, ssd will rise due to shortages.
-
Is it really so far fetched to imagine that most people would most likely be "fine" with renting their full computing solutions from companies like Microsoft and Amazon?
no, esp not if it's either or (like for some FB is internet access). But fully locked down devices and settings will get a liability quickly too that people can't ignore.
-
-
human.libretexts.org human.libretexts.org
-
Revising literally means “to see again” not just once but multiple times. Revision has two types of processes: To look at the larger problems such as content and organization To look at the smaller problems such as sentence structure, word choice, and formatting Part of revising may include asking others to read drafts and make revision recommendations. Ultimately, it’s always up to the writer whether those revision recommendations will be implemented into the final draft.
Revising means re-vision, to see again because everything looks different the second time.
-
Cooling means setting aside the document, at least 24-48 hours before revising begins for short pieces of work. This allows writers to have a break from the content and a new perspective when entering the revision stage. To do this, writers need to be organized and time managers. The first draft must be done early enough to set it aside for the recommended cooling time. Authors of books have even longer cooling periods. It may be weeks, months, and sometimes even years, depending on the writer’s preference and the deadline for the publication of the book.
After writing, put the story aside and take time to forget about what you've written and then revise with un-biased eyes.
-
Drafting involves writing the first draft of a document. Some writers write their first draft with a pen and a notebook. Other writers write directly on a laptop or computer. The choice depends on the preference of the writer. A short piece of writing can be drafted in one sitting. The goal is to get everything down on paper before it is lost. If a piece cannot be drafted in one sitting because it is too long, writers generally stop at a place where they know what they will write next. This prevents writer’s block, the inability to write the next day. When drafting, writers are encouraged to not pay attention to spelling, punctuation, grammar, etc. Revising while writing causes writers to lose the original flow of the idea. Spelling, punctuation, grammar, etc. can be addressed in the final revision.
Choice of instrument is up to you (computer, typewriter, pen) but make sure you get everything on paper before you forget and leave revision for the end.
-
Writers make several decisions in the prewriting stage as well. They will answer questions like the following: What is the topic? Who are the readers? What genre (type of writing) works best as the vehicle of communication? What point of view (perspective) will this piece be told from? What kind of research needs to be completed before drafting begins?
Pre-writing is for determining the topic, audience, genre, POV and research topics
-
Prewriting writing begins with what draws the writer to write. The writer may be inspired by nature, people, animals, life events, etc. Some writers keep a writing journal, a record of lists and notes, maybe even drawings or photographs, that initially caught their attention. Writers generally are strong observers who record what they see, hear, taste, touch, and smell because it may become part of a story, a poem, a non-fiction essay, a play, etc. Writers may carry a small notebook with them throughout the day and set it on the nightstand next to their bed at night. Then, it is readily available when an idea–an inspiration–grabs their attention.
Writers employ many methods of observation to color their stories with vivid descriptions.
-
Every piece of writing goes through a process of stages: prewriting (also sometimes called planning), drafting, cooling, revising, and publishing. These steps do not always follow one another in succession. Instead, they are recursive, meaning a step can occur again at any point in the process. For instance, while revising an historically-based short story, a writer may discover he/she needs to do additional research about the time period that the story is set, which takes the writer back to the prewriting stage. See Figure 1.4.
The writing process is recursive which means any step can be done out of order and repeated
-
Most writers are somewhere between these two extreme types of drafters, and that’s the best place to be. See Figure 1.3 which illustrates these two types of drafters. If you are an extreme think-write writer, cultivate some of the traits of the write-write writer, and if you are an extreme write-write writer, try some of the traits of the think-write writer. Attempting both styles of writing will help writers avoid writer’s block.
Writing style is a spectrum and the best place to be is between the two extremes
-
They have a hard time knowing when a draft is finished, and they sometimes over revise. They are often writing under pressure–a deadline. They are often referred to as the messy writers, and the revision of their work takes a long time.
As you can imagine, revision is a long process and they tend to miss deadlines.
-
They are willing to try multiple ideas to see what will work best. They can easily leave sentence and grammar errors to be edited later in the revision stage. They embrace revision as it is part of their drafting process.
They just get the words on paper so they can get to the revision stage to fix everything
-
Other writers are write-write writers. They write, cut, copy, and reorganize their work as well as throw away and start again—sometimes multiple times. They are constantly prewriting, planning, and revising as they go. They sometimes struggle with finishing a final draft, and they have even been known to delete some of their best work. These writers need to remember to save all drafts, so that the best work is never lost. See Figure 1.2 for a list of advantages and disadvantages of being an extreme write-write writer.
A write writer is more haphazard and tends to plow forward with writing and deal with revision later.
-
They need time to think; they can’t write under command or time pressure. Starting the opening paragraph can be difficult because they are still thinking. Revising their work is difficult because from their perspective a lot of the revision decisions were made in the thinking process.
But they need a lot of time to plan and aren't eager to edit or revise.
-
Advantages Once they’ve start writing, they finish the draft easily. The first draft can feel like a polished final draft to the writer. They usually finish drafts on time or earlier than the deadline.
A think writer finishes first drafts easily and on time.
-
Some are think-write writers. They need to think and think and think some more until they can write their first draft. When they write their first draft, they need a large block of time to get it down on paper. Their first drafts feel like a finished product to the writer because they’ve done most of their prewriting and revising in the thinking process. However, these writers need to remember that the first draft is just that—a first draft. Revision is necessary. See Figure 1.1 for a list of the advantages and disadvantages of being an extreme think-write writer.
The think-write writers do a lot of pre-planning so they think they don't need a second draft, but they are wrong.
-
Each writer has his/her own preferences when drafting a document. Whether a person is writing a story, a poem, a journal entry, a letter, or a creative non-fiction piece, the writing approach is idiosyncratic, meaning that it is distinctive to the person who is writing.
Writing style is as diverse as the writers themselves.
-
-
raypodder.medium.com raypodder.medium.com
-
ESG
https://bafybeihbbq7hyuv6t5atovt2y5pfj6w4ixp55vco345xj2ozojdlxc635y.ipfs.dweb.link?filename=esg%20-%20Brave%20Search%20(15_01_2026%2016%EF%BC%9A07%EF%BC%9A10).html / 💻/ asus/ 🧊/ ♖/ hyperpost/ ~/ gyuri/ 🏛️/ 20/ 26/ 01/ 15 https://bafybeihbbq7hyuv6t5atovt2y5pfj6w4ixp55vco345xj2ozojdlxc635y.ipfs.dweb.link?filename=esg%20-%20Brave%20Search%20(15_01_2026%2016%EF%BC%9A07%EF%BC%9A10).html
-
machine
didn’t just replace workers — it replaced wages.
-
morals
O will give them =
-
the flow
is just getting started.
-
next world order
isn’t another empire
???
-
filling
no one’s.
???
-
O gives it
context.
-
AI gave us
power without purpose.
-
-
saalck.pressbooks.pub saalck.pressbooks.pub
-
What outcome do you want to achieve?
I would want to quickly and efficiently inform my boss that I would not be able to make it to work that morning/ with an apology. I would do it as quickly as possible Sunday evening so that my boss hopefully view's it and has the proper time to take specific measures to rearrange anything. Furthermore, I would explain to them what I have already done to get ready and what still needs to be completed.
-
-
www.biorxiv.org www.biorxiv.org
-
eLife Assessment
This manuscript presents useful insights into the molecular basis underlying the positive cooperativity between the co-transported substrates (galactoside sugar and sodium ion) in the melibiose transporter MelB. Building on years of previous studies, this convincing study improves on the resolution of previously published structures and reports the presence of a water molecule in the sugar binding site that would appear to be key for its recognition, introduces further structures bound to different substrates, and utilizes binding and transport assays, as well as HDX-MS and molecular dynamics simulations to further understand the positive cooperativity between sugar and the co-transported sodium cation. The work will be of interest to biologists and biochemists working on cation-coupled symporters, which mediate the transport of a wide range of solutes across cell membranes.
-
Reviewer #1 (Public review):
While the structure of the melibiose permease in both outward and inward-facing forms has been solved previously, there remains unanswered questions regarding its mechanism. Hariharan et al set out to address this with further crystallographic studies complemented with ITC and hydrogen deuterium exchange (HDX) mass spectrometry. They first report 4 different crystal structures of galactose derivatives to explore molecular recognition showing that the galactose moiety itself is the main source of specificity. Interestingly, they observe a water-mediated hydrogen bonding interaction with the protein and suggest that this water molecule may be important in binding.
The results from the crystallography appear sensible, though the resolution of the data is low with only the structure with NPG better than 3Å. Support for the conclusion of the water molecule in the binding site, as interpreted from the density, is given by MD studies.
The HDX also appears to be well done and is explained reasonably well in the revision.
-
Reviewer #3 (Public review):
Summary:
The melibiose permease from Salmonella enterica serovar Typhimurium (MelBSt) is a member of the Major Facilitator Superfamily (MFS). It catalyzes the symport of a galactopyranoside with Na⁺, H⁺, or Li⁺, and serves as a prototype model system for investigating cation-coupled transport mechanisms. In cation-coupled symporters, a coupling cation typically moves down its electrochemical gradient to drive the uphill transport of a primary substrate; however, the precise role and molecular contribution of the cation in substrate binding and translocation remain unclear. In a prior study, the authors showed that the binding affinity for melibiose is increased in the presence of Na+ by about 8-fold, but the molecular basis for the cooperative mechanism remains unclear. The objective of this study was to better understand the allosteric coupling between the Na+ and melibiose binding sites. To verify the sugar-recognition specific determinants, the authors solved the outward-facing crystal structures of a uniport mutant D59C with four sugar ligands containing different numbers of monosaccharide units (α-NPG, melibiose, raffinose, or α-MG). The structure with α-NPG bound has improved resolution (2.7 Å) compared to a previously published structure and to those with other sugars. These structures show that the specificity is clearly directed toward the galactosyl moiety. However, the increased affinity for α-NPG involves its hydrophobic phenyl group, positioned at 4 Å-distance from the phenyl group of Tyr26 forms a strong stacking interaction. Moreover, a water molecule bound to OH-4 in the structure with α-NPG was proposed to contribute to the sugar recognition and appears on the pathway between the two specificity-determining pockets. Next, the authors analyzed by hydrogen-to-deuterium exchange coupled to mass spectrometry (HDX-MS) the changes in structural dynamics of the transporter induced by melibiose, Na+, or both. The data support the conclusion that the binding of the coupling cation at a remote location stabilizes the sugar-binding residues to switch to a higher-affinity state. Therefore, the coupling cation in this symporter was proposed to be an allosteric activator.
Strengths:
(1) The manuscript is generally well written.
(2) This study builds on the authors' accumulated knowledge of the melibiose permease and integrates structural and HDX-MS analyses to better understand the communication between the sodium ion and sugar binding sites. A high sequence coverage was obtained for the HDX-MS data (86-87%), which is high for a membrane protein.
The revised manuscript shows clear improvement, and the authors have addressed my concerns in a satisfactory manner. Of note, I noticed two mistakes that should be corrected:
- page 11. Unless I am mistaken, the sentence "In contrast, Na+ alone or with melibiose primarily caused deprotections" should be corrected with "protections". The authors may wish to verify this sentence and also the previous one in the main text.
- Figure 8 displays two cytoplasmic gates (one of them should be periplasmic)
-
Author response:
The following is the authors’ response to the original reviews
eLife Assessment
This manuscript presents useful insights into the molecular basis underlying the positive cooperativity between the co-transported substrates (galactoside sugar and sodium ion) in the melibiose transporter MelB. Building on years of previous studies, this work improves on the resolution of previously published structures and reports the presence of a water molecule in the sugar binding site that would appear to be key for its recognition, introduces further structures bound to different substrates, and utilizes HDX-MS to further understand the positive cooperativity between sugar and the co-transported sodium cation. Although the experimental work is solid, the presentation of the data lacks clarity, and in particular, the HDX-MS data interpretation requires further explanation in both methodology and discussion, as well as a clearer description of the new insight that is obtained in relation to previous studies. The work will be of interest to biologists and biochemists working on cation-coupled symporters, which mediate the transport of a wide range of solutes across cell membranes.
We express our gratitude to the associate editor, review editor, and reviewers for their favorable evaluation of this manuscript, as well as their constructive comments and encouragement. Their feedback has been integrated to fortify the evidence, refine the data analysis, and elevate the presentation of the results, thereby enhancing the overall quality and clarity of the manuscript.
A brief summary of the modifications in this revision:
(a) We performed four new experiments: 1) intact cell [<sup>3</sup>H]raffinose transport assay; 2) intact cell p-nitrophenol detection to demonstrate α-NPG transport; 3) ITC binding assay for the D59C mutant; and 4) molecular dynamics to simulate the water-1 in sugar-binding site and the dynamics of side chains in the Na<sup>+</sup>- and melibiose-binding pockets. All data consistently support the conclusion draw in this article.
(b) We have added a new figure to show the apo state dynamics (the new Fig. 5a,b) and annotated the amino acid residue positions and marked positions in sugar- or Na<sup>+</sup>-binding pockets.
(c) As suggested by reviewer-3, we have moved the individual mapping of ligand effects on HDX data to the main figure, combined with the residual plots, and marked the amino-acid residue positions.
(d) We have added more deuterium uptake plots to cover all residues in the sugar- or Na<sup>+</sup>-binding pockets in the current figure 7 (previously figure 6).
(e) We have added a new figure 8 showing the positions at the well-studied cytoplasmic gating salt-bridge network and other loops likely important for conformational changes, along with a membrane topology marked with the HDX data. We have added a new figure 9 from MD simulations.
Reviewer #1:
While the structure of the melibiose permease in both outward and inward-facing forms has been solved previously, there remain unanswered questions regarding its mechanism. Hariharan et al set out to address this with further crystallographic studies complemented with ITC and hydrogen-deuterium exchange (HDX) mass spectrometry.
(1) They first report 4 different crystal structures of galactose derivatives to explore molecular recognition, showing that the galactose moiety itself is the main source of specificity. Interestingly, they observe a water-mediated hydrogen bonding interaction with the protein and suggest that this water molecule may be important in binding.
We thank you for understanding what we've presented in this manuscript.
(2) The results from the crystallography appear sensible, though the resolution of the data is low, with only the structure with NPG better than 3Å. However, it is a bit difficult to understand what novel information is being brought out here and what is known about the ligands. For instance, are these molecules transported by the protein or do they just bind? They measure the affinity by ITC, but draw very few conclusions about how the affinity correlates with the binding modes. Can the protein transport the trisaccharide raffinose?
The four structures with bound sugars of different sizes were used to identify the binding motif on both the primary substrate (sugar) and the transporter (MelB<sub>St</sub>). Although the resolutions of the structures complexed with melibiose, raffinose, or a-MG are relatively low, the size and shape of the densities at each structure are consistent with the corresponding sugar molecules, which provide valuable data for confirming the pose of the bound sugar proposed previously. In this revision, we further refine the α-NPG-bound structure to 2.60 Å. The identified water-1 in this study further confirms the orientation of C4-OH. Notably, this transporter does not recognize or transport glucosides in which the orientation of the C4-OH at the glucopyranosyl ring is opposite. To verify the water in the sugar-binding site, we initiated a new collaborative study using MD simulations. Results showed that Wat-1 exhibited nearly full occupancy when melibiose was present, regardless of whether Na<sup>+</sup> was bound at the cation-binding site.
As detailed in the Summary, we added two additional sets of transport assays and confirmed that raffinose and α-NPG are transportable substrates of MelB<sub>St</sub>. For α-NPG transport, we measured the end products of the process—enzyme hydrolysis and membrane diffusion of p-nitrophenol released from intracellular α-NPG.
As a bonus, based on the WT-like downhill α-NPG transport activity by the D59C uniporter mutant that failed in active transport against a sugar concentration gradient, we further emphasized that the sugar translocation pathway is isolated from the cation-binding site. The new data strongly support the allosteric effects of cation binding on sugar-binding affinity. Thank you for this helpful suggestion.
A meaningful analysis of ITC data heavily depends on the quality of the data. My laboratory has extensive experience with ITC and has gained rich, insightful mechanistic knowledge of MelB<sub>St</sub>. Because of the low affinity in raffinose and a-MG, unfortunately, no further information can be convincingly obtained. Therefore, we did not dissect the enthalpic and entropic contributions but focused on the Kd value and binding stoichiometry.
(3) The HDX also appears to be well done; however, in the manuscript as written, it is difficult to understand how this relates to the overall mechanism of the protein and the conformational changes that the protein undergoes.
We are sorry for not presenting our data clearly in the initial submission. In this revised manuscript, we have made numerous improvements, as described in the Summary. These enhancements in the HDX data analysis provided new mechanistic insights into the allosteric effects, leading us to conclude that protein dynamics and conformational transitions are coupled with sugar-binding affinity. Na<sup>+</sup> binding restricts protein conformational flexibility, thereby increasing sugar-binding affinity. The HDX study revealed that the major dynamic region includes a sugar-binding residue, Arg149, which also plays a gating role. Structurally, this dual-function residue undergoes significant displacement during the sugar-affinity-coupled conformational transition, thereby coupling the sugar binding and structural dynamics.
Reviewer #2:
This manuscript from Hariharan, Shi, Viner, and Guan presents x-ray crystallographic structures of membrane protein MelB and HDX-MS analysis of ligand-induced dynamics. This work improves on the resolution of previously published structures, introduces further sugar-bound structures, and utilises HDX to explore in further depth the previously observed positive cooperatively to cotransported cation Na<sup>+</sup>. The work presented here builds on years of previous study and adds substantial new details into how Na<sup>+</sup> binding facilitates melibiose binding and deepens the fundamental understanding of the molecular basis underlying the symport mechanism of cation-coupled transporters. However, the presentation of the data lacks clarity, and in particular, the HDX-MS data interpretation requires further explanation in both methodology and discussion.
We appreciate this reviewer's time in reading our previous articles related to this manuscript.
Comments on Crystallography and biochemical work:
(1) It is not clear what Figure 2 is comparing. The text suggests this figure is a comparison of the lower resolution structure to the structure presented in this work; however, the figure legend does not mention which is which, and both images include a modelled water molecule that was not assigned due to poor resolution previously, as stated by the authors, in the previously generated structure. This figure should be more clearly explained.
This figure is a stereo view of a density map created in cross-eye style. In this revision, we changed this figure to Fig. 3 and showed only the density for sugar and water-1.
(2) It is slightly unclear what the ITC measurements add to this current manuscript. The authors comment that raffinose exhibiting poor binding affinity despite having more sugar units is surprising, but it is not surprising to me. No additional interactions can be mapped to these units on their structure, and while it fits into the substrate binding cavity, the extra bulk of additional sugar units is likely to reduce affinity. In fact, from their listed ITC measurements, this appears to be the trend. Additionally, the D59C mutant utilised here in structural determination is deficient in sodium/cation binding. The reported allostery of sodium-sugar binding will likely influence the sugar binding motif as represented by these structures. This is clearly represented by the authors' own ITC work. The ITC included in this work was carried out on the WT protein in the presence of Na<sup>+</sup>. The authors could benefit from clarifying how this work fits with the structural work or carrying out ITC with the D59C mutant, or additionally, in the absence of sodium.
Thank this reviewer for your helpful suggestions. We have performed the suggested ITC measurements with the D59C mutant. The purpose of the ITC experiments was to demonstrate that MelB<sub>St</sub> can bind raffinose and α-MG to support the crystal structures.
Comments on HDX-MS work:
While the use of HDX-MS to deepen the understanding of ligand allostery is an elegant use of the technique, this reviewer advises the authors to refer to the Masson et al. (2019) recommendations for the HDX-MS article (https://doi.org/10.1038/s41592-019-0459-y) on how to best present this data. For example:
All authors value this reviewer's comments and suggestions, which have been included in this revision.
(1) The Methodology includes a lipid removal step. Based on other included methods, I assumed that the HDX-MS was being carried out in detergent-solubilised protein samples. I therefore do not see the need for a lipid removal step that is usually included for bilayer reconstituted samples. I note that this methodology is the same as previously used for MelB. It should be clarified why this step was included, if it was in fact used, aka, further details on the sample preparation should be included.
Yes, a lipid/detergent removal step was included in this study and previous ones, and this information was clearly described in the Methods.
(2) A summary of HDX conditions and results should be given as recommended, including the mean peptide length and average redundancy per state alongside other included information such as reaction temperature, sequence coverage, etc., as prepared for previous publications from the authors, i.e., Hariharan et al., 2024.
We have updated the Table S2 and addressed the reviewer’ request for the details of HDX experiments.
(3) Uptake plots per peptide for the HDX-MS data should be included as supporting information outside of the few examples given in Figure 6.
We have prepared and presented deuterium uptake time-course plots for any peptides with ΔD > threshold in Fig. S5a-c.
(4) A reference should be given to the hybrid significance testing method utilised. Additionally, as stated by Hageman and Weis (2019) (doi:10.1021/acs.analchem.9b01325), the use of P < 0.05 greatly increases the likelihood of false positive ΔD identifications. While the authors include multiple levels of significance, what they refer to as high and lower significant results, this reviewer understands that working with dynamic transporters can lead to increased data variation; a statement of why certain statistical criteria were chosen should be included, and possibly accompanied by volcano plots. The legend of Figure 6 should include what P value is meant by * and ** rather than statistically significant and highly statistically significant.
We appreciate this comment and have cited the suggested article on the hybrid significance method. We fully acknowledge that using a cutoff of P < 0.05 can increase the likelihood of false-positive identifications. By applying multiple levels of statistical testing, we determined that P < 0.05 is an appropriate threshold for this study. The threshold values were presented in the residual plots and explained in the text. For the previous Fig. 6 (renamed Fig. S4b in the current version), we have reported the P value. *, < 0.05; **, < 0.01. (The text for 0.01 was not visible in the previous version. Sorry for the confusion.)
(5) Line 316 states a significant difference in seen in dynamics, how is significance measured here? There is no S.D. given in Table S4. Can the authors further comment on the potential involvement in solvent accessibility and buried helices that might influence the overall dynamics outside of their role in sugar vs sodium binding? An expected low rate of exchange suggests that dynamics are likely influenced by solvent accessibility or peptide hydrophobicity. The increased dynamics at peptides covering the Na binding site on overall more dynamic helices suggests that there is no difference between the dynamics of each site.
The current Table S3 (combined from previous Tables S3 and S4 as suggested) was prepared to provide an overall view of the dynamic regions with SD values provided. For other questions, if we understand correctly, this reviewer asked us to comment on the effects of solvent accessibility or hydrophobic regions on the overall dynamics outside the binding residues of the peptides that cover them. Since HDX rates are influenced by two linked factors: solvent accessibility and hydrogen-bonding interactions that reflect structural dynamics, poor solvent accessibility in buried regions should result in low deuterium uptakes. The peptides in our dataset that include the Na<sup>+</sup>-binding site showed lower HDX, likely due to limited solvent accessibility and lower structural stability. It is unclear what this reviewer meant by "increased dynamics at peptides covering the Na binding site on overall more dynamic helices." We did not observe increased dynamics in peptides covering the Na<sup>+</sup>-binding site; instead, all Na<sup>+</sup>-binding residues and nearby sugar-binding residues have lower degrees of deuteriation.
(6) Previously stated HDX-MS results of MelB (Hariharan et al., 2024) state that the transmembrane helices are less dynamic than polypeptide termini and loops with similar distributions across all transmembrane bundles. The previous data was obtained in the presence of sodium. Does this remove the difference in dynamics in the sugar-binding helices and the cation-binding helices? Including this comparison would support the statement that the sodium-bound MelB is more stable than the Apo state, along with the lack of deprotection observed in the differential analysis.
Thanks for this suggestion. The previous datasets were collected in the presence of Na<sup>+</sup>. In the current study, we also have two Na<sup>+</sup>-containing datasets. Both showed similar results: the multiple overlapping peptides covering the sugar-binding residues on helices I and V have higher HDX rates than those peptides covering the Na<sup>+</sup>-binding residues, even when Na<sup>+</sup> was present.
(7) Have the authors considered carrying out an HDX-MS comparison between the WT and the D59C mutant? This may provide some further information on the WT structure (particularly a comparison with sugar-bound). This could be tied into a nice discussion of their structural data.
Thank you for this suggestion. Comparing HDX-MS between the WT and the D59C mutant is certainly interesting, especially with the increasing amount of structural, biochemical, and biophysical data now available for this mutant. However, due to limited resources, we might consider it later.
(8) Have the authors considered utilising Li<sup>+</sup> to infer how cation selectivity impacts the allostery? Do they expect similar stabilisation of a higher-affinity sugar binding state with all cations?
We have shown that Li<sup>+</sup> also works positively with melibiose. Li<sup>+</sup> binds to MelB<sub>St</sub> with a higher affinity than Na<sup>+</sup> and modifies MelB<sub>St</sub> differently. It is important to study this thoroughly and separately. To answer the second question, H<sup>+</sup> is a weak coupling cation with little effect on melibiose binding. Since its pKa is around 6.5, only a small population of MelB<sub>St</sub> is protonated at pH 7.5. The order of sugar-binding cooperativity is highest with Na<sup>+</sup>, then Li<sup>+</sup>, and finally H<sup>+</sup>.
(9) MD of MelB suggests all transmembrane helices are reorientated during substrate translocation, yet substrate and cotransporter ligand binding only significantly impacts a small number of helices. Can the authors comment on the ensemble of states expected from each HDX experiment? The data presented here instead shows overall stabilisation of the transporter. This data can be compared to that of HDX on MFS sugar cation symporter XylE, where substrate binding induces a transition to the OF state. There is no discussion of how this HDX data compares to previous MFS sugar transporter HDX. The manuscript could benefit from this comparison rather than a comparison to LacY. It is unlikely that there are universal mechanisms that can be inferred even from these model proteins. Highlighting differences between these transport systems provides broader insights into this protein class. Doi: 10.1021/jacs.2c06148 and 10.1038/s41467-018-06704-1.
The sugar translocation free-energy landscape simulations showed that both helix bundles move relative to the membrane plane. This analysis aimed to clarify a hypothesis in the field—that the MFS transporter can use an asymmetric mode to perform the conformational transition between inward- and outward-facing states. In the case of MelB<sub>St</sub>, we clearly demonstrated that both domains move and each helix bundle moves as a unit. So only a small number of helices and loops showed labeling changes. Thanks for the suggestion about comparing with XylE. We have included that in the discussion.
(10) Additionally, the recent publication of SMFS data (by the authors: doi:10.1016/j.str.2022.11.011) states the following: "In the presence of either melibiose or a coupling Na<sup>+</sup>-cation, however, MelB increasingly populates the mechanically less stable state which shows a destabilized middle-loop C3." And "In the presence of both substrate and co-substrate, this mechanically less stable state of MelB is predominant.". It would benefit the authors to comment on these data in contrast to the HDX obtained here. Additionally, is the C3 loop covered, and does it show the destabilization suggested by these studies? HDX can provide a plethora of results that are missing from the current analysis on ligand allostery. The authors instead chose to reference CD and thermal denaturation methods as comparisons.
Thank this reviewer for reading the single-molecule force spectroscopy (SMFS) study on MelB<sub>St</sub>. The C3 loop mentioned in this SMFS article is partially covered in the dataset Mel or Mel plus Na<sup>+</sup> vs. apo, and there is more coverage in the Na<sup>+</sup> vs. apo dataset. In either condition, no deprotection was detected. The labeling time point might not be long enough to detect it.
Reviewer #3:
Summary:
The melibiose permease from Salmonella enterica serovar Typhimurium (MelB<sub>St</sub>) is a member of the Major Facilitator Superfamily (MFS). It catalyzes the symport of a galactopyranoside with Na<sup>+</sup>, H<sup>+</sup>, or Li<sup>+</sup>, and serves as a prototype model system for investigating cation-coupled transport mechanisms. In cation-coupled symporters, a coupling cation typically moves down its electrochemical gradient to drive the uphill transport of a primary substrate; however, the precise role and molecular contribution of the cation in substrate binding and translocation remain unclear. In a prior study, the authors showed that the binding affinity for melibiose is increased in the presence of Na<sup>+</sup> by about 8-fold, but the molecular basis for the cooperative mechanism remains unclear. The objective of this study was to better understand the allosteric coupling between the Na<sup>+</sup> and melibiose binding sites. To verify the sugar-recognition specific determinants, the authors solved the outward-facing crystal structures of a uniport mutant D59C with four sugar ligands containing different numbers of monosaccharide units (α-NPG, melibiose, raffinose, or α-MG). The structure with α-NPG bound has improved resolution (2.7 Å) compared to a previously published structure and to those with other sugars. These structures show that the specificity is clearly directed toward the galactosyl moiety. However, the increased affinity for α-NPG involves its hydrophobic phenyl group, positioned at 4 Å-distance from the phenyl group of Tyr26, which forms a strong stacking interaction. Moreover, a water molecule bound to OH-4 in the structure with α-NPG was proposed to contribute to the sugar recognition and appears on the pathway between the two specificity-determining pockets. Next, the authors analyzed by hydrogen-to-deuterium exchange coupled to mass spectrometry (HDX-MS) the changes in structural dynamics of the transporter induced by melibiose, Na<sup>+</sup>, or both. The data support the conclusion that the binding of the coupling cation at a remote location stabilizes the sugar-binding residues to switch to a higher-affinity state. Therefore, the coupling cation in this symporter was proposed to be an allosteric activator.
Strengths:
(1) The manuscript is generally well written.
(2) This study builds on the authors' accumulated knowledge of the melibiose permease and integrates structural and HDX-MS analyses to better understand the communication between the sodium ion and sugar binding sites. A high sequence coverage was obtained for the HDX-MS data (86-87%), which is high for a membrane protein.
Thank this reviewer for your positive comments.
Weaknesses:
(1) I am not sure that the resolution of the structure (2.7 Å) is sufficiently high to unambiguously establish the presence of a water molecule bound to OH-4 of the α-NPG sugar. In Figure 2, the density for water 1 is not obvious to me, although it is indeed plausible that water mediates the interaction between OH4/OH6 and the residues Q372 and T373.
A water molecule can be modeled at a resolution ranging from 2.4 to 3.2 Å, and the quality of the model depends on the map quality and water location. In this revision, we refined the resolution to 2.6 Å using the same dataset and also performed all-atom MD simulations. All results support the occupancy of water-1 in the sugar-bound MelB<sub>St</sub>.
(2) Site-directed mutagenesis could help strengthen the conclusions of the authors. Would the mutation(s) of Q372 and/or T373 support the water hypothesis by decreasing the affinity for sugars? Mutations of Thr121, Arg 295, combined with functional and/or HDX-MS analyses, may also help support some of the claims of the authors regarding the allosteric communication between the two substrate-binding sites.
The authors thank this reviewer for the thoughtful suggestions. MelB<sub>St</sub> has been subjected to Cys-scanning mutagenesis (https://doi.org/10.1016/j.jbc.2021.101090). Placing a Cys residue at Gln372 significantly decreased the transport initial rate, accumulation, and melibiose fermentation, with minimal effect on protein expression, as shown in Figure 2 of this JBC article, which could support its role in the binding pocket. The T373C mutant retained most of the WT's activities. Our previous studies showed that Thr121 is only responsible for Na<sup>+</sup> binding in MelB<sub>St</sub>, and mutations decreased protein stability; now, HDX reveals that this is the rigid position. Additionally, our previous studies indicated that Arg295 is another conformationally important residue. In this version, we have added more HDX analysis to explore the relationship between the two substrate-binding sites with conformational dynamics, especially focusing on the gating salt-bridge network including Arg295, which has provided meaningful new insights.
(3) The main conclusion of the authors is that the binding of the coupling cation stabilizes those dynamic sidechains in the sugar-binding pocket, leading to a high-affinity state. This is visible when comparing panels c and a from Figure S5. However, there is both increased protection (blue, near the sugar) and decreased protection in other areas (red). The latter was less commented, could the increased flexibility in these red regions facilitate the transition between inward- and outward-facing conformations? The HDX changes induced by the different ligands were compared to the apo form (see Figure S5). It might be worth it for data presentation to also analyze the deuterium uptake difference by comparing the conditions sodium ion+melibiose vs melibiose alone. It would make the effect of Na<sup>+</sup> on the structural dynamics of the melibiose-bound transporter more visible. Similarly, the deuterium uptake difference between sodium ion+melibiose vs sodium ion alone could be analyzed too, in order to plot the effect of melibiose on the Na<sup>+</sup>-bound transporter.
Thanks for this important question. We have added more discussion of the deprotected data and prepared a new Fig. 8b to highlight the melibiose-binding-induced flexibility in several loops, especially the gating area on both sides of the membrane. We also proposed that these changes might facilitate the formation of the transition-competent state. The overall effects induced by substrate binding are relatively small, and the datasets for apo and Na were collected separately, so comparing melibiose&Na<sup>+</sup> versus Na<sup>+</sup> might not be as precise. In fact, the Na<sup>+</sup> effects on the sugar-binding site can be clearly seen in the deuterium uptake plots shown in Figures 7-8, by comparing the first and last panels.
(4) For non-specialists, it would be beneficial to better introduce and explain the choice of using D59C for the structural analyses.
Asp59 is the only site that responds to the binding of all coupling cations: Na<sup>+</sup>, Li<sup>+</sup>, or H<sup>+</sup>. Notably, this thermostable mutant D59C selectively abolishes all cation binding and associated cotransport activities, but it maintains intact sugar binding and exhibits conformational transition as the WT, as demonstrated by electroneutral transport reactions including α-NPG transport showed in this articles, and melibiose exchange and fermentation showed previously. Therefore, the structural data derived from this mutant are significant and offer important mechanistic insights into sugar transport, which supports the conclusion that the Na<sup>+</sup> functions as allosteric activator.
(5) In Figure 5a, deuterium changes are plotted as a function of peptide ID number. It is hardly informative without making it clearer which regions it corresponds to. Only one peptide is indicated (213-226). I would recommend indicating more of them in areas where deuterium changes are substantial.
We appreciate this comment and have modified the plots by marking the residue position as well as labeled several peptides of significant HDX in the Fig 5b. We also provided a deuteriation map based on peptide coverage (Fig. 5a).
(6) From prior work of the authors, melibiose binding also substantially increases the affinity of the sodium ion. Can the authors interpret this observation based on the HDX data?
This is an intriguing mechanistic question. In this HDX study, we found that the cation-binding pocket and nearby sugar-binding residues are conformationally rigid, while some sugar-binding residues farther from the cation-binding pocket are flexible. We concluded that conformational dynamics regulate sugar-binding affinity, but the increase in Na-binding affinity caused by melibiose is not related to protein dynamics. Our previous interpretation based on structural data remains our preferred explanation; therefore, the bound melibiose physically prevents the release of Na<sup>+</sup> or Li<sup>+</sup> from the cation-binding pocket. We also proposed the mechanism of intracellular NA<sup>+</sup> release in the 2024 JBC paper (https://doi.org/10.1016/j.jbc.2024.107427); after sugar release, the rotamer change of Asp55 will help NA<sup>+</sup> exit the cation pocket into the empty sugar pocket, and the negative membrane potential inside the cell will further facilitate movement from MelB<sub>St</sub> to the cytosol.
Recommendations for the authors:
Reviewing Editor Comments:
(1) It would help the reader if the previous work were introduced more clearly, and if the results of the experiments reported in this manuscript were put into the context of the previous work. Lines 283-296 discuss observations that are similar to previous reported structures as well as novel interpretations. It would help the reader to be clearer about what the new observations are.
Thank you for the important comment. We have revised accordingly by adding related citations and words “as showed previously” when we stated our previous observations.
(2) The affinity by ITC is measured for various ligands, but very few conclusions are drawn about how the affinity correlates with the binding modes. Are the other ligands that are investigated in this study transported by the protein, or do they just bind? Can the protein transport the trisaccharide raffinose? The authors comment that raffinose exhibiting poor binding affinity despite having more sugar units is surprising, but this is not surprising to me. No additional interactions can be mapped to these units on their structure, and while it fits into the substrate binding cavity, the extra bulk of additional sugar units is likely to reduce affinity. In fact, from their listed ITC measurements, this appears to be the trend.
Additionally, the D59C mutant utilized here in structural determination is deficient in sodium/cation binding. The reported allostery of sodium-sugar binding will likely influence the sugar binding motif as represented by these structures. This is clearly represented by the authors' own ITC work. The ITC included in this work was carried out on the WT protein in the presence of Na<sup>+</sup>. The authors could benefit from clarifying how this work fits with the structural work or carrying out ITC with the D59C mutant, or additionally, in the absence of sodium. For non-specialists, please better introduce and explain the choice of using D59C for the structural analyses.
Thank you for the meaningful comments. We have comprehensively addressed all the concerns and suggestions as listed in the summary of this revision. Notably, the D59C mutant does not catalyze any electrogenic melibiose transport involved in a cation transduction but catalyze downhill transport location of the galactosides, as shown by the downhill α-NPG transport assay in Fig. 1a. The intact downhill transport results from D59C mutant further supports the allosteric coupling between the cation- and sugar-binding sites.
The binding isotherm and poor affinity of the ITC measurements do not support to further analyze the binding mode since none showed sigmoidal curve, so the enthalpy change cannot be accurately determined. But authors thank this comment.
(3) It is not clear what Figure 2 is comparing. The text suggests this figure is a comparison of the lower resolution structure to the structure presented in this work; however, the figure legend does not mention which is which, and both images include a modelled water molecule that was not assigned due to poor resolution previously, as stated by the authors, in the previously generated structure. This figure should be more clearly explained.
We have addressed these concerns in the response to the Public Reviews at reviewer-2 #1.
(4) I am not sure that the resolution of the structure (2.7 Å) is sufficiently high to unambiguously establish the presence of a water molecule bound to OH-4 of the α-NPG sugar. In Figure 2, the density for water 1 is not obvious to me, although it is indeed plausible that water mediates the interaction between OH4/OH6 and the residues Q372 and T373. Please change line 278 to state "this OH-4 water molecule is likely part of sugar binding".
We have addressed these concerns in the response to the Public Reviews at reviewer-3 #1.
(5) Line 290-296: The Thr121 is not represented in any figures, while the Lys377 is. Their relative positioning between sugar water and sodium is not made clear by any figure.
Thanks for this comment. This information has been clearly presented in the Figs. 7-8. Lys377 is closer to the cation site and related far from the sugar-binding site.
(6) Methodology includes a lipid removal step. Based on other included methods, I assumed that the HDX-MS was being carried out in detergent-solubilized protein samples. I therefore do not see the need for a lipid removal step that is usually included for bilayer reconstituted samples. I note that this methodology is the same as previously used for MelB. It should be clarified why this step was included, if it was in fact used, aka, further details on the sample preparation should be included.
(7) A summary of HDX conditions and results should be given as recommended, including the mean peptide length and average redundancy per state alongside other included information such as reaction temperature, sequence coverage, etc., as prepared for previous publications from the authors, i.e., Hariharan et al., 2024.
We have addressed these concerns in the response to the Public Reviews at reviewer-2 #4.
(8) Uptake plots per peptide for the HDX-MS data should be included as supporting information outside of the few examples given in Figure 6.
We have addressed these concerns in the response to the Public Reviews at reviewer-2 #4.
(9) A reference should be given to the hybrid significance testing method utilised. Additionally, as stated by Hageman and Weis (2019) (doi:10.1021/acs.analchem.9b01325), the use of P < 0.05 greatly increases the likelihood of false positive ΔD identifications. While the authors include multiple levels of significance, what they refer to as high and lower significant results, and this reviewer understands that working with dynamic transporters can lead to increased data variation, a statement of why certain statistical criteria were chosen should be included, and possibly accompanied by volcano plots. The legend of Figure 6 should include what P value is meant by * and ** rather than statistically significant and highly statistically significant.
We have addressed these concerns in the response to the Public Reviews at reviewer-2 #4.
(10) The table (S3) and figure (S4) showing uncovered residues is an unclear interpretation of the data; this would be better given as a peptide sequence coverage heat map. This would also be more informative for the redundancy in covered regions, too. In this way, S3 and S4 can be combined.
We have addressed these concerns in the response to the Public Reviews at reviewer-2 #4.
(11) Residual plots in Figure 5 could be improved by a topological map to indicate how peptide number resembles the protein amino acid sequence.
Thanks for the request, due to the figure 6 is big so that we add a transmembrane topology plot colored with the HDX results in Fig. 8c.
(12) The presentation of data in S5 could be clarified. Does the number of results given in the brackets indicate overlapping peptides? What are the lengths of each of these peptides? Classical HDX data presentation utilizes blue for protection and red for deprotection. The use of yellow ribbons to show protection in non-sugar binding residues takes some interpretation and could be clarified by also depicting in a different blue. I also don't see the need to include ribbon and cartoon representation when also using colors to depict protection and deprotection. The authors should change or clarify this choice.
We have moved this figure into the current Fig. 6b as suggested by Reviewer-3. To address your questions listed in the figure legend, the number of results shown in brackets indeed indicates overlapping peptides. What are the lengths of each of these peptides? The sequences of each peptide are shown in Figures 7-8 and are also included in Supplemental Figure S5. Regarding the use of color, both blue and green were used to distinguish peptides protecting the substrate-binding site from other regions. The ribbon and cartoon representations are provided for clarity, as the cartoon style hides many helices.
(13) In Table S5, the difference between valid points and protection is unclear. And what is indicated by numbers in brackets or slashes? Additionally, it should be highlighted again here that single-residue information is inferred from peptide-level data. By value, are the authors referring to peptide-level differential data?
Please review our responses in the Public Reviews at reviewer-2 #5.
(14) Line 316 states a significant difference in seen in dynamics, how is significance measured here? There is no S.D. given in Table S4. Can the authors further comment on the potential involvement in solvent accessibility and buried helices that might influence the overall dynamics outside of their role in sugar vs sodium binding? An expected low rate of exchange suggests that dynamics are likely influenced by solvent accessibility or peptide hydrophobicity? The increased dynamics at peptides covering the Na binding site on overall more dynamic helices suggests that there isn't a difference between the dynamics of each site.
Please review our responses in the Public Reviews at reviewer-2 #5.
(15) Previously stated HDX-MS results of MelB (Hariharan et al., 2024) state that the transmembrane helices are less dynamic than polypeptide termini and loops with similar distributions across all transmembrane bundles. The previous data was obtained in the presence of sodium. Does this remove the difference in dynamics in the sugar-binding helices and the cation-binding helices? Including this comparison would support the statement that the sodium-bound MelB is more stable than the Apo state, along with the lack of deprotection observed in the differential analysis.
Please review our responses in the Public Reviews.
(16) MD of MelB suggests all transmembrane helices are reorientated during substrate translocation, yet substrate and cotransporter ligand binding only significantly impacts a small number of helices. Can the authors comment on the ensemble of states expected from each HDX experiment? The data presented here instead shows overall stabilisation of the transporter. This data can be compared to that of HDX on MFS sugar cation symporter XylE, where substrate binding induces a transition to the OF state. There is no discussion of how this HDX data compares to previous MFS sugar transporter HDX. The manuscript could benefit from this comparison rather than a comparison to LacY. It is unlikely that there are universal mechanisms that can be inferred even from these model proteins. Highlighting differences instead between these transport systems provides broader insights into this protein class. Doi: 10.1021/jacs.2c06148 and 10.1038/s41467-018-06704-1.
Please review our responses in the Public Reviews.
(17) Additionally, the recent publication of SMFS data (by the authors: doi:10.1016/j.str.2022.11.011) states the following: "In the presence of either melibiose or a coupling Na<sup>+</sup>-cation, however, MelB increasingly populates the mechanically less stable state which shows a destabilized middle-loop C3." And "In the presence of both substrate and co-substrate this mechanically less stable state of MelB is predominant.". It would benefit the authors to comment on these data in contrast to the HDX obtained here. Additionally, is the C3 loop covered, and does it show the destabilization suggested by these studies? HDX can provide a plethora of results that are missing from the current analysis on ligand allostery. The authors instead chose to reference CD and thermal denaturation methods as comparisons.
Please review our responses in the Public Reviews.
(18) The main conclusion of the authors is that the binding of the coupling cation stabilizes those dynamic sidechains in the sugar-binding pocket, leading to a high-affinity state. This is visible when comparing panels c and a from Figure S5. However, there is both increased protection (blue, near the sugar) and decreased protection in other areas (red). The latter was less commented, could the increased flexibility in these red regions facilitate the transition between inward- and outward-facing conformations? The HDX changes induced by the different ligands were compared to the apo form (see Figure S5). It might be worth it for data presentation more visible to also analyze the deuterium uptake difference by comparing the conditions sodium ion+melibiose vs melibiose alone. You would make the effect of Na<sup>+</sup> on the structural dynamics of the melibiose-bound transporter. Similarly, the deuterium uptake difference between sodium ion+melibiose vs sodium ion alone could be analyzed too, in order to plot the effect of melibiose on the Na<sup>+</sup>-bound transporter.
Please review our responses in the Public Reviews.
(19) In Figure 5a, deuterium changes are plotted as a function of peptide ID number. It is hardly informative without making it clearer which regions it corresponds to. Only one peptide is indicated (213-226); I would recommend indicating more of them, in areas where deuterium changes are substantial.
Please review our responses in the Public Reviews.
(20) Figure 6, please indicate in the legend what the black and blue lines are (I assume black is for the apo?)
We are sorry that we did not make it clear. Yes, the black was used for apo state and blue was used for all bound states
(21) From prior work of the authors, melibiose binding also substantially increases the affinity of the sodium ion. Can the authors interpret this observation based on the HDX data?
Please review our responses in the Public Reviews.
Addressing the following three points would strengthen the manuscript, but also involve a significant amount of additional experimental work. If the authors decide not to carry out the experiments described below, they can still improve the assessment by focusing on points (1-21) described above.
(22) Have the authors considered carrying out an HDX-MS comparison between the WT and the D59C mutant? This may provide some further information on the WT structure (particularly a comparison with sugar-bound). This could be tied into a nice discussion of their structural data.
Please review our responses in the Public Reviews.
(23) Have the authors considered utilising Li<sup>+</sup> to infer how cation selectivity impacts the allostery? Do they expect similar stabilisation of a higher-affinity sugar binding state with all cations?
Please review our responses in the Public Reviews.
(24) Site-directed mutagenesis could help strengthen the conclusions. Would the mutation(s) of Q372 and/or T373 support the water hypothesis by decreasing the affinity for sugars? Mutations of Thr 121 and Arg 295, combined with functional and/or HDX-MS analyses, may also help support some of the authors' claims regarding allosteric communication between the two substrate-binding sites.
Please review our responses in the Public Reviews.
-
-
pressbooks.online.ucf.edu pressbooks.online.ucf.edu
-
Our mothers told us that the whites were killing everybody and eating them.
This may be a reference to the Donner Party: https://www.britannica.com/topic/Donner-party
-
-
Local file Local file
-
Coastal resorts were, then, often to be found in symbiosis with fishing and commercial ports, evenwith associated manufacturing and import processing industries, and each function could benefit fromthe presence of the other, although the more exclusive resort interests were sometimes reluctant torecognise this.
Highly slay quote!
-
-
www.biorxiv.org www.biorxiv.org
-
Note: This response was posted by the corresponding author to Review Commons. The content has not been altered except for formatting.
Learn more at Review Commons
Reply to the reviewers
Reviewer #1
Evidence, reproducibility and clarity
__Summary
Köver et al. examine the genetic and environmental underpinnings of multicellular-like phenotypes (MLPs) in fission yeast, studying 57 natural isolates of Schizosaccharomyces pombe. They uncover that a noteworthy subset of these isolates can develop MLPs, with the extent of these phenotypes varying according to growth media. Among these, two strains demonstrate pronounced MLP across a range of conditions. By genetically manipulating one strain with an MLP phenotype (distinct from the previously mentioned two strains), they provide evidence that genes such as MBX2 and SRB11 play a direct role in MLP formation, strengthening their genetic mapping findings. The study also reveals that while some key genes and their phenotypic effects are strikingly similar between budding and fission yeast, other aspects of MLP formation are not conserved, which is an intriguing finding.
Overall, the manuscript is well-written, dense yet logically structured, and the figures are well presented. The combination of phenotypic, genetic, and bioinformatics analyses, particularly from wet lab experiments, is commendable. The study addresses a significant gap in our understanding, primarily explored in budding yeast, by providing comprehensive data on MLP diversity in fission yeast and the interplay of genetic and environmental factors.
In summary, I enjoyed reading the manuscript and have only a few minor suggestions to strengthen the paper:
Minor revisions:
- Although this may seem like a minor revision, but it is a crucial point. Please make sure that all raw data used to generate figures, run stats, sequence data, and scripts used to run data analysis are made publicly available. Provide relevant accession numbers and links to public data repositories. It is important that others can download the various types of data that went into the major conclusions of this paper in order to replicate your analysis or expand upon the scope of this work. I am not sure if the journal has a policy regarding this, but it should be followed to allow for transparency and reproducibility of the research.__
Reply: We very much agree with the reviewer that sharing raw data and scripts is an essential part of open science. All code and data are deposited to Github (https://github.com/BKover99/S.-Pombe-MLPs) and Figshare (https://figshare.com/articles/software/S_-Pombe-MLPs/25750980), which have now been updated to reflect our revisions. Additionally, the sequenced genomes have been deposited to ENA (PRJEB69522). Where external data was used, it was properly referenced and specifically included in Supplementary Table 3.
Two out of 57 strains exhibit strong and consistent MLP across multiple environments. Providing more information on these strains (JB914 and JB953), such as their natural habitats and distinct appearances of their MLP phenotypes under varying conditions, would provide valuable insights.
First, a brief discussion highlighting what differentiates these two strains from the rest would be helpful for readers (e.g. insight into their unique genetic and environmental background that might be linked to the MLP phenotype).
Additionally, culture tube and microscopy images of these strains, similar to those presented for JB759 in Figure 2A, can be included in the supplementary materials. My reasoning is that these images could help illustrate variation or lack thereof in aggregative group size across different media.
Reply: We thank the reviewer for highlighting this issue. Our further investigation into these strains has added additional interesting insights. JB914 and JB953 were isolated from molasses in Jamaica and the exudate of Eucalyptus in Australia, respectively, though it remains unclear whether these environments are related or even selective for the ability of these strains to form MLPs. We note that the environment from which a strain is isolated is an incomplete way of assessing its ecology. Indeed, recent research suggests that the primary habitat of S. pombe is honeybee honey and suggests that bees, which may be attracted to a number of sugary substances, may be a vector by which fission yeast are transported (1). Therefore, isolation from a particular nectar or food production environment might not reflect significant ecological differences. We now refer to the location of strain isolation in the manuscript text (lines 208-209).
However, there is more to learn from the genetic backgrounds of these two strains. We found that JB914 possesses the same variant in srb11 causally related to MLPs as JB759, the MLP-forming parental strain for our QTL analysis. To understand whether the appearance of this variant in these two strains derived from a single mutation event or was a case of convergent evolution, we analysed homology between the genomes of JB759 and JB914, focusing specifically on that variant. We found an approximately 20kb region of homology between JB759 and JB914 surrounding the srb11 truncation variant, in contrast to the majority of the genome, which does not share homology between those two strains (New Supplementary Figure 9A, B)). This result suggests that, while the two strains are largely unrelated, that specific region shares a recent common ancestor and is likely a result of interbreeding across strains.
Importantly, this analysis further emphasizes the point that the srb11 variant segregates with the MLP-forming phenotype. We conclude this because none of the other strains similar to JB759 (either across the whole genome, or specifically in the region surrounding srb11) exhibit MLPs (New Supplementary Figure 9C). This thereby further complements our QTL analysis on the significance of this variant. We have added this analysis to the manuscript text (lines 337-349).
Furthermore, we searched other strains which exhibited MLPs in our experiments (e.g. JB953) for frame shifts, insertions or deletions in any other genes in the CKM module or in the genes that were identified in our deletion library screen as adhesive, and did not identify any severe mutations falling into coding regions (other than the srb11 truncation in JB914 and JB759). This indicates that MLPs in these other strains may be caused by differences in regulatory regions surrounding these genes, or variants in other genes that were not identified in our screen. We have added this analysis to our manuscript (lines 424-425) and Supplementary Table 13.
We agree that microscopy and culture tube images of JB914 and JB953 may give insight into the nature of the MLPs exhibited by those strains. We have included such images of cultures grown in YES, EMM and EMM-Phosphate media in our revision (Lines 207-208, Supplementary Figures 4 and 5). These images are consistent with our adhesion assay screen and show that JB914 and JB953 are adhesive at the microscopic level in the relevant conditions (EMM or EMM-Phosphate).
The phenotypic outcome of overexpressing MXB2 is striking, as shown in Supplementary Figure 4C. Incorporating at least one of the culture tube images depicting large flocs into the main text, perhaps adjacent to Figure 3 panel D, would improve the visual appeal and highlight this key finding (at the moment those images are only shown in the supplementary materials).
Reply: We thank the reviewer for this suggestion. In response to Reviewer 2's suggestion to overexpress mbx2 in YES, we created new mbx2 overexpression strains that could overexpress mbx2 in YES, which was not possible in our previous strain in which mbx2 overexpression was triggered by removal of thymine from the media. We have replaced our original data from Figure 3D with data from the new mbx2 overexpression experiment, including flask images.
I know that the authors discuss the knowledge gap in the intro and results, but the abstract does not mention this critical gap. Please stress this critical gap (i.e., MLPs understudied in fission yeast) with a brief sentence in the abstract. Similarly, please consider writing a brief concluding sentence summarizing the paper's most significant finding referring to the knowledge gap would provide a clearer takeaway message for the reader - the abstract ends abruptly without any conclusion.
Reply: We agree and have now emphasized the critical gap in our abstract:
"As MLP formation remains understudied in fission yeast compared to budding yeast, we aimed to narrow this gap." at lines 18-19.
Additionally, we added the following final sentence to give the reader a clearer takeaway message:
"Our findings provide a comprehensive genetic survey of MLP formation in fission yeast, and a functional description of a causal mutation that drives MLP formation in nature." at lines 31-32.
- The observation that strains with adhesive phenotypes have a lower growth rate compared to non-adhesive strains is a noteworthy point (lines 532-535). This represents yet another example of this classical trade-off. This point could be emphasized in the Discussion or alongside the relevant result, with a brief speculative explanation for this phenomenon.
Reply: We agree that the nature of the trade-off between MLP formation is an interesting discussion point that could arise from our work. Understanding this trade-off is made more complicated by the fact that growth is always condition-dependent, and measuring growth in strains exhibiting MLPs is non-trivial, as adhesion to labware and thick clumps of cells separated by regions of cell-free media can add variability. Nonetheless, there has been some previous work on this problem. In S. cerevisiae, it was shown that larger group size correlates with slower growth rate (3), and that flocculating cells grow more slowly (4). In S. cerevisiae, cAMP, a signalling molecule heavily involved in regulating growth in response to nutrient availability, also regulates filamentation (5). However, the relationship between flocculation and slow growth is not consistent in the literature. In some settings overexpressing the flocculins FLO8, FLO5, and FLO10 results in slower growth (6), while in others it does not (7). In addition, ethanol production has been shown to improve for biofilms (7).
Furthermore, in S. cerevisiae, MLP-forming cells grow better in low sucrose concentrations (8) and under various stress conditions (4). Flocculating cells have also shown faster fermentation in media containing common industrial bioproduction inhibitors, despite slower fermentation than non-flocculating cells in non-inhibitory media (9). However, any consequence of this possible advantage on growth has not been characterised.
In S. pombe, there is less work on this topic; however, it has been shown that deletions of rpl3201 and rpl3202, which code for ribosomal proteins, cause flocculation and slow growth (10). In that case, it is not clear if there is any causal relationship between slow growth and flocculation or if they are both parallel consequences of the ribosomal pathway disruption. We have added some of these points to the portion of the discussion that discusses this tradeoff (Lines 477-499).
To get a better understanding of this tradeoff in our system, we took several approaches. First, we added a supporting analysis (New Supplementary Figure 12B), using published growth data based on measurements on agar plates for the S. pombe gene deletion library (11). There, the authors defined a set of deletion strains that grow more slowly on EMM than the wild-type lab strain. We found that our MLP hit strains were significantly enriched in this "EMM-slow" category. This information is now included in the manuscript (Lines 409-413, New Supplementary Figure 12B).
It is, however, possible that for the assays from that work, the appearance of slow growth on solid agar in adhesive cells could be partially artifactual. Indeed, we have observed that adhesive cells tend to stick to flasks and, when grown on agar plates, cells in the same colony can stick to one another rather than to inoculation loops or pin pads. Both of these dynamics can reduce initial inoculation densities. This is less of a concern for our adhesion assay and Figures 2E, 5B, and 5F, because our before-wash intensity was done with a 7x7 pinned square about 10x10 mm2. Nonetheless, as we wanted to make a point about srb10 and srb11 mutants growing faster than other deletion mutants that exhibit MLP-formation, we also conducted growth assays in liquid media (New Figure 5F).
We observed that srb10Δ and srb11Δ strains (which exhibit MLPs in EMM) show growth curves similar to wild-type cells in minimal (EMM) and rich media (YES). On the other hand, other strains that grow similarly to wild type cells in YES, such as tlg2Δ and rpa12Δ, grow much more slowly in EMM when they clump together. There are also some strains, mus7Δ and kgd2Δ, that grow more slowly in both YES and EMM but are only adhesive in EMM.
The text mentions two lab strains, JB22 and JB50, displaying strong adhesion under phosphate starvation (lines 525-526), yet the data point for JB22 in Figure 2C is not labeled.
Reply: We agree that highlighting JB22 on the figure is crucial, given that it was mentioned in the main text. JB22 is now highlighted in green on Fig 2C.
- Although I generally avoid commenting on formatting, I found the manuscript to be dense. As mentioned above, I truly enjoyed reading it! But I couldn't help but think of ways to make the manuscript more concise for readers. The Results section spans nine pages (excluding figure captions), and the Discussion is five pages long. The main text contains 6 figures with approximately 27 panels and 32 plots and Venn diagrams, while the supplementary material has 11 figures with 22 panels and about 59 plots. Altogether, the manuscript comprises 17 figures, 49 panels, and roughly 91 plots and Venn diagrams! While I will not request any changes, I encourage the authors to consider streamlining the text/data where possible to focus on the core theme of the study.
We thank the reviewer for these suggestions and have reorganised some of our figures and text to appear less dense. We have also added several figures and panels in response to reviewer comments. While we endeavor to make our points clear and concise in the main figures, we believe that it is important to retain key supplementary figures so that an interested reader can evaluate the data in more detail:
A summary of our major changes to the figures is below, and we also provide a manuscript with changes tracked for the reviewers' convenience:
Fig 2:
Added Panel E in response to reviewer comments. Fig 3:
Removed axes for pfl3 and pfl7 from Fig 3C, as the point was made by the other genes displayed (mbx2, pfl8 and gsf2) Replaced Fig 3D with similar data from an improved experiment in response to reviewer comments. Added New Fig 3F from Original Supp Fig 5 Fig 5:
Moved Original Fig 5A to New Supp Fig 10A. Added New Fig 5F in response to reviewer comments. Original Supp Fig 4 / New Supp Fig 6:
Removed mbx2 overexpression images from Original Fig 4C, to be replaced by new overexpression data and images in New Fig 3D. Added flask images for srb10 and srb11 deletion mutants from Original Supp Fig 5A to New Supp Fig 6C. Added microscope image for srb11 deletion mutant from Ooriginal Supp Fig 5A to New Supp Fig 6C. Added adhesion assay results from Original Supp Fig 5C to New Supp Fig 6C. Added New Supp Fig 6D in response to review Original Supp Fig 5
Removed this figure. Original Supp Fig 5A and 5B were moved to New Supp Fig 6. Original Supp Fig 5B was removed to make the manuscript more concise. Original Supp Figs 6, 7 and 8 were combined into New Supp Fig 8.
Original Supp Fig 6A and 6B are now New Supp Fig 8A and 8B. Original Supp Fig 7 is now New Supp Fig 8C. Original Supp Fig 8A is now New Supp Fig 8D and 8E. Original Supp Fig 8B is now New Supp Fig 8F Original Supp Fig 9/New Supp Fig 10
Added Original Fig 5A as new Supp Fig 10A. Original Supp Fig 11/New Supp Fig 12
Removed Original Fig 11B and the relevant text to make the manuscript more concise. Added New Supp Fig 12B in response to reviewer comments. New Supplementary Figures added in response to reviewer comments:
New Supp Fig 4: Microscopy images of natural isolates. New Supp Fig 5: Flask images of natural isolates New Supp Fig 7: Microscopy and flask images of mbx2 overexpression strains. New Supp Fig 9: Genomic comparisons between JB759 and the MLP-forming wild isolate, JB914. Removed some less relevant points from our discussion, to reduce the length.
Added new Supplementary Tables:
Supplementary Table 13: Variants in candidate genes. Added in response to reviewer comments Supplementary Table 14: List of plasmids used in the study.
**Referees cross-commenting**
There are many useful recommendations from all the other reviewers that will help improve the final product. Once those points are revised, I think this will be a nice paper of interest to folks interested in natural variation in MLPs and its genetic background.
Significance
My expertise: evolutionary genetics, evolution of multicellularity, yeast genetics, experimental evolution
Overall, the manuscript is well-written, dense yet logically structured, and the figures are well presented. The combination of phenotypic, genetic, and bioinformatics analyses, particularly from wet lab experiments, is commendable. The study addresses a significant gap in our understanding, primarily explored in budding yeast, by providing comprehensive data on MLP diversity in fission yeast and the interplay of genetic and environmental factors.
In summary, I enjoyed reading the manuscript and have only a few minor suggestions to strengthen the paper.
Reviewer #2
Evidence, reproducibility and clarity
REVIEWER COMMENTS
Yeast species, including fission yeast and budding yeast, could form multicellular-like phenotypes (MLP). In this work, Kӧvér and colleagues found most proteins involved in MLP formation are not functionally conserved between S. pombe and budding yeast by bioinformatic analysis. The authors analyzed 57 natural S. pombe isolates and found MLP formation to widely vary across different nutrient and drug conditions. The authors demonstrate that MLP formation correlated with expression levels of the transcription factor gene mbx2 and several flocculins. The authors also show that Cdk8 kinase module and srub11 deletions also resulted in MLP formation. The experimental design is logic, the manuscript is well-written and organized. I have a few concerns that should be addressed before the publication.
Major points:
1) Line 61-62, how did the authors grow yeast cells in the liquid medium? Shaking or static? If shaking, the nutrient should be even distributed in the medium.
If static culture, most single yeast cells could precipitate on the bottom, how do you address the advantage of flocculation for increasing the sedimentation? In addition, under static culture, the bottom will have less air than the up medium, how to balance the air and nutrients?
Reply: In line 61-62 we stated that "Similarly, flocculation could increase sedimentation in liquid media, thereby assisting the search for more nutrient-rich or less stressful environments (4)".
Our intent was to speculate on the advantages of multicellular-like growth, and cited a review article which has mentioned sedimentation. After further consideration, we decided that this is a minor point and is rather speculative, and removed it altogether from the manuscript.
In response to the Reviewer's question about how cells were grown in liquid medium, throughout the paper we used shaking cultures for our flocculation assays and for pre-cultures. We have made this more clear in the text where it was ambiguous (e.g. line 189, throughout the methods section, and in the legend of Fig. 2A).
2) Line 555, it will be interesting to test whether overexpression of mbx2 could cause flocculation in YES medium. In Figure 3D, the authors use two control strains, but only one mbx2 OE strain, mbx2 OE should be tested in both strains. In addition, did the authors transform empty plasmid into the control strains, please indicate in the figure.
In this experiment, mbx2 was overexpressed using a thiamine-repressible nmt1 promoter, which is a standard construct in fission yeast studies. Assaying MLP formation was not feasible in YES with this strain, because YES is a rich media made up of yeast extract which contains thiamine. Thus, we could not remove thiamine from the media to trigger mbx2 overexpression.
In order to test the influence of mbx2 overexpression in YES, we constructed strains in which mbx2 was integrated into the genome and expression was driven by the rpl2102 promoter, which has been shown to provide constitutive moderate expression levels (12). We observed strong flocculation in both EMM and YES (Fig 3D, New Supplementary Figure 7) . We did not see strong flocculation in a control in which GFP was expressed under the rpl2102 promoter. The flocculation phenotype was so strong that our original adhesion assay protocol required modification for this experiment, including resuspension in 10 mM EDTA before repinning (Methods). We observed strong adhesion for the mbx2 overexpression strains (Fig 3D), but not for control strains in YES. We could not check adhesion in EMM for those strains because cells pinned on EMM did not survive resuspension in EDTA.
We performed these experiments in two backgrounds, 968 h90 (JB50), which is one of the parental strains of the segregant library analysed in Figure 3 and 972 h- (JB22), which is an appropriate background for the gene deletion collection.
We have replaced the data from the original Figure 3D with the new adhesion assay and added New Supplementary Figure 7 to the manuscript (Lines 236-244).
This result also helped us to further refine our model for the pathway. We can now say that the repression of MLPs in rich media must act via Mbx2, as overexpression of mbx2 is sufficient to abolish it, and is likely to act transcriptionally (if it acted on the protein level, the mild overexpression would likely not have led to the phenotype) (Figure 6, Lines 554-556 in the discussion)
3) Line 600-601, the authors may do the backcross of srb11Δ::Kan to exclude the possibility caused by other mutations.
Reply: We thank the reviewer for noticing our concern about suppressor mutations arising in the srb11Δ strain obtained from our deletion library. This initial concern arose following the observation that while qualitatively the srb11Δ::Kan and srb11Δ(CRISPR) strains were both strongly adhesive, there was a minor quantitative difference in their adhesion.
As we obtained this strain from an h+ deletion library strain backcrossed with a prototrophic h- strain (JB22) in order to restore auxotrophies (13), the chances for a suppressor mutation to arise are very low. We have therefore removed that language from our text. We now suspect that a more likely explanation for this small difference could be the strain background, as our CRISPR engineered strain was made in a JB50 background which has the h90 mating type, while the deletion library strains are h- without auxotrophic markers.
We would like to emphasize, however, that despite this quantitative difference in the adhesion phenotype between the two srb11Δ strains, they both have a large increase in the adhesion phenotype relative to the respective wild-type strains. To address this point, we have removed the unnecessary statistical comparison of these two deletion strains and focused on their qualitatively high levels of adhesion in the text (lines 267-269) and in our Revised Supplementary Figure 6D.
Minor points:
1) Line 506, what are the growth conditions of cells in Figure 2A? Did the authors use the liquid or solid medium? Please mention in the Methods or figure legends.
Reply: We have updated the manuscript to include the relevant details in the text (line 189), figure caption for Fig. 2A and in the methods section (lines 829-831).
2) Line 533-535, please explain why the strains exhibiting strong adhesion have a decreased growth rate. Is there any related research? Please add some references.
Reply: Please see reply to Reviewer 1, comment 5.
**Referees cross-commenting**
I agree with most of the comments from other reviewers. This publication may indeed be of interest to a minor area. But the results and the interpretations of the data are interesting and warranted, the findings are scientifically important.
Significance
The authors did many large-scale screens and bioinformatic analyses. The experiments in the manuscript are generally logical and sound. This study is useful for deciphering the mechanism of multicellular-like phenotype formation in the fission yeast, with some implications for some other organisms.
Reviewer #3 (Evidence, reproducibility and clarity (Required)):
Summary: Using a variety of targeted and genome wide analyses, the authors investigate the basis for "multicellular-like phenotypes" in S. pombe. Authors developed several methodologies to detect and quantify "multicellular-like phenotypes" (flocculation, aggregation...) and defined genes involved in these processes in laboratory and wild S. pombe.
SECTION A - Evidence, reproducibility and clarity
This is a very solid manuscript that is well-written and supported by convincing data. While one can imagine many additional experiments, the manuscript stands on its own and presents a quite exhaustive analysis of the area. I commend the author for their rigorous work and clear presentation. They are only a few minor points that warrant comments or corrections: - Supplementary Figure 1 is a typical example of the "necessity" to have statistics and P-values everywhere. The data are convincing but what is the evidence that the Filtering assay and the Plate-reader assay values should be linearly related? Lets imagine that Plate-reader assay value is proportional to the square of the Filtering assay value. What would be the Pearson R and P-value in this case? What is most appropriate? Why would one use a linear correlation? What is the "real" significance?
Reply: We thank the reviewer for pointing out that the data in Supplementary Figure 1 does not appear to be linear and, therefore, reporting the Pearson correlation coefficient may not be the best way to represent the relationship between the two assays. The nonlinear nature of this data could indicate that
The filtering assay saturates before the plate reader assay, and is less able to distinguish between strains that flocculate strongly and The filtering assay may be more sensitive for strains that show lower levels of flocculation. In general, we observed fewer strains with intermediate phenotypes for both assays, making it difficult to ascertain the true relationship between them; however, we believe that the key result is that the strains with the highest level of flocculation have the highest values in both assays. To capture this aspect of the data, we now report the Spearman correlation which is non-parametric and indicates how similar the ranking of each strain is based on both assays. With the alternative hypothesis being that the correlation is > 0, we report a Spearman correlation coefficient of 0.24 and a P-value of 0.04 (lines 823-826)
- Minor points: * They are several "personal communications" in the manuscript (page 11, page 18, page 23). It should be checked whether this is accepted in the journal that publishes this manuscript.
Reply: We thank the reviewer for highlighting this issue. We had three instances of "personal communications" in our original submission.
The first instance was an acknowledgement for advice on our DNA extraction protocol from Dan Jeffares. We now include this in the Acknowledgements section instead.
The second communication with Angad Garg described that they observed flocculation while growing cells in phosphate starvation conditions, which was not reported in their publication (14). Though we appreciate their willingness to share unpublished data with us, we have removed this observation from our manuscript and instead rely only on our own observations and arguments based on their published RNA-seq data to make our point.
The third personal communication with Olivia Hillson supplements a minor hypothesis, namely that deletion of SPNCRNA.781 might cause MLP formation by affecting the promoter of hsr1, for which we had access to unpublished ChIP-seq data, showing its binding to flocculins. Recently published work from a different group (15) also suggests this link between hsr1 and flocculation and is now discussed in our manuscript instead of the result based on unpublished data obtained from personal communication at Lines 397-398.
* Page 4 check "a few regulators"
Reply: For clarity, this has now been changed to "several regulatory proteins" at Line 108. The specific proteins we are referring to are highlighted in Figure 1C.
* Page 19, line 567: "remaining 8 strains" may be confusing as Material and Methods states "remaining 10 strains".
Reply: Two of the 10 strains were found to be redundant after sequencing as explained in the Methods (Lines 930-934). Therefore, we only added 8 new strains to the analysis. We thank the reviewer for highlighting this as a potential source of misunderstanding, and clarified this point in the text (Lines 247-250 and in the methods).
**Referees cross-commenting**
I concur with most comments. Overall, the reviewers agree that this is a solid piece of work that could benefit from minor modifications and should be published. I reiterate that, for me, despite its quality, this publication will only be of interest to specialists.
Reviewer #3 (Significance (Required)):
A limited number of studies have investigated "multicellular-like phenotypes" in S. pombe. This manuscript brings therefore new and solid information. Yet, despite an impressive amount of work, our conceptual advance in understanding this process and its phylogenetic conservation remains limited. This is probably best illustrated in the figure 6 that summarize the study and contains 3 question marks and an additional unknown mechanism. (Most of the solid arrows in this figure correspond to interactions within the Mediator complex that were well known before this study.) In addition, while only few studies have been published in this area, the authors' findings are often only bringing additional support to already published observations. Overall, while this manuscript will be of interest to a restricted group of aficionados, it will most likely not attract the attention of a wide readership.
__ Reviewer #4 (Evidence, reproducibility and clarity (Required)):__
In this manuscript, the authors explore how multicellular-like phenotypes (MLPs) arise in the fission yeast S. pombe. Although yeasts are characterized as unicellular fungi, diverse species show MLPs, including filamentous growth on agar plates and flocculation in liquid media. MLPs may provide certain advantages in nutritionally poor conditions and protection against external challenges, upon which natural selection can then act. Previous work on MLPs has mostly been carried out in the budding yeasts S. cerevisiae and C. albicans, and little was known about these behaviors in S. pombe. The authors thus set out to investigate both genetic and environmental regulators of MLP formation.
First, their analysis of published data revealed a limited number of shared regulators of MLP between S. pombe, S. cerevisiae, and C. albicans, although the cell adhesion proteins themselves are largely not conserved. Next, the authors screened a set of non-clonal natural isolates using two high-throughput assays that they developed and found that MLPs vary in strains and depending on nutrient conditions. Focusing on a natural isolate that showed both adhesion on agar plates and flocculation in liquid medium, they then analyzed a segregant library generated from this and a laboratory strain using their assays. Using QTL analysis, they uncovered a frameshift in the srb11 gene, which encodes a subunit of the Mediator complex, as the likely causal inducer of MLP. This was confirmed by additional analyses of strains lacking srb11 or other members of Mediator. Furthermore, the authors showed that loss of srb11 function resulted in the upregulation of the Mbx2 transcription factor, which was both necessary and sufficient for MLP formation in this background. Finally, screening of two additional yeast strain collections (gene and long intergenic non-coding RNA deletion) identified both known and novel regulators representing different pathways that may be involved in MLP formation.
Altogether, this study provides new perspectives into our understanding of the diverse inputs that regulate multicellular-like phenotypes in yeast.
Major comments:
- The methods for screening for adhesion and flocculation are well described, with representative figures that show plates and flasks. However, there are few microscopy images of cells, and it would be interesting and helpful for the reader to have an idea of how cells look when they exhibit MLPs. For instance, are there any differences in cell shape or size when strains present different degrees of adhesion or flocculation? In addition, the authors mention that mutants with strong adhesion generally had lower colony density and are likely to be slower growing. Although their analyses suggest otherwise (page 22), this has a potential for introducing error in their observations, and including images of the adhesion/flocculation phenotypes may provide further support for their conclusions. I suggest that the authors present microscopy images 1) similar to what is shown for JB759 in Figure 2A and 2) of cells growing on agar in the adhesion assay. This could be included for the different Mediator subunit deletions that they tested, where there appear to be varying phenotypes. It could also be informative for a subset of the 31 high-confidence candidates that they identified in their screen.
Reply: We thank the reviewer for highlighting the need for further microscopic characterisation of MLP forming strains. We therefore now include images of JB914, JB953 (New Supplementary Figures 4, Figure 2E) in liquid media in EMM, EMM-Phosphate, and YES; an srb11 deletion strain (Figure 3F), and mbx2 overexpression strains (New Supplementary Figure 7).
- Upon identifying a frameshift in srb11 that is responsible for the MLP, the authors assessed whether deletion of other Mediator subunits would result in the same phenotype. They found that srb10 and srb11 deletions both flocculate and show adhesion, while other mutants had milder phenotypes. However, the authors also found that a new deletion of srb11 that they generated had a stronger adhesion phenotype than the srb11 deletion from the prototrophic deletion library, which was attributed this the accumulation of suppressor mutations in the strains of the deletion collection. As the authors make clear distinctions between the phenotypes of different Mediator mutants, I suggest generating and analyzing "clean" deletions of the 6 other subunits that they tested. This would strengthen their conclusion and help to rule out accumulated suppressors as the cause of the differences in the observed phenotypes.
Reply: We thank the reviewer for noticing our concern about suppressor mutations in the manuscript. As we describe above in response to a similar question from reviewer 2, as the prototrophic deletion library from which we extracted the Mediator deletion strains had been backcrossed during its construction (13), we no longer suspect that small difference between the srb11Δ::Kan strain from the deletion library and the newly created srb11Δ (CRISPR) strains is due to suppressor mutations. Rather, we think they may be a result of the difference in genetic background and possibly mating type between the two strains. We also want to emphasize that this difference is small compared to the difference between the adhesion ratios of the srb11Δ strains and their respective control strains.
Nevertheless, we made clean, independent Mediator mutants for 5 out of 6 Mediator genes tested (med10Δ, med13Δ, med19Δ, med27Δ, and srb10Δ) as well as an additional mutant that we didn't have in our library, med12Δ (Figure R9). When running the assay on these new strains we got an overall lower dynamic range, possibly due to variations in the water flow rate relative to the first assay. However, we saw a strong phenotype for both library and our own srb10Δ and CRISPR srb11Δ strains. We did not see a significant increase in adhesion for the other Mediator deletion mutants in EMM relative to wild type with the exception of for med10Δ in both the library strain and for our clean mutant, for which we did not observe a phenotype in our previous experiment. We included the experiment for the newly created mutants as New Supplementary Figure S6E and described them in lines 276-281 in our revised manuscript.
Minor comments:
- One point that recurs in the manuscript is the idea that mutations that give rise to strong MLPs also generally lead to slower growth, representing a potential trade-off. This idea could be reinforced with measurements of growth rate or generation time by optical density or cell number, for instance, rather than comparisons of colony density. Also, it would be interesting to mention if the slow growth phenotype is only observed in MLP-inducing conditions or also in rich medium.
Reply: As described above in response to item 5 from Reviewer 1, we have conducted growth assays in liquid media for srb10Δ, srb11Δ, and other mutants from our adhesion screen (tlg2Δ, rpa12Δ, mus7Δ and kgd2Δ) that showed a similar phenotype to those genes in both minimal (EMM) and rich (YES) media. We observe that in rich media, srb10Δ and srb11Δ cells grow similarly to control strains, and they exhibit a lower decrease in growth rate than the other similarly adhesive strains. Both mus7Δ and kgd2Δ cells grow more slowly, even in rich media.
We have also added data on the tradeoff between growth and adhesion based on growth on solid media from (11) for all mutants identified in our screen (New Supp Fig 12B)).
Thus, the relationship between slow growth and clumpiness depends on the mutation, and specifically, mutations of the Mediator, including those to srb11 and srb10, seem to decrease the impact of any tradeoff between growth and adhesion.
- The authors show that the MLPs of the srb10 and srb11 deletions occur through mbx2 upregulation. Do the varying strengths of the phenotypes of the strains lacking different Mediator subunits correlate with mbx2 levels in these backgrounds?
Reply: There is some evidence from previous work that the relationship between the strength of the MLPs and the expression of mbx2 may not be perfectly proportional. In (16), med12Δ had a higher (though qualitatively comparable) level of mbx2 upregulation than srb10Δ (New Supp Fig 8E), even though that paper reported a milder phenotype for med12Δ than for srb10Δ cells. We did not observe a significant increase in adhesion in our med12Δ strain (New Supp Fig 6D). This suggests that in the case of these mutants, it is not simply the level of mbx2 that controls MLP formation, but that there are likely additional regulatory mechanisms. We have added some discussion on this context in the manuscript (lines 545-547).
**Referees cross-commenting**
I agree overall with the comments and suggestions from the other reviewers. The revision would require only minor modifications. The paper is interesting both for the combination of methodologies used and its findings, and I believe that it would benefit a growing community of researchers.
Reviewer #4 (Significance (Required)):
This study employed a variety of methods that allowed the authors to uncover previously unknown regulators of MLPs. Taking advantage of the diversity of natural fission yeast isolates as well as the constructed gene and non-coding RNA deletion collections, the authors identified novel genetic determinants that give rise to MLPs, opening new avenues into this exciting area of research. The overall conclusions of the work are solid and supported by the reported results and analyses. This study will be appreciated by a broad audience of readers who are interested in understanding how organisms respond to environmental challenges as well as how MLPs may result in emergent properties that play key roles in these responses. Some of the limitations of the work are described above, with recommendations for addressing these points.
Keywords for my field of expertise: fission yeast, cell cycle, transcription, replication.
References for Response to Reviews
- Brysch-Herzberg M, Jia GS, Seidel M, Assali I, Du LL. Insights into the ecology of Schizosaccharomyces species in natural and artificial habitats. Antonie Van Leeuwenhoek. 2022 May 1;115(5):661-95.
- Jeffares DC, Rallis C, Rieux A, Speed D, Převorovský M, Mourier T, et al. The genomic and phenotypic diversity of Schizosaccharomyces pombe. Nat Genet. 2015 Mar;47(3):235-41.
- Ratcliff WC, Denison RF, Borrello M, Travisano M. Experimental evolution of multicellularity. Proc Natl Acad Sci. 2012 Jan 31;109(5):1595-600.
- Smukalla S, Caldara M, Pochet N, Beauvais A, Guadagnini S, Yan C, et al. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell. 2008 Nov 14;135(4):726-37.
- Lorenz MC, Heitman J. Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog. EMBO J. 1997 Dec 1;16(23):7008-18.
- Ignacia DGL, Bennis NX, Wheeler C, Tu LCL, Keijzer J, Cardoso CC, et al. Functional analysis of Saccharomyces cerevisiae FLO genes through optogenetic control. FEMS Yeast Res. 2025 Sept 24;25:foaf057.
- Wang Z, Xu W, Gao Y, Zha M, Zhang D, Peng X, et al. Engineering Saccharomyces cerevisiae for improved biofilm formation and ethanol production in continuous fermentation. Biotechnol Biofuels Bioprod. 2023 July 31;16(1):119.
- Koschwanez JH, Foster KR, Murray AW. Improved use of a public good selects for the evolution of undifferentiated multicellularity. eLife. 2013 Apr 2;2:e00367.
- Westman JO, Mapelli V, Taherzadeh MJ, Franzén CJ. Flocculation Causes Inhibitor Tolerance in Saccharomyces cerevisiae for Second-Generation Bioethanol Production. Appl Environ Microbiol. 2014 Nov;80(22):6908-18.
- Li R, Li X, Sun L, Chen F, Liu Z, Gu Y, et al. Reduction of Ribosome Level Triggers Flocculation of Fission Yeast Cells. Eukaryot Cell. 2013 Mar;12(3):450-9.
- Rodríguez-López M, Bordin N, Lees J, Scholes H, Hassan S, Saintain Q, et al. Broad functional profiling of fission yeast proteins using phenomics and machine learning. Marston AL, James DE, editors. eLife. 2023 Oct 3;12:RP88229.
- Hebra T, Smrčková H, Elkatmis B, Převorovský M, Pluskal T. POMBOX: A Fission Yeast Cloning Toolkit for Molecular and Synthetic Biology. ACS Synth Biol. 2024 Feb 16;13(2):558-67.
- Malecki M, Bähler J. Identifying genes required for respiratory growth of fission yeast. Wellcome Open Res. 2016 Nov 15;1:12.
- Garg A, Sanchez AM, Miele M, Schwer B, Shuman S. Cellular responses to long-term phosphate starvation of fission yeast: Maf1 determines fate choice between quiescence and death associated with aberrant tRNA biogenesis. Nucleic Acids Res. 2023 Feb 16;51(7):3094-115.
- Ohsawa S, Schwaiger M, Iesmantavicius V, Hashimoto R, Moriyama H, Matoba H, et al. Nitrogen signaling factor triggers a respiration-like gene expression program in fission yeast. EMBO J. 2024 Oct 15;43(20):4604-24.
- Linder T, Rasmussen NN, Samuelsen CO, Chatzidaki E, Baraznenok V, Beve J, et al. Two conserved modules of Schizosaccharomyces pombe Mediator regulate distinct cellular pathways. Nucleic Acids Res. 2008 May;36(8):2489-504.
-
Note: This preprint has been reviewed by subject experts for Review Commons. Content has not been altered except for formatting.
Learn more at Review Commons
Referee #4
Evidence, reproducibility and clarity
In this manuscript, the authors explore how multicellular-like phenotypes (MLPs) arise in the fission yeast S. pombe. Although yeasts are characterized as unicellular fungi, diverse species show MLPs, including filamentous growth on agar plates and flocculation in liquid media. MLPs may provide certain advantages in nutritionally poor conditions and protection against external challenges, upon which natural selection can then act. Previous work on MLPs has mostly been carried out in the budding yeasts S. cerevisiae and C. albicans, and little was known about these behaviors in S. pombe. The authors thus set out to investigate both genetic and environmental regulators of MLP formation.
First, their analysis of published data revealed a limited number of shared regulators of MLP between S. pombe, S. cerevisiae, and C. albicans, although the cell adhesion proteins themselves are largely not conserved. Next, the authors screened a set of non-clonal natural isolates using two high-throughput assays that they developed and found that MLPs vary in strains and depending on nutrient conditions. Focusing on a natural isolate that showed both adhesion on agar plates and flocculation in liquid medium, they then analyzed a segregant library generated from this and a laboratory strain using their assays. Using QTL analysis, they uncovered a frameshift in the srb11 gene, which encodes a subunit of the Mediator complex, as the likely causal inducer of MLP. This was confirmed by additional analyses of strains lacking srb11 or other members of Mediator. Furthermore, the authors showed that loss of srb11 function resulted in the upregulation of the Mbx2 transcription factor, which was both necessary and sufficient for MLP formation in this background. Finally, screening of two additional yeast strain collections (gene and long intergenic non-coding RNA deletion) identified both known and novel regulators representing different pathways that may be involved in MLP formation.
Altogether, this study provides new perspectives into our understanding of the diverse inputs that regulate multicellular-like phenotypes in yeast.
Major comments:
- The methods for screening for adhesion and flocculation are well described, with representative figures that show plates and flasks. However, there are few microscopy images of cells, and it would be interesting and helpful for the reader to have an idea of how cells look when they exhibit MLPs. For instance, are there any differences in cell shape or size when strains present different degrees of adhesion or flocculation? In addition, the authors mention that mutants with strong adhesion generally had lower colony density and are likely to be slower growing. Although their analyses suggest otherwise (page 22), this has a potential for introducing error in their observations, and including images of the adhesion/flocculation phenotypes may provide further support for their conclusions. I suggest that the authors present microscopy images 1) similar to what is shown for JB759 in Figure 2A and 2) of cells growing on agar in the adhesion assay. This could be included for the different Mediator subunit deletions that they tested, where there appear to be varying phenotypes. It could also be informative for a subset of the 31 high-confidence candidates that they identified in their screen.
- Upon identifying a frameshift in srb11 that is responsible for the MLP, the authors assessed whether deletion of other Mediator subunits would result in the same phenotype. They found that srb10 and srb11 deletions both flocculate and show adhesion, while other mutants had milder phenotypes. However, the authors also found that a new deletion of srb11 that they generated had a stronger adhesion phenotype than the srb11 deletion from the prototrophic deletion library, which was attributed this the accumulation of suppressor mutations in the strains of the deletion collection. As the authors make clear distinctions between the phenotypes of different Mediator mutants, I suggest generating and analyzing "clean" deletions of the 6 other subunits that they tested. This would strengthen their conclusion and help to rule out accumulated suppressors as the cause of the differences in the observed phenotypes.
Minor comments:
- One point that recurs in the manuscript is the idea that mutations that give rise to strong MLPs also generally lead to slower growth, representing a potential trade-off. This idea could be reinforced with measurements of growth rate or generation time by optical density or cell number, for instance, rather than comparisons of colony density. Also, it would be interesting to mention if the slow growth phenotype is only observed in MLP-inducing conditions or also in rich medium.
- The authors show that the MLPs of the srb10 and srb11 deletions occur through mbx2 upregulation. Do the varying strengths of the phenotypes of the strains lacking different Mediator subunits correlate with mbx2 levels in these backgrounds?
Referees cross-commenting
I agree overall with the comments and suggestions from the other reviewers. The revision would require only minor modifications. The paper is interesting both for the combination of methodologies used and its findings, and I believe that it would benefit a growing community of researchers.
Significance
This study employed a variety of methods that allowed the authors to uncover previously unknown regulators of MLPs. Taking advantage of the diversity of natural fission yeast isolates as well as the constructed gene and non-coding RNA deletion collections, the authors identified novel genetic determinants that give rise to MLPs, opening new avenues into this exciting area of research. The overall conclusions of the work are solid and supported by the reported results and analyses. This study will be appreciated by a broad audience of readers who are interested in understanding how organisms respond to environmental challenges as well as how MLPs may result in emergent properties that play key roles in these responses. Some of the limitations of the work are described above, with recommendations for addressing these points.
Keywords for my field of expertise: fission yeast, cell cycle, transcription, replication.
-
Note: This preprint has been reviewed by subject experts for Review Commons. Content has not been altered except for formatting.
Learn more at Review Commons
Referee #3
Evidence, reproducibility and clarity
Summary:
Using a variety of targeted and genome wide analyses, the authors investigate the basis for "multicellular-like phenotypes" in S. pombe. Authors developed several methodologies to detect and quantify "multicellular-like phenotypes" (flocculation, aggregation...) and defined genes involved in these processes in laboratory and wild S. pombe.
SECTION A - Evidence, reproducibility and clarity
This is a very solid manuscript that is well-written and supported by convincing data. While one can imagine many additional experiments, the manuscript stands on its own and presents a quite exhaustive analysis of the area. I commend the author for their rigorous work and clear presentation. They are only a few minor points that warrant comments or corrections:
- Supplementary Figure 1 is a typical example of the "necessity" to have statistics and P-values everywhere. The data are convincing but what is the evidence that the Filtering assay and the Plate-reader assay values should be linearly related? Lets imagine that Plate-reader assay value is proportional to the square of the Filtering assay value. What would be the Pearson R and P-value in this case? What is most appropriate? Why would one use a linear correlation? What is the "real" significance?
Minor points:
- They are several "personal communications" in the manuscript (page 11, page 18, page 23). It should be checked whether this is accepted in the journal that publishes this manuscript.
- Page 4 check "a few regulators"
- Page 19, line 567: "remaining 8 strains" may be confusing as Material and Methods states "remaining 10 strains".
Referees cross-commenting
I concur with most comments. Overall, the reviewers agree that this is a solid piece of work that could benefit from minor modifications and should be published. I reiterate that, for me, despite its quality, this publication will only be of interest to specialists.
Significance
A limited number of studies have investigated "multicellular-like phenotypes" in S. pombe. This manuscript brings therefore new and solid information. Yet, despite an impressive amount of work, our conceptual advance in understanding this process and its phylogenetic conservation remains limited. This is probably best illustrated in the figure 6 that summarize the study and contains 3 question marks and an additional unknown mechanism. (Most of the solid arrows in this figure correspond to interactions within the Mediator complex that were well known before this study.) In addition, while only few studies have been published in this area, the authors' findings are often only bringing additional support to already published observations. Overall, while this manuscript will be of interest to a restricted group of aficionados, it will most likely not attract the attention of a wide readership.
-
Note: This preprint has been reviewed by subject experts for Review Commons. Content has not been altered except for formatting.
Learn more at Review Commons
Referee #2
Evidence, reproducibility and clarity
Yeast species, including fission yeast and budding yeast, could form multicellular-like phenotypes (MLP). In this work, Kӧvér and colleagues found most proteins involved in MLP formation are not functionally conserved between S. pombe and budding yeast by bioinformatic analysis. The authors analyzed 57 natural S. pombe isolates and found MLP formation to widely vary across different nutrient and drug conditions. The authors demonstrate that MLP formation correlated with expression levels of the transcription factor gene mbx2 and several flocculins. The authors also show that Cdk8 kinase module and srub11 deletions also resulted in MLP formation. The experimental design is logic, the manuscript is well-written and organized. I have a few concerns that should be addressed before the publication.
Major points:
- Line 61-62, how did the authors grow yeast cells in the liquid medium? Shaking or static? If shaking, the nutrient should be even distributed in the medium. If static culture, most single yeast cells could precipitate on the bottom, how do you address the advantage of flocculation for increasing the sedimentation? In addition, under static culture, the bottom will have less air than the up medium, how to balance the air and nutrients?
- Line 555, it will be interesting to test whether overexpression of mbx2 could cause flocculation in YES medium. In Figure 3D, the authors use two control strains, but only one mbx2 OE strain, mbx2 OE should be tested in both strains. In addition, did the authors transform empty plasmid into the control strains, please indicate in the figure.
- Line 600-601, the authors may do the backcross of srb11Δ::Kan to exclude the possibility caused by other mutations.
Minor points:
- Line 506, what are the growth conditions of cells in Figure 2A? Did the authors use the liquid or solid medium? Please mention in the Methods or figure legends.
- Line 533-535, please explain why the strains exhibiting strong adhesion have a decreased growth rate. Is there any related research? Please add some references.
Referees cross-commenting
I agree with most of the comments from other reviewers. This publication may indeed be of interest to a minor area. But the results and the interpretations of the data are interesting and warranted, the findings are scientifically important.
Significance
The authors did many large-scale screens and bioinformatic analyses. The experiments in the manuscript are generally logical and sound. This study is useful for deciphering the mechanism of multicellular-like phenotype formation in the fission yeast, with some implications for some other organisms.
-
Note: This preprint has been reviewed by subject experts for Review Commons. Content has not been altered except for formatting.
Learn more at Review Commons
Referee #1
Evidence, reproducibility and clarity
Summary
Köver et al. examine the genetic and environmental underpinnings of multicellular-like phenotypes (MLPs) in fission yeast, studying 57 natural isolates of Schizosaccharomyces pombe. They uncover that a noteworthy subset of these isolates can develop MLPs, with the extent of these phenotypes varying according to growth media. Among these, two strains demonstrate pronounced MLP across a range of conditions. By genetically manipulating one strain with an MLP phenotype (distinct from the previously mentioned two strains), they provide evidence that genes such as MBX2 and SRB11 play a direct role in MLP formation, strengthening their genetic mapping findings. The study also reveals that while some key genes and their phenotypic effects are strikingly similar between budding and fission yeast, other aspects of MLP formation are not conserved, which is an intriguing finding.
Overall, the manuscript is well-written, dense yet logically structured, and the figures are well presented. The combination of phenotypic, genetic, and bioinformatics analyses, particularly from wet lab experiments, is commendable. The study addresses a significant gap in our understanding, primarily explored in budding yeast, by providing comprehensive data on MLP diversity in fission yeast and the interplay of genetic and environmental factors.
In summary, I enjoyed reading the manuscript and have only a few minor suggestions to strengthen the paper:
Minor revisions:
- Although this may seem like a minor revision, but it is a crucial point. Please make sure that all raw data used to generate figures, run stats, sequence data, and scripts used to run data analysis are made publicly available. Provide relevant accession numbers and links to public data repositories. It is important that others can download the various types of data that went into the major conclusions of this paper in order to replicate your analysis or expand upon the scope of this work. I am not sure if the journal has a policy regarding this, but it should be followed to allow for transparency and reproducibility of the research.
- Two out of 57 strains exhibit strong and consistent MLP across multiple environments. Providing more information on these strains (JB914 and JB953), such as their natural habitats and distinct appearances of their MLP phenotypes under varying conditions, would provide valuable insights.
First, a brief discussion highlighting what differentiates these two strains from the rest would be helpful for readers (e.g. insight into their unique genetic and environmental background that might be linked to the MLP phenotype).
Additionally, culture tube and microscopy images of these strains, similar to those presented for JB759 in Figure 2A, can be included in the supplementary materials. My reasoning is that these images could help illustrate variation or lack thereof in aggregative group size across different media. 3. The phenotypic outcome of overexpressing MXB2 is striking, as shown in Supplementary Figure 4C. Incorporating at least one of the culture tube images depicting large flocs into the main text, perhaps adjacent to Figure 3 panel D, would improve the visual appeal and highlight this key finding (at the moment those images are only shown in the supplementary materials). 4. I know that the authors discuss the knowledge gap in the intro and results, but the abstract does not mention this critical gap. Please stress this critical gap (i.e., MLPs understudied in fission yeast) with a brief sentence in the abstract. Similarly, please consider writing a brief concluding sentence summarizing the paper's most significant finding referring to the knowledge gap would provide a clearer takeaway message for the reader - the abstract ends abruptly without any conclusion. 5. The observation that strains with adhesive phenotypes have a lower growth rate compared to non-adhesive strains is a noteworthy point (lines 532-535). This represents yet another example of this classical trade-off. This point could be emphasized in the Discussion or alongside the relevant result, with a brief speculative explanation for this phenomenon. 6. The text mentions two lab strains, JB22 and JB50, displaying strong adhesion under phosphate starvation (lines 525-526), yet the data point for JB22 in Figure 2C is not labeled. 7. Although I generally avoid commenting on formatting, I found the manuscript to be dense. As mentioned above, I truly enjoyed reading it! But I couldn't help but think of ways to make the manuscript more concise for readers. The Results section spans nine pages (excluding figure captions), and the Discussion is five pages long. The main text contains 6 figures with approximately 27 panels and 32 plots and Venn diagrams, while the supplementary material has 11 figures with 22 panels and about 59 plots. Altogether, the manuscript comprises 17 figures, 49 panels, and roughly 91 plots and Venn diagrams! While I will not request any changes, I encourage the authors to consider streamlining the text/data where possible to focus on the core theme of the study.
Referees cross-commenting
There are many useful recommendations from all the other reviewers that will help improve the final product. Once those points are revised, I think this will be a nice paper of interest to folks interested in natural variation in MLPs and its genetic background.
Significance
My expertise: evolutionary genetics, evolution of multicellularity, yeast genetics, experimental evolution
Overall, the manuscript is well-written, dense yet logically structured, and the figures are well presented. The combination of phenotypic, genetic, and bioinformatics analyses, particularly from wet lab experiments, is commendable. The study addresses a significant gap in our understanding, primarily explored in budding yeast, by providing comprehensive data on MLP diversity in fission yeast and the interplay of genetic and environmental factors.
In summary, I enjoyed reading the manuscript and have only a few minor suggestions to strengthen the paper.
-
-
www.biorxiv.org www.biorxiv.org
-
eLife Assessment
This important study uses standard single-cell RNA-seq analyses combined with methods from the social sciences to reduce heterogeneity in gene expression in Drosophila imaginal wing disc cells treated with 4000 rads of ionizing radiation. The use of this methodology from social sciences is novel in Drosophila and allows them to identify a subpopulation of cells that is disproportionately responsible for much of the radiation-induced gene expression. Their compelling analyses reveal genes that are expressed regionally after irradiation, including ligands and transcription factors that have been associated with regeneration, as well as others whose roles in response to irradiation are unknown. This paper would be of interest to researchers in the field of DNA damage responses, regeneration, and development.
-
Reviewer #1 (Public review):
Summary:
The authors analyze transcription in single cells before and after 4000 rads of ionizing radiation. They use Seuratv5 for their analyses, which allows them to show that most of the genes cluster along the proximal-distal axis. Due to the high heterogeneity in the transcripts, they use the Herfindahl-Hirschman index (HHI) from Economics, which measures market concentration. Using the HHI, they find that genes involved in several processes (like cell death, response to ROS, DNA damage response (DDR)) are relatively similar across clusters. However, ligands activating the JAK/STAT, Pvr, and JNK pathways and transcription factors Ets21C and dysf are upregulated regionally. The JAK/STAT ligands Upd1,2,3 require p53 for their upregulation after irradiation, but the normal expression of Upd1 in unirradiated discs is p53-independent. This analysis also identified a cluster of cells that expressed tribbles, encoding a factor that downregulates mitosis-promoting String and Twine, that appears to be G2/M arrested and expressed numerous genes involved in apoptosis, DDR, the aforementioned ligands and TFs. As such, the tribbles-high cluster contains much of the heterogeneity.
Strengths:
(1) The authors have used robust methods for rearing Drosophila larvae, irradiating wing discs and analyzing the data with Seurat v5 and HHI.<br /> (2) These data will be informative for the field.<br /> (3) Most of the data is well-presented.<br /> (4) The literature is appropriately cited.
Weaknesses
The authors have addressed my concerns in the revised article.
-
Reviewer #2 (Public review):
This manuscript investigates the question of cellular heterogeneity using the response of Drosophila wing imaginal discs to ionizing radiation as a model system. A key advance here is the focus on quantitatively expressing various measures of heterogeneity, leveraging single-cell RNAseq approaches. To achieve this goal, the manuscript creatively uses a metric from the social sciences called the HHI to quantify the spatial heterogeneity of expression of individual genes across the identified cell clusters. Inter- and intra-regional levels of heterogeneity are revealed. Some highlights include identification of spatial heterogeneity in expression of ligands and transcription factors after IR. Expression of some of these genes shows dependence on p53. An intriguing finding, made possible by using an alternative clustering method focusing on cell cycle progression, was the identification of a high-trbl subset of cells characterized by concordant expression of multiple apoptosis, DNA damage repair, ROS related genes, certain ligands and transcription factors, collectively representing HIX genes. This high-trbl set of cells may correspond to an IR-induced G2/M arrested cell state.
Overall, the data presented in the manuscript are of high quality but are largely descriptive. This study is therefore perceived as a resource that can serve as an inspiration for the field to carry out follow-up experiments.
The authors responded well to my suggestions for improvement, which were incorporated in the revised version of the manuscript.
-
Reviewer #3 (Public review):
Summary:
Cruz and colleagues report a single cell RNA sequencing analysis of irradiated Drosophila larval wing discs. This is a pioneering study because prior analyses used bulk RNAseq analysis so differences at single cell resolution were not discernable. To quantify heterogeneity in gene expression, the authors make clever use of a metric used to study market concentration, the Herfindahl-Hirschman Index. They make several important observations including region-specific gene expression coupled with heterogeneity within each region and the identification of a cell population (high Trbl) that seems disproportionately responsible for radiation-induced gene expression.
Strengths:
Overall, the manuscript makes a compelling case for heterogeneity in gene expression changes that occurs in response to uniform induction of damage by X-rays in a single layer epithelium. This is an important finding that would be of interest to researchers in the field of DNA damage responses, regeneration and development.
Weaknesses:
The authors have addressed my concerns adequately with changes made in the revised version.
-
Author response:
The following is the authors’ response to the original reviews
Reviewing Editor Comment:
The reviewers felt that the study could be improved by (1) better integrating the results with the existing literature in the field
(1) In the Introduction and Results section of the manuscript, we had made every attempt to cite the relevant literature. (Reviewer 1 stated that “The literature is appropriately cited”). We agree with the Reviewing Editor that rather than simply cite the relevant literature, we could have done a better job of integrating our findings with what has been previously discovered by others. We have attempted to do this in the revised manuscript. Also, we have included many additional citations in the Introduction and in the first section of the Results where work by others has provided a framework for interpreting our single-cell studies.
and (2) manipulating Trib expression and analyzing the expression of 1-2 HIX genes.
(2) We are grateful for this suggestion. As suggested by the Reviewing Editor we have attempted to increase and decrease trbl expression and assess the effect on expression of two genes, Swim and CG15784.
We increased trbl levels in the wing pouch using rn-Gal4, tub-Gal80<sup>ts</sup> and UAS-trbl. By transferring larvae for 24 h from 18oC to 31oC, we were able to induce trbl expression in the wing pouch. When these larvae were irradiated at 4000 rad, we found reduced levels of apoptosis in the wing pouch of discs that overexpressed trbl (Figure 7-figure supplement 1). This indicated that upregulation of trbl is radioprotective. Consistent with our findings, others have previously shown that upregulation of trbl and stalling in the G2 phase of the cells cycle protects cells from JNK-induced apoptosis (Cosolo et al., 2019, PMID:30735120) or that downregulating the G2/M progression promoting factor string protects cells from X-ray radiation induced apoptosis (Ruiz-Losada et al., 2021, PMID:34824391).
As suggested by the Reviewing Editor, we also examined the effect of trbl overexpression on the induction of two “highly induced by X-ray irradiation (HIX)” gene, Swim and CG15784. Increasing trbl expression had no effect on the induction of Swim and only a modest decrease in the induction of CG15784 (Figure 7-figure supplement 2). Thus, increasing trbl expression, is in itself, insufficient to promote HIX gene expression indicating that other factors are necessary for HIX gene induction.
We also attempted to reduce trbl expression, using three different RNAi lines. While some of these lines have been used previously by others to reduce trbl expression under unirradiated conditions (Cosolo et al., 2019, PMID:30735120), we nevertheless wanted to check if they reduced trbl induction following irradiation. For each of the three lines, we observed no obvious reduction in trbl RNA following irradiation when visualized using HCR (Author response image 1). Thus, any effects on gene expression that we observe could not be attributed to a decrease in trbl expression. We have therefore included the images showing a lack of knockdown in this Response to Reviews document but not included these experiments in the revised manuscript.
Author response image 1.
RNA in situ hybridizations using the hybridization chain reaction performed using probes to trbl. In A-F, the RNAi is expressed using nubbin-Gal4. In G-I the RNAi is expressed using rn-Gal4, tub-Gal80<sup>ts</sup>. white-RNAi was used as a control (A, B, G, H). Three different RNAi lines directed against trbl were tested: Vienna lines VDRC 106774 (C, D) and VDRC 22113 (E, F), and Bloomington line BL42523. In no case was a reduction in trbl RNA upregulation in the wing pouch following 4000 rad observed, except for one disc (n = 6) of VDRC 106774 crossed to nubbin-gal4.
Reviewer #1 (Public review):
Summary:
The authors analyze transcription in single cells before and after 4000 rads of ionizing radiation. They use Seuratv5 for their analyses, which allows them to show that most of the genes cluster along the proximal-distal axis. Due to the high heterogeneity in the transcripts, they use the Herfindahl-Hirschman index (HHI) from Economics, which measures market concentration. Using the HHI, they find that genes involved in several processes (like cell death, response to ROS, DNA damage response (DDR)) are relatively similar across clusters. However, ligands activating the JAK/STAT, Pvr, and JNK pathways and transcription factors Ets21C and dysf are upregulated regionally. The JAK/STAT ligands Upd1,2,3 require p53 for their upregulation after irradiation, but the normal expression of Upd1 in unirradiated discs is p53-independent. This analysis also identified a cluster of cells that expressed tribbles, encoding a factor that downregulates mitosis-promoting String and Twine, that appears to be G2/M arrested and expressed numerous genes involved in apoptosis, DDR, the aforementioned ligands, and TFs. As such, the tribbles-high cluster contains much of the heterogeneity.
Strengths:
(1) The authors have used robust methods for rearing Drosophila larvae, irradiating wing discs, and analyzing the data with Seurat v5 and HHI.
(2) These data will be informative for the field.
(3) Most of the data is well-presented
(4) The literature is appropriately cited.
We thank the reviewer for these comments.
Weaknesses:
(1) The data in Figure 1 are single-image representations. I assume that counting the number of nuclei that are positive for these markers is difficult, but it would be good to get a sense of how representative these images are and how many discs were analyzed for each condition in B-M.
For each condition at least 5 discs were imaged but we imaged up to 15 discs in some cases. We tried to choose a representative disc for each condition after looking at all of them. All discs imaged under each condition are shown below; the disc chosen for the figure is indicated with an asterisk. All scale bars are 100 mm.
Author response image 2.
Images for discs shown in Manuscript Figure 1panels B, C
Author response image 3.
Images for discs shown in Manuscript Figure 1panels D, E
Author response image 4.
Images used in Manuscript Figure 1, F, G
Author response image 5.
Images used in Manuscript Figure 1H, I
Author response image 6.
Images used in Manuscript Figure 1J, K
Author response image 7.
Images used in Manuscript Figure 1L, M
(2) Some of the figures are unclear.
It is unclear to us exactly which figures the Reviewer is referring to. Perhaps this is the same issue mentioned below in “Recommendations for the authors”. We address it below.
Reviewer #1 (Recommendations for the authors):
(1) Regarding Figure 1, what is stained in blue? Is it DAPI? If so, this should be added to the figure legend.
Thank you for pointing out this omission. This has been addressed in the revised manuscript.
It is very difficult to see blue on black, so could the authors please outline the discs?
Alternatively, they could show DAPI in green and the markers (pH2Av, etc) in magenta.
We used DAPI (blue) as a way of outlining the discs. While we appreciate the reviewer’s concern, after reviewing the images, we found that the blue is clearly visible when the document is viewed on the screen. It is less obvious if the document is printed on some kinds or printers. Since boosting this channel would make the signal from the channels more difficult to see, we left the images as they were.
(2) Figure 3, Figure Supplement 2, panel B. It is not possible to read the gene names in the panel's current form. Please break this up into 4 lines (as much as possible from the current 2).
Thank you for this suggestion. We have done this in the revised manuscript.
Reviewer #2 (Public review):
This manuscript investigates the question of cellular heterogeneity using the response of Drosophila wing imaginal discs to ionizing radiation as a model system. A key advance here is the focus on quantitatively expressing various measures of heterogeneity, leveraging single-cell RNAseq approaches. To achieve this goal, the manuscript creatively uses a metric from the social sciences called the HHI to quantify the spatial heterogeneity of expression of individual genes across the identified cell clusters. Inter- and intra-regional levels of heterogeneity are revealed. Some highlights include the identification of spatial heterogeneity in the expression of ligands and transcription factors after IR. Expression of some of these genes shows dependence on p53. An intriguing finding, made possible by using an alternative clustering method focusing on cell cycle progression, was the identification of a high-trbl subset of cells characterized by concordant expression of multiple apoptosis, DNA damage repair, ROS-related genes, certain ligands, and transcription factors, collectively representing HIX genes. This high-trbl set of cells may correspond to an IR-induced G2/M arrested cell state.
Overall, the data presented in the manuscript are of high quality but are largely descriptive. This study is therefore perceived as a resource that can serve as an inspiration for the field to carry out follow-up experiments.
Thank you for your assessment of the work.
Reviewer #2 (Recommendations for the authors):
I suggest two major points for improvement:
(1) It is important to test whether manipulation of trbl levels (i.e., overexpression, knockdown, mutation) would result in measurable biological outcomes after IR, such as altered HIX gene expression, altered cell cycle progression, or both. This may help disentangle the question of whether high trbl expression and correlated HIX gene expression are a cause or consequence of G2/M stalling.
We have described these experiments at the beginning of this Response to Reviews document when addressing the comments made by the Reviewing Editor. Please see Figure 7, figure supplements 1 and 2. These experiments suggest that upregulation of trbl offers some protection from radiation-induced death, yet it is itself insufficient to induce expression of two HIX genes tested. As we have also described earlier, three different RNAi lines tested did not reduce trbl upregulation after irradiation.
(2) A more extensive characterization of the high-trbl cell state would also be appropriate, particularly in terms of their relationship to the cell cycle.
We attempted to address this issue in two ways. First, we used the expression of a trbl-gfp transgene and RNA in-situ hybridization experiments to visualize the distribution of the high-trbl cells (shown in new manuscript figure, Figure 6-figure supplement 3). When examining trbl RNA in irradiated discs, there is no obvious demarcation between cells that express high levels of trbl and other cells. This is also apparent in the UMAP shown in Figure 6A and A’. Most cells seem to express trbl; cells in the “high trbl” cluster simply express more trbl than others. We observed cells expressing trbl and PCNA as well as cells expressing only one of those two genes at detectable levels. Thus, it was not possible to distinguish the “high trbl” cells from other cells by this approach.
We decided instead to focus on examining the expression of other cell-cycle genes in the high-trbl cluster. We have added a paragraph in the Results section that details our findings. Many transcriptional changes are indeed consistent with stalling in G2 such as high levels of trbl and low levels of string (stg). Additionally, that the cells are likely in G2 is consistent with reduced levels of genes that are normally expressed at other stages of the cell cycle: G1 genes such as E2f1 and Dp, S-phase genes such as several Mcm genes, PCNA and RnrS, and genes that encode mitotic proteins such as polo, Incenp and claspin. There are however, several anomalies such as slightly increased expression of the early-G1 cyclin, CycD, and the retinoblastoma ortholog Rbf. Thus, at least as assessed by the transcriptome, this cluster may not correspond to a cell state that is found under normal physiological conditions.
(3) Minor: p. 12, line 3. Figure 5A is mentioned, but it seems that it should be 4A instead.
Thank you for pointing this out. We have addressed this in our revisions.
Reviewer #3 (Public review):
Strengths:
Overall, the manuscript makes a compelling case for heterogeneity in gene expression changes that occur in response to uniform induction of damage by X-rays in a single-layer epithelium. This is an important finding that would be of interest to researchers in the field of DNA damage responses, regeneration, and development.
Weaknesses:
This work would be more useful to the field if the authors could provide a more comprehensive discussion of both the impact and the limitations of their findings, as explained below.
Propidium iodide staining was used as a quality control step to exclude cells with a compromised cell membrane. But this would exclude dead/dying cells that result from irradiation. What fraction of the total do these cells represent? Based on the literature, including works cited by the authors, up to 85% of cells die at 4000R, but this likely happens over a longer period than 4 hours after irradiation. Even if only half of the 85% are PI-positive by 4 hr, this still removes about 40% of the cell population from analysis. The remaining cells that manage to stay alive (excluding PI) at 4 hours and included in the analysis may or may not be representative of the whole disc. More relevant time points that anticipate apoptosis at 4 hr may be 2 hr after irradiation, at which time pro-apoptotic gene expression peaks (Wichmann 2006). Can the authors rule out the possibility that there is heterogeneity in apoptosis gene expression, but cells with higher expression are dead by 4 hours, and what is left behind (and analyzed in this study) may be the ones with more uniform, lower expression? I am not asking the authors to redo the study with a shorter time point, but to incorporate the known schedule of events into their data interpretation.
We thank the reviewer for these important comments. The generation of single-cell RNA-seq data from irradiated cells is tricky. Many cells have already died. Even those that do not incorporate propidium iodide are likely in early stages of apoptosis or are physiologically unhealthy and likely made it through our FACS filters. Indeed, in irradiated samples up to 57% of sequenced cells were not included in our analysis since their RNA content seemed to be of low quality. It is therefore likely that our data are biased towards cells that are less damaged. As advised by the reviewer, we will include a clearer discussion of these issues as well as the time course of events and how our analysis captures RNA levels only at a single time point.
If cluster 3 is G1/S, cluster 5 is late S/G2, and cluster 4 is G2/M, what are clusters 0, 1, and 2 that collectively account for more than half of the cells in the wing disc? Are the proportions of clusters 3, 4, and 5 in agreement with prior studies that used FACS to quantify wing disc cells according to cell cycle stage?
Work by others (Ruiz-Losada et al., 2021, PMID:34824391) has shown that almost 80% of cells have a 4C DNA content 4 h after 4,000 rad X-ray irradiation. The high-trbl cluster accounts for only 18% of cells and can therefore account for a minority of cells with a 4C DNA content.
Thus clusters 0, 1 and 2 could potentially contain other populations that also have a 4C DNA content. Importantly, similar proportions of cells in these clusters are also observed in unirradiated discs.
We expect that clusters 1 and 2 are largely comprised of cells in G2/M. Together, these clusters are marked by some genes previously found to be higher in FACS separated G2 cells compared to G1 cells (Liang et al., 2014, PMID: 24684830). These genes include Det, aurA, and ana1. Strangely, cluster 0 is not strongly marked by any of the 175 cell cycle genes used in our clustering (eff being the strongest marker) and has a lower-than-average expression of 165/175 cell cycle genes. Cluster 0 is however marked by the genes ac and sc, which are known to be expressed in proneuronal cell clusters interspersed throughout the disc that stall in G2 and form mitotically quiescent domains (Usui & Kimura 1992, Development, 116 (1992), pp. 601-610 (no PMID); Nègre et al., 2003, PMID: 12559497). Given these observations, we hypothesize that cluster 0 is largely comprised of stalled G2 cells like those found in ac/sc-expressing proneural clusters.
The EdU data in Figure 1 is very interesting, especially the persistence in the hinge. The authors speculate that this may be due to cells staying in S phase or performing a higher level of repair-related DNA synthesis. If so, wouldn't you expect 'High PCNA' cells to overlap with the hinge clusters in Figures 6G-G'? Again, no new experiments are needed. Just a more thorough discussion of the data.
We have found that the locations of elevated PCNA expression do not always correlate with the location of EdU incorporation either by examining scRNA-seq data or by using HCR to detect PCNA. PCNA expression is far more widespread as we now show in Figure 6-figure supplement 3.
Trbl/G2/M cluster shows Ets21C induction, while the pattern of Ets21C induction as detected by HCR in Figures 5H-I appears in localized clusters. I thought G2/M cells are not spatially confined. Are Ets21C+ cells in Figure 5 in G2/M? Can the overlap be confirmed, for example, by co-staining for Trbl or a G2/M marker with Ets21C?
The data show that the high-trbl cells are higher in Ets21C transcripts relative to other cell-cycle-based clusters after irradiation. This does not imply that high-trbl-cells in all regions of the disc upregulate Ets21C equally. Ets21C expression is likely heterogeneous in both ways – by location in the disc and by cell-cycle state.
Induction of dysf in some but not all discs is interesting. What were the proportions? Any possibility of a sex-linked induction that can be addressed by separating male and female larvae?
We can separate the cells in our dataset into male and female cells by expression of lncRNA:roX1/2. When we do this, we see X-ray induced dysf expressed similarly in both male and female cells. We think that it is therefore unlikely that this difference in expression can be attributed to cell sex. Another possibility is that dysf upregulation might be acutely sensitive to the developmental stage of the disc. This would require experiments with very precisely-staged larvae. We have not investigated this further as it is not a central issue in our paper.
Reviewer #3 (Recommendations for the authors):
Please check the color-coding in Figure 1A. The region marked as pouch appears to include hinge folds that express Zfh2 (a hinge marker) in Figure 2A (even after accounting for low Zfh2 expression in part of the pouch).
We have corrected this and have marked the pouch region based on the analysis of expression of different hinge and pouch markers by Ayala-Camargo et al. 2013 (PMID 2398534).
The statement 'Furthermore, within tissues, stem cells are most sensitive while differentiated cells are relatively radioresistant' needs to be qualified, as there are differences in radiosensitivity of adult versus embryonic stem cells (e.g., PMID: 30588339)
We thank the reviewer for bringing this point to our attention and for pointing us to an article that addresses this issue in detail. We appreciate that our statement was rather simplistic – we have modified it and added two additional references.
-
-
internationaldataspaces.org internationaldataspaces.org
-
IDSA on the ISO efforts on a DS standard. ISO/IEC 20151 (what's the IEC bit?)
-